An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List


The impact of the COVID-19 pandemic on scientific research in the life sciences
Massimo riccaboni.
1 AXES, IMT School for Advanced Studies Lucca, Lucca, Italy
Luca Verginer
2 Chair of Systems Design D-MTEC, ETH Zürich, Zurich, Switzerland
Associated Data
The processed data, instructions on how to process the raw PubMed dataset as well as all code are available via Zenodo at https://doi.org/10.5281/zenodo.5121216 .
The COVID-19 outbreak has posed an unprecedented challenge to humanity and science. On the one side, public and private incentives have been put in place to promptly allocate resources toward research areas strictly related to the COVID-19 emergency. However, research in many fields not directly related to the pandemic has been displaced. In this paper, we assess the impact of COVID-19 on world scientific production in the life sciences and find indications that the usage of medical subject headings (MeSH) has changed following the outbreak. We estimate through a difference-in-differences approach the impact of the start of the COVID-19 pandemic on scientific production using the PubMed database (3.6 Million research papers). We find that COVID-19-related MeSH terms have experienced a 6.5 fold increase in output on average, while publications on unrelated MeSH terms dropped by 10 to 12%. The publication weighted impact has an even more pronounced negative effect (-16% to -19%). Moreover, COVID-19 has displaced clinical trial publications (-24%) and diverted grants from research areas not closely related to COVID-19. Note that since COVID-19 publications may have been fast-tracked, the sudden surge in COVID-19 publications might be driven by editorial policy.
Introduction
The COVID-19 pandemic has mobilized the world scientific community in 2020, especially in the life sciences [ 1 , 2 ]. In the first three months after the pandemic, the number of scientific papers about COVID-19 was fivefold the number of articles on H1N1 swine influenza [ 3 ]. Similarly, the number of clinical trials related to COVID-19 prophylaxis and treatments skyrocketed [ 4 ]. Thanks to the rapid mobilization of the world scientific community, COVID-19 vaccines have been developed in record time. Despite this undeniable success, there is a rising concern about the negative consequences of COVID-19 on clinical trial research, with many projects being postponed [ 5 – 7 ]. According to Evaluate Pharma, clinical trials were one of the pandemic’s first casualties, with a record number of 160 studies suspended for reasons related to COVID-19 in April 2020 [ 8 , 9 ] reporting a total of 1,200 trials suspended as of July 2020. As a consequence, clinical researchers have been impaired by reduced access to healthcare research infrastructures. Particularly, the COVID-19 outbreak took a tall on women and early-career scientists [ 10 – 13 ]. On a different ground, Shan and colleagues found that non-COVID-19-related articles decreased as COVID-19-related articles increased in top clinical research journals [ 14 ]. Fraser and coworker found that COVID-19 preprints received more attention and citations than non-COVID-19 preprints [ 1 ]. More recently, Hook and Porter have found some early evidence of ‘covidisation’ of academic research, with research grants and output diverted to COVID-19 research in 2020 [ 15 ]. How much should scientists switch their efforts toward SARS-CoV-2 prevention, treatment, or mitigation? There is a growing consensus that the current level of ‘covidisation’ of research can be wasteful [ 4 , 5 , 16 ].
Against this background, in this paper, we investigate if the COVID-19 pandemic has induced a shift in biomedical publications toward COVID-19-related scientific production. The objective of the study is to show that scientific articles listing covid-related Medical Subject Headings (MeSH) when compared against covid-unrelated MeSH have been partially displaced. Specifically, we look at several indicators of scientific production in the life sciences before and after the start of the COVID-19 pandemic: (1) number of papers published, (2) impact factor weighted number of papers, (3) opens access, (4) number of publications related to clinical trials, (5) number of papers listing grants, (6) number of papers listing grants existing before the pandemic. Through a natural experiment approach, we analyze the impact of the pandemic on scientific production in the life sciences. We consider COVID-19 an unexpected and unprecedented exogenous source of variation with heterogeneous effects across biomedical research fields (i.e., MeSH terms).
Based on the difference in difference results, we document the displacement effect that the pandemic has had on several aspects of scientific publishing. The overall picture that emerges from this analysis is that there has been a profound realignment of priorities and research efforts. This shift has displaced biomedical research in fields not related to COVID-19.
The rest of the paper is structured as follows. First, we describe the data and our measure of relatedness to COVID-19. Next, we illustrate the difference-in-differences specification we rely on to identify the impact of the pandemic on scientific output. In the results section, we present the results of the difference-in-differences and network analyses. We document the sudden shift in publications, grants and trials towards COVID-19-related MeSH terms. Finally, we discuss the findings and highlight several policy implications.
Materials and methods
The present analysis is based primarily on PubMed and the Medical Subject Headings (MeSH) terminology. This data is used to estimate the effect of the start of the COVID 19 pandemic via a difference in difference approach. This section is structured as follows. We first introduce the data and then the econometric methodology. This analysis is not based on a pre-registered protocol.
Selection of biomedical publications
We rely on PubMed, a repository with more than 34 million biomedical citations, for the analysis. Specifically, we analyze the daily updated files up to 31/06/2021, extracting all publications of type ‘Journal Article’. For the principal analysis, we consider 3,638,584 papers published from January 2019 to December 2020. We also analyze 11,122,017 papers published from 2010 onwards to identify the earliest usage of a grant and infer if it was new in 2020. We use the SCImago journal ranking statistics to compute the impact factor weighted number (IFWN) of papers in a given field of research. To assign the publication date, we use the ‘electronically published’ dates and, if missing, the ‘print published’ dates.
Medical subject headings
We rely on the Medical Subject Headings (MeSH) terminology to approximate narrowly defined biomedical research fields. This terminology is a curated medical vocabulary, which is manually added to papers in the PubMed corpus. The fact that MeSH terms are manually annotated makes this terminology ideal for classification purposes. However, there is a delay between publication and annotation, on the order of several months. To address this delay and have the most recent classification, we search for all 28 425 MeSH terms using PubMed’s ESearch utility and classify paper by the results. The specific API endpoint is https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi , the relevant scripts are available with the code. For example, we assign the term ‘Ageusia’ (MeSH ID D000370) to all papers listed in the results of the ESearch API. We apply this method to the whole period (January 2019—December 2020) and obtain a mapping from papers to the MeSH terms. For every MeSH term, we keep track of the year they have been established. For instance, COVID-19 terms were established in 2020 (see Table 1 ): in January 2020, the WHO recommended 2019-nCoV and 2019-nCoV acute respiratory disease as provisional names for the virus and disease. The WHO issued the official terms COVID-19 and SARS-CoV-2 at the beginning of February 2020. By manually annotating publications, all publications referring to COVID-19 and SARS-CoV-2 since January 2020 have been labelled with the related MeSH terms. Other MeSH terms related to COVID-19, such as coronavirus, for instance, have been established years before the pandemic (see Table 2 ). We proxy MeSH term usage via search terms using the PubMed EUtilities API; this means that we are not using the hand-labelled MeSH terms but rather the PubMed search results. This means that the accuracy of the MeSH term we assign to a given paper is not perfect. In practice, this means that we have assigned more MeSH terms to a given term than a human annotator would have.
The list contains only terms with at least 100 publications in 2020.
Clinical trials and publication types
We classify publications using PubMed’s ‘PublicationType’ field in the XML baseline files (There are 187 publication types, see https://www.nlm.nih.gov/mesh/pubtypes.html ). We consider a publication to be related to a clinical trial if it lists any of the following descriptors:
- D016430: Clinical Trial
- D017426: Clinical Trial, Phase I
- D017427: Clinical Trial, Phase II
- D017428: Clinical Trial, Phase III
- D017429: Clinical Trial, Phase IV
- D018848: Controlled Clinical Trial
- D065007: Pragmatic Clinical Trial
- D000076362: Adaptive Clinical Trial
- D000077522: Clinical Trial, Veterinary
In our analysis of the impact of COVID-19 on publications related to clinical trials, we only consider MeSH terms that are associated at least once with a clinical trial publication over the two years. We apply this restriction to filter out MeSH terms that are very unlikely to be relevant for clinical trial types of research.
Open access
We proxy the availability of a journal article to the public, i.e., open access, if it is available from PubMed Central. PubMed Central archives full-text journal articles and provides free access to the public. Note that the copyright license may vary across participating publishers. However, the text of the paper is for all effects and purposes freely available without requiring subscriptions or special affiliation.
We infer if a publication has been funded by checking if it lists any grants. We classify grants as either ‘old’, i.e. existed before 2019, or ‘new’, i.e. first observed afterwards. To do so, we collect all grant IDs for 11,122,017 papers from 2010 on-wards and record their first appearance. This procedure is an indirect inference of the year the grant has been granted. The basic assumption is that if a grant number has not been listed in any publication since 2010, it is very likely a new grant. Specifically, an old grant is a grant listed since 2019 observed at least once from 2010 to 2018.
Note that this procedure is only approximate and has a few shortcomings. Mistyped grant numbers (e.g. ‘1234-M JPN’ and ‘1234-M-JPN’) could appear as new grants, even though they existed before, or new grants might be classified as old grants if they have a common ID (e.g. ‘Grant 1’). Unfortunately, there is no central repository of grant numbers and the associated metadata; however, there are plans to assign DOI numbers to grants to alleviate this problem (See https://gitlab.com/crossref/open_funder_registry for the project).
Impact factor weighted publication numbers (IFWN)
In our analysis, we consider two measures of scientific output. First, we simply count the number of publications by MeSH term. However, since journals vary considerably in terms of impact factor, we also weigh the number of publications by the impact factor of the venue (e.g., journal) where it was published. Specifically, we use the SCImago journal ranking statistics to weigh a paper by the impact factor of the journal it appears in. We use the ‘citation per document in the past two years’ for 45,230 ISSNs. Note that a journal may and often has more than one ISSN, i.e., one for the printed edition and one for the online edition. SCImago applies the same score for a venue across linked ISSNs.
For the impact factor weighted number (IFWN) of publication per MeSH terms, this means that all publications are replaced by the impact score of the journal they appear in and summed up.
COVID-19-relatedness
To measure how closely related to COVID-19 is a MeSH term, we introduce an index of relatedness to COVID-19. First, we identify the focal COVID-19 terms, which appeared in the literature in 2020 (see Table 1 ). Next, for all other pre-existing MeSH terms, we measure how closely related to COVID-19 they end up being.
Our aim is to show that MeSH terms that existed before and are related have experienced a sudden increase in the number of (impact factor weighted) papers.
We define a MeSH term’s COVID-19 relatedness as the conditional probability that, given its appearance on a paper, also one of the focal COVID-19 terms listed in Table 1 are present. In other words, the relatedness of a MeSH term is given by the probability that a COVID-19 MeSH term appears alongside. Since the focal COVID-19 terms did not exist before 2020, we estimate this measure only using papers published since January 2020. Formally, we define COVID-19-relatedness ( σ ) as in Eq (1) , where C is the set of papers listing a COVID-19 MeSH term and M i is the set of papers listing MeSH term i .
Intuitively we can read this measure as: what is the probability in 2020 that a COVID-19 MeSH term is present given that we chose a paper with MeSH term i ? For example, given that in 2020 we choose a paper dealing with “Ageusia” (i.e., Complete or severe loss of the subjective sense of taste), there is a 96% probability that this paper also lists COVID-19, see Table 1 .
Note that a paper listing a related MeSH term does not imply that that paper is doing COVID-19 research, but it implies that one of the MeSH terms listed is often used in COVID-19 research.
In sum, in our analysis, we use the following variables:
- Papers: Number of papers by MeSH term;
- Impact: Impact factor weighted number of papers by MeSH term;
- PMC: Papers listed in PubMed central by MeSH term, as a measure of Open Access publications;
- Trials: number of publications of type “Clinical Trial” by MeSH term;
- Grants: number of papers with at least one grant by MeSH term;
- Old Grants: number of papers listing a grant that has been observed between 2010 and 2018, by MeSH term;
Difference-in-differences
The difference-in-differences (DiD) method is an econometric technique to imitate an experimental research design from observation data, sometimes referred to as a quasi-experimental setup. In a randomized controlled trial, subjects are randomly assigned either to the treated or the control group. Analogously, in this natural experiment, we assume that medical subject headings (MeSH) have been randomly assigned to be either treated (related) or not treated (unrelated) by the pandemic crisis.
Before the COVID, for a future health crisis, the set of potentially impacted medical knowledge was not predictable since it depended on the specifics of the emergency. For instance, ageusia (loss of taste), a medical concept existing since 1991, became known to be a specific symptom of COVID-19 only after the pandemic.
Specifically, we exploit the COVID-19 as an unpredictable and exogenous shock that has deeply affected the publication priorities for biomedical scientific production, as compared to the situation before the pandemic. In this setting, COVID-19 is the treatment, and the identification of this new human coronavirus is the event. We claim that treated MeSH terms, i.e., MeSH terms related to COVID-19, have experienced a sudden increase in terms of scientific production and attention. In contrast, research on untreated MeSH terms, i.e., MeSH terms not related to COVID-19, has been displaced by COVID-19. Our analysis compares the scientific output of COVID-19 related and unrelated MeSH terms before and after January 2020.
Consider the simple regression model in Eq (2) . We have an outcome Y and dummy variable P identifying the period as before the event P = 0 and P = 1 as after the event. Additionally, we have a dummy variable identifying an observation belonging to the treated group ( G = 1) or control ( G = 0) group.
In our case, some of the terms turn out to be related to COVID-19 in 2020, whereas most of the MeSH terms are not closely related to COVID-19.
Thus β 1 identifies the overall effect on the control group after the event, β 2 the difference across treated and control groups before the event (i.e. the first difference in DiD) and finally the effect on the treated group after the event, net of the first difference, β 3 . This last parameter identifies the treatment effect on the treated group netting out the pre-treatment difference.
For the DiD to have a causal interpretation, it must be noted that pre-event, the trends of the two groups should be parallel, i.e., the common trend assumption (CTA) must be satisfied. We will show that the CTA holds in the results section.
To specify the DiD model, we need to define a period before and after the event and assign a treatment status or level of exposure to each term.
Before and after
The pre-treatment period is defined as January 2019 to December 2019. The post-treatment period is defined as the months from January 2020 to December 2020. We argue that the state of biomedical research was similar in those two years, apart from the effect of the pandemic.
Treatment status and exposure
The treatment is determined by the COVID-19 relatedness index σ i introduced earlier. Specifically, this number indicates the likelihood that COVID-19 will be a listed MeSH term, given that we observe the focal MeSH term i . To show that the effect becomes even stronger the closer related the subject is, and for ease of interpretation, we also discretize the relatedness value into three levels of treatment. Namely, we group MeSH terms with a σ between, 0% to 20%, 20% to 80% and 80% to 100%. The choice of alternative grouping strategies does not significantly affect our results. Results for alternative thresholds of relatedness can be computed using the available source code. We complement the dichotomized analysis by using the treatment intensity (relatedness measure σ ) to show that the result persists.
Panel regression
In this work, we estimate a random effects panel regression where the units of analysis are 28 318 biomedical research fields (i.e. MeSH terms) observed over time before and after the COVID-19 pandemic. The time resolution is at the monthly level, meaning that for each MeSH term, we have 24 observations from January 2019 to December 2020.
The basic panel regression with continuous treatment follows a similar setup as Eq (2) but with MeSH term random effects and monthly fixed effects.
The outcome variable Y it identifies the outcome at time t (i.e., month), for MeSH term i . As before, P t identifies the period with P t = 0 if the month is before January 2020 and P t = 1 if it is on or after this date. In (3) , the treatment level is measure by the relatedness to COVID-19 ( σ i ), where again the γ 1 identifies pre-trend (constant) differences and δ 1 the overall effect.
As mentioned before, to highlight that the effect is not linear but increases with relatedness, we split σ into three groups: from 0% to 20%, 20% to 80% and 80% to 100%. In the three-level treatment specification, the number of treatment levels ( G i ) is 3; hence we have two γ parameters. Note that I (⋅) is the indicator function, which is 1 if the argument is true, and 0 otherwise.
In total, we estimate six coefficients. As before, the δ l coefficient identifies the DiD effect.
Verifying the Common Trend Assumption (CTA)
To show that the pre-event trends are parallel and that the effect on publication activity is only visible from January 2020, we estimate a panel regression with each month modelled as a different event. Specifically, we estimate the following model.
We show that the CTA holds for this model by comparing the pre-event trends of the control group to the treated groups (COVID-19 related MeSH terms). Namely, we show that the pre-event trends of the control group are the same as the pre-event trends of the treated group.
Co-occurrence analysis
To investigate if the pandemic has caused a reconfiguration of research priorities, we look at the MeSH term co-occurrence network. Precisely, we extract the co-occurrence network of all 28,318 MeSH terms as they appear in the 3.3 million papers. We considered the co-occurrence networks of 2018, 2019 and 2020. Each node represents a MeSH term in these networks, and a link between them indicates that they have been observed at least once together. The weight of the edge between the MeSH terms is given by the number of times those terms have been jointly observed in the same publications.
Medical language is hugely complicated, and this simple representation does not capture the intricacies, subtle nuances and, in fact, meaning of the terms. Therefore, we do not claim that we can identify how the actual usage of MeSH terms has changed from this object, but rather that it has. Nevertheless, the co-occurrence graph captures rudimentary relations between concepts. We argue that absent a shock to the system, their basic usage patterns, change in importance (within the network) would essentially be the same from year to year. However, if we find that the importance of terms changes more than expected in 2020, it stands to reason that there have been some significant changes.
To show that that MeSH usage has been affected, we compute for each term in the years 2018, 2019 and 2020 their PageRank centrality [ 17 ]. The PageRank centrality tells us how likely a random walker traversing a network would be found at a given node if she follows the weights of the empirical edges (i.e., co-usage probability). Specifically, for the case of the MeSH co-occurrence network, this number represents how often an annotator at the National Library of Medicine would assign that MeSH term following the observed general usage patterns. It is a simplistic measure to capture the complexities of biomedical research. Nevertheless, it captures far-reaching interdependence across MeSH terms as the measure uses the whole network to determine the centrality of every MeSH term. A sudden change in the rankings and thus the position of MeSH terms in this network suggests that a given research subject has risen as it is used more often with other important MeSH terms (or vice versa).
To show that COVID-19-related research has profoundly impacted the way MeSH terms are used, we compute for each MeSH term the change in its PageRank centrality ( p it ).
We then compare the growth for each MeSH i term in g i (2019), i.e. before the the COVID-19 pandemic, with the growth after the event ( g i (2020)).
Publication growth
To estimate growth in scientific output, we compute the year over year growth in the number of the impact weighted number of publications per MeSH. Specifically, we measure the year by year growth as defined below, where m is the impact weighted number of publications at time t .
Changes in output and COVID-19 relatedness
Before we show the regression results, we provide descriptive evidence that publications from 2019 to 2020 have drastically increased. By showing that this growth correlates strongly with a MeSH term’s COVID-19 relatedness ( σ ), we demonstrate that (1) σ captures an essential aspect of the growth dynamics and (2) highlight the meteoric rise of highly related terms.
We look at the year over year growth in the number of the impact weighted number of publications per MeSH term from 2018 to 2019 and 2019 to 2020 as defined in the methods section.
Fig 1 shows the yearly growth of the impact weighted number of publications per MeSH term. By comparing the growth of the number of publications from the years 2018, 2019 and 2020, we find that the impact factor weighted number of publications has increased by up to a factor of 100 compared to the previous year for Betacoronavirus, one of the most closely related to COVID-19 MeSH term.

Each dot represents, a MeSH term. The y axis (growth) is in symmetric log scale. The x axis shows the COVID-19 relatedness, σ . Note that the position of the dots on the x-axis is the same in the two plots. Below: MeSH term importance gain (PageRank) and their COVID-19 relatedness.
Fig 1 , first row, reveals how strongly correlated the growth in the IFWN of publication is to the term’s COVID-19 relatedness. For instance, we see that the term ‘Betacoronavirus’ skyrocketed from 2019 to 2020, which is expected given that SARS-CoV-2 is a species of the genus. Conversely, the term ‘Alphacoronavirus’ has not experienced any growth given that it is twin a genus of the Coronaviridae family, but SARS-CoV-2 is not one of its species. Note also the fast growth in the number of publications dealing with ‘Quarantine’. Moreover, MeSH terms that grew significantly from 2018 to 2019 and were not closely related to COVID-19, like ‘Vaping’, slowed down in 2020. From the graph, the picture emerges that publication growth is correlated with COVID-19 relatedness σ and that the growth for less related terms slowed down.
To show that the usage pattern of MeSH terms has changed following the pandemic, we compute the PageRank centrality using graph-tool [ 18 ] as discussed in the Methods section.
Fig 1 , second row, shows the change in the PageRank centrality of the MeSH terms after the pandemic (2019 to 2020, right plot) and before (2018 to 2019, left plot). If there were no change in the general usage pattern, we would expect the variance in PageRank changes to be narrow across the two periods, see (left plot). However, PageRank scores changed significantly more from 2019 to 2020 than from 2018 to 2019, suggesting that there has been a reconfiguration of the network.
To further support this argument, we carry out a DiD regression analysis.
Common trends assumption
As discussed in the Methods section, we need to show that the CTA assumption holds for the DiD to be defined appropriately. We do this by estimating for each month the number of publications and comparing it across treatment groups. This exercise also serves the purpose of a placebo test. By assuming that each month could have potentially been the event’s timing (i.e., the outbreak), we show that January 2020 is the most likely timing of the event. The regression table, as noted earlier, contains over 70 estimated coefficients, hence for ease of reading, we will only show the predicted outcome per month by group (see Fig 2 ). The full regression table with all coefficients is available in the S1 Table .

The y axis is in log scale. The dashed vertical line identifies January 2020. The dashed horizontal line shows the publications in January 2019 for the 0–20% group before the event. This line highlights that the drop happens after the event. The bands around the lines indicate the 95% confidence interval of the predicted values. The results are the output of the Stata margins command.
Fig 2 shows the predicted number per outcome variable obtained from the panel regression model. These predictions correspond to the predicted value per relatedness group using the regression parameters estimated via the linear panel regression. The bands around the curves are the 95% confidence intervals.
All outcome measures depict a similar trend per month. Before the event (i.e., January 2020), there is a common trend across all groups. In contrast, after the event, we observe a sudden rise for the outcomes of the COVID-19 related treated groups (green and red lines) and a decline in the outcomes for the unrelated group (blue line). Therefore, we can conclude that the CTA assumption holds.
Regression results
Table 3 shows the DiD regression results (see Eq (3) ) for the selected outcome measures: number of publications (Papers), impact factor weighted number of publications (Impact), open access (OA) publications, clinical trial related publications, and publications with existing grants.
t statistics in parentheses, Std. Err. adjusted by MeSH-id. All outcome variables are in natural log.
* p < 0.05,
** p < 0.01,
*** p < 0.001
Table 3 shows results for the discrete treatment level version of the DiD model (see Eq (4) ).
Note that the outcome variable is in natural log scale; hence to get the effect of the independent variable, we need to exponentiate the coefficient. For values close to 0, the effect is well approximated by the percentage change of that magnitude.
In both specifications we see that the least related group, drops in the number of publications between 10% and 13%, respectively (first row of Tables Tables3 3 and and4, 4 , exp(−0.102) ≈ 0.87). In line with our expectations, the increase in the number of papers published by MeSH term is positively affected by the relatedness to COVID-19. In the discrete model (row 2), we note that the number of documents with MeSH terms with a COVID-19 relatedness between 20 and 80% grows by 18% and highly related terms by a factor of approximately 6.6 (exp(1.88)). The same general pattern can be observed for the impact weighted publication number, i.e., Model (2). Note, however, that the drop in the impact factor weighted output is more significant, reaching -19% for COVID-19 unrelated publications, and related publications growing by a factor of 8.7. This difference suggests that there might be a bias to publish papers on COVID-19 related subjects in high impact factor journals.
t statistics in parentheses, Std. Err. adjusted by MeSH-id. All outcome variables are in natural log. σ is the MeSH term relatedness to COVID-19.
By looking at the number of open access publications (PMC), we note that the least related group has not been affected negatively by the pandemic. However, the number of COVID-19 related publications has drastically increased for the most COVID-19 related group by a factor of 6.2. Note that the substantial increase in the number of papers available through open access is in large part due to journal and editorial policies to make preferentially COVID research immediately available to the public.
Regarding the number of clinical trial publications, we note that the least related group has been affected negatively, with the number of publications on clinical trials dropping by a staggering 24%. At the same time, publications on clinical trials for COVID-19-related MeSH have increased by a factor of 2.1. Note, however, that the effect on clinical trials is not significant in the continuous regression. The discrepancy across Tables Tables3 3 and and4 4 highlights that, especially for trials, the effect is not linear, where only the publications on clinical trials closely related to COVID-19 experiencing a boost.
It has been reported [ 19 ] that while the number of clinical trials registered to treat or prevent COVID-19 has surged with 179 new registrations in the second week of April 2020 alone. Only a few of these have led to publishable results in the 12 months since [ 20 ]. On the other hand, we find that clinical trial publications, considering related MeSH (but not COVID-19 directly), have had significant growth from the beginning of the pandemic. These results are not contradictory. Indeed counting the number of clinical trial publications listing the exact COVID-19 MeSH term (D000086382), we find 212 publications. While this might seem like a small number, consider that in 2020 only 8,485 publications were classified as clinical trials; thus, targeted trials still made up 2.5% of all clinical trials in 2020 . So while one might doubt the effectiveness of these research efforts, it is still the case that by sheer number, they represent a significant proportion of all publications on clinical trials in 2020. Moreover, COVID-19 specific Clinical trial publications in 2020, being a delayed signal of the actual trials, are a lower bound estimate on the true number of such clinical trials being conducted. This is because COVID-19 studies could only have commenced in 2020, whereas other studies had a head start. Thus our reported estimates are conservative, meaning that the true effect on actual clinical trials is likely larger, not smaller.
Research funding, as proxied by the number of publications with grants, follows a similar pattern, but notably, COVID-19-related MeSH terms list the same proportion of grants established before 2019 as other unrelated MeSH terms, suggesting that grants which were not designated for COVID-19 research have been used to support COVID-19 related research. Overall, the number of publications listing a grant has dropped. Note that this should be because the number of publications overall in the unrelated group has dropped. However, we note that the drop in publications is 10% while the decline in publications with at least one grant is 15%. This difference suggests that publications listing grants, which should have more funding, are disproportionately COVID-19 related papers. To further investigate this aspect, we look at whether the grant was old (pre-2019) or appeared for the first time in or after 2019. It stands to reason that an old grant (pre-2019) would not have been granted for a project dealing with the pandemic. Hence we would expect that COVID-19 related MeSH terms to have a lower proportion of old grants than the unrelated group. In models (6) in Table 4 we show that the number of old grants for the unrelated group drops by 13%. At the same time, the number of papers listing old grants (i.e., pre-2019) among the most related group increased by a factor of 3.1. Overall, these results suggest that COVID-19 related research has been funded largely by pre-existing grants, even though a specific mandate tied to the grants for this use is unlikely.
The scientific community has swiftly reallocated research efforts to cope with the COVID-19 pandemic, mobilizing knowledge across disciplines to find innovative solutions in record time. We document this both in terms of changing trends in the biomedical scientific output and the usage of MeSH terms by the scientific community. The flip side of this sudden and energetic prioritization of effort to fight COVID-19 has been a sudden contraction of scientific production in other relevant research areas. All in all, we find strong support to the hypotheses that the COVID-19 crisis has induced a sudden increase of research output in COVID-19 related areas of biomedical research. Conversely, research in areas not related to COVID-19 has experienced a significant drop in overall publishing rates and funding.
Our paper contributes to the literature on the impact of COVID-19 on scientific research: we corroborate previous findings about the surge of COVID-19 related publications [ 1 – 3 ], partially displacing research in COVID-19 unrelated fields of research [ 4 , 14 ], particularly research related to clinical trials [ 5 – 7 ]. The drop in trial research might have severe consequences for patients affected by life-threatening diseases since it will delay access to new and better treatments. We also confirm the impact of COVID-19 on open access publication output [ 1 ]; also, this is milder than traditional outlets. On top of this, we provide more robust evidence on the impact weighted effect of COVID-19 and grant financed research, highlighting the strong displacement effect of COVID-19 on the allocation of financial resources [ 15 ]. We document a substantial change in the usage patterns of MeSH terms, suggesting that there has been a reconfiguration in the way research terms are being combined. MeSH terms highly related to COVID-19 were peripheral in the MeSH usage networks before the pandemic but have become central since 2020. We conclude that the usage patterns have changed, with COVID-19 related MeSH terms occupying a much more prominent role in 2020 than they did in the previous years.
We also contribute to the literature by estimating the effect of COVID-19 on biomedical research in a natural experiment framework, isolating the specific effects of the COVID-19 pandemic on the biomedical scientific landscape. This is crucial to identify areas of public intervention to sustain areas of biomedical research which have been neglected during the COVID-19 crisis. Moreover, the exploratory analysis on the changes in usage patterns of MeSH terms, points to an increase in the importance of covid-related topics in the broader biomedical research landscape.
Our results provide compelling evidence that research related to COVID-19 has indeed displaced scientific production in other biomedical fields of research not related to COVID-19, with a significant drop in (impact weighted) scientific output related to non-COVID-19 and a marked reduction of financial support for publications not related to COVID-19 [ 4 , 5 , 16 ]. The displacement effect is persistent to the end of 2020. As vaccination progresses, we highlight the urgent need for science policy to re-balance support for research activity that was put on pause because of the COVID-19 pandemic.
We find that COVID-19 dramatically impacted clinical research. Reactivation of clinical trials activities that have been postponed or suspended for reasons related to COVID-19 is a priority that should be considered in the national vaccination plans. Moreover, since grants have been diverted and financial incentives have been targeted to sustain COVID-19 research leading to an excessive entry in COVID-19-related clinical trials and the ‘covidisation’ of research, there is a need to reorient incentives to basic research and otherwise neglected or temporally abandoned areas of biomedical research. Without dedicated support in the recovery plans for neglected research of the COVID-19 era, there is a risk that more medical needs will be unmet in the future, possibly exacerbating the shortage of scientific research for orphan and neglected diseases, which do not belong to COVID-19-related research areas.
Limitations
Our empirical approach has some limits. First, we proxy MeSH term usage via search terms using the PubMed EUtilities API. This means that the accuracy of the MeSH term we assign to a given paper is not fully validated. More time is needed for the completion of manually annotated MeSH terms. Second, the timing of publication is not the moment the research has been carried out. There is a lead time between inception, analysis, write-up, review, revision, and final publication. This delay varies across disciplines. Nevertheless, given that the surge in publications happens around the alleged event date, January 2020, we are confident that the publication date is a reasonable yet imperfect estimate of the timing of the research. Third, several journals have publicly declared to fast-track COVID-19 research. This discrepancy in the speed of publication of COVID-19 related research and other research could affect our results. Specifically, a surge or displacement could be overestimated due to a lag in the publication of COVID-19 unrelated research. We alleviate this bias by estimating the effect considering a considerable time after the event (January 2020 to December 2020). Forth, on the one hand, clinical Trials may lead to multiple publications. Therefore we might overestimate the impact of COVID-19 on the number of clinical trials. On the other hand, COVID-19 publications on clinical trials lag behind, so the number of papers related COVID-19 trials is likely underestimated. Therefore, we note that the focus of this paper is scientific publications on clinical trials rather than on actual clinical trials. Fifth, regarding grants, unfortunately, there is no unique centralized repository mapping grant numbers to years, so we have to proxy old grants with grants that appeared in publications from 2010 to 2018. Besides, grant numbers are free-form entries, meaning that PubMed has no validation step to disambiguate or verify that the grant number has been entered correctly. This has the effect of classifying a grant as new even though it has appeared under a different name. We mitigate this problem by using a long period to collect grant numbers and catch many spellings of the same grant, thereby reducing the likelihood of miss-identifying a grant as new when it existed before. Still, unless unique identifiers are widely used, there is no way to verify this.
So far, there is no conclusive evidence on whether entry into COVID-19 has been excessive. However, there is a growing consensus that COVID-19 has displaced, at least temporally, scientific research in COVID-19 unrelated biomedical research areas. Even though it is certainly expected that more attention will be devoted to the emergency during a pandemic, the displacement of biomedical research in other fields is concerning. Future research is needed to investigate the long-run structural consequences of the COVID-19 crisis on biomedical research.
Supporting information
Full regression table with all controls and interactions.
Funding Statement
The author(s) received no specific funding for this work.
Data Availability
- Life Sciences
We're working on groundbreaking research aiming to revolutionize the field of life sciences. We're solving some of the most important issues humanity faces with artificial intelligence, developing novel and unconventional computing structures, as well as mathematical and computational modeling.
AI transformers shed light on the brain’s mysterious astrocytes
- Foundation Models
- Machine Learning
- Natural Language Processing
IBM Research and JDRF continue to advance biomarker discovery research
An ai foundation model that learns the grammar of molecules.
- Accelerated Discovery
- Materials Discovery
How we react to smells could unlock how we form conscious thoughts

Crowdsourcing to trace cell lineages
Neural networks take titan-sized step toward t cell specificity prediction.
- See more of our work on Life Sciences
Publications
- Jenna Reinen
- Pablo Polosecki
- npj Schizophrenia
- Yves Gaetan Nana Teukam
- Loic Kwate Dassi
- Computational And Structural Biotechnology Journal
- Katja Ovchinnikova
- Jannis Born
- npj Precision Oncology
- James Hedrick
- Nathaniel Park
- MRS Spring Meeting 2024
- Sara Capponi
- Shangying Wang
- Biophysical Journal
Related topics
Computer science, healthcare and life sciences, mathematical sciences, physical sciences.
Alert Banner
Ncbiotech's move program wins federal grant to expand biopharma job opportunities for nc’s military community. .

What are Life Sciences?
The simplest way to define life sciences is the study of living organisms and life processes..
At NCBiotech, we see it as science involving cells and their components, products and processes. Biology, medicine and agriculture are the most obvious examples of the discipline. However, in recent years, a convergence is underway that requires a multi-discipline approach. For example, biologists, chemists, and robotics and automation engineers will work together to develop new solutions.
What is Biotechnology?
Biotechnology, the most prominent component of the life sciences, is a toolbox that leverages our understanding of the natural sciences to create solutions for our world's problems. We use biotechnology to grow food to feed families and to make medicines and vaccines to fight diseases. We even turn to biotechnology for new innovations, such as leveraging plants to manufacture medicines and create alternative fuels.
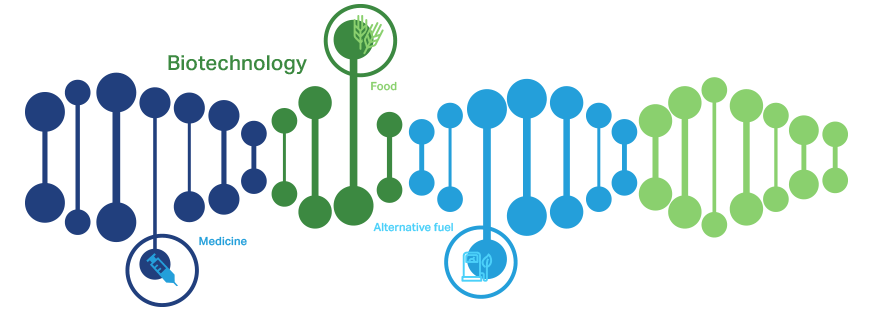
How Biotechnology Works
Biotechnology is grounded in the pure biological sciences of genetics, microbiology, animal cell cultures, molecular biology, embryology and cell biology. Biotechnology discoveries are intimately entwined in the life sciences industry sectors for development in agricultural biotechnology , biomanufacturing , human health , precision medicine and medical devices and diagnostics. For example, biomedical researchers use their understanding of genes, cells and proteins to pinpoint the differences between diseased and healthy cells. Once they discover how diseased cells are altered, researchers can more easily develop new medical diagnostics, devices and therapies to treat diseases and chronic conditions.
History of Biotech
Biotech has led us to the greatest innovations. Since 1984, North Carolina has nurtured its life sciences assets, firmly establishing itself as a leading U.S. life sciences hub characterized by steady growth of companies and talent statewide. Diverse, specialized subsectors, serve a variety of needs globally. The state pivoted from its deep roots in agriculture and furniture manufacturing to focus on biotechnology. In the early 2000s, as more biotech products gained regulatory approval, companies expanded production capacity. This led to the state's growing demand for skilled biopharmaceutical manufacturing workers. Workforce development programs continue to fuel our state's talent pool. This talent pool, in turn, helps to recruit new life sciences companies and supports local company growth.
The Future of Biotech
Today, North Carolina is home to more than 830 life sciences companies, a talent pool of 75,000 skilled workers and an additional 2,500 companies that support the sector. According to the 2022 TEConomy Report , despite the COVID-19 pandemic and economic challenges, the state's life sciences growth has outpaced national growth, placing itself among the top-tier life sciences hubs.
North Carolina has long invested in scientific infrastructure to fuel innovation. With three top-tier research universities, scientific innovations are seeding new spinouts and advancing technologies to the next level.
Through our strengths in research and development, talent, training and scientific infrastructure, North Carolina will remain at the forefront of biotechnology and life sciences.
251+ Life Science Research Topics [Updated]
Life science research is like peering into the intricate workings of the universe, but instead of stars and galaxies, it delves into the mysteries of life itself. From unraveling the secrets of our genetic code to understanding ecosystems and everything in between, life science research encompasses a vast array of fascinating topics. In this blog post, we’ll embark on a journey through some of the most captivating life science research topics within the realm of life science research.
What is research in life science?
Table of Contents
Research in life science involves the systematic investigation and study of living organisms, their interactions, and their environments. It encompasses a wide range of disciplines, including biology, genetics, ecology, microbiology, neuroscience, and more.
Life science research aims to expand our understanding of the fundamental principles governing life processes, uncover new insights into biological systems, develop innovative technologies and therapies, and address pressing challenges in areas such as healthcare, agriculture, and conservation.
251+ Life Science Research Topics: Category Wise
Genetics and genomics.
- Genetic basis of inherited diseases
- Genome-wide association studies
- Epigenetics and gene regulation
- Evolutionary genomics
- CRISPR/Cas9 gene editing technology
- Pharmacogenomics and personalized medicine
- Population genetics
- Functional genomics
- Comparative genomics across species
- Genetic diversity and conservation
Biotechnology and Bioengineering
- Biopharmaceutical production
- Metabolic engineering for biofuel production
- Synthetic biology applications
- Bioremediation techniques
- Nanotechnology in drug delivery
- Tissue engineering and regenerative medicine
- Biosensors for environmental monitoring
- Bioprocessing optimization
- Biodegradable plastics and sustainable materials
- Agricultural biotechnology for crop improvement
Ecology and Environmental Biology
- Biodiversity hotspots and conservation strategies
- Ecosystem services and human well-being
- Climate change impacts on ecosystems
- Restoration ecology techniques
- Urban ecology and biodiversity
- Marine biology and coral reef conservation
- Habitat fragmentation and species extinction
- Ecological modeling and forecasting
- Wildlife conservation genetics
- Microbial ecology in natural environments
Neuroscience and Cognitive Science
- Brain mapping techniques (fMRI, EEG, etc.)
- Neuroplasticity and learning
- Neural circuitry underlying behavior
- Neurodegenerative diseases (Alzheimer’s, Parkinson’s, etc.)
- Neural engineering for prosthetics
- Consciousness and the mind-body problem
- Psychiatric genetics and mental health disorders
- Neuroimaging in psychiatric research
- Developmental cognitive neuroscience
- Neural correlates of consciousness
Evolutionary Biology
- Mechanisms of speciation
- Molecular evolution and phylogenetics
- Coevolutionary dynamics
- Evolution of antibiotic resistance
- Cultural evolution and human behavior
- Evolutionary consequences of climate change
- Evolutionary game theory
- Evolutionary medicine and infectious diseases
- Evolutionary psychology and human cognition
- Paleogenomics and ancient DNA analysis
Cell Biology and Physiology
- Cell cycle regulation and cancer biology
- Stem cell biology and regenerative medicine
- Organelle dynamics and intracellular transport
- Cellular senescence and aging
- Ion channels and neuronal excitability
- Metabolic pathways and cellular energetics
- Cell signaling pathways in development and disease
- Autophagy and cellular homeostasis
- Mitochondrial function and disease
- Cell adhesion and migration in development and cancer
Microbiology and Immunology
- Microbiome composition and function
- Antibiotic resistance mechanisms
- Host-microbe interactions in health and disease
- Viral pathogenesis and vaccine development
- Microbial biotechnology for waste treatment
- Immunotherapy approaches for cancer treatment
- Microbial diversity in extreme environments
- Antimicrobial peptides and drug discovery
- Microbial biofilms and chronic infections
- Host immune responses to viral infections
Biomedical Research and Clinical Trials
- Translational research in oncology
- Precision medicine approaches
- Clinical trials for gene therapies
- Biomarker discovery for disease diagnosis
- Stem cell-based therapies for regenerative medicine
- Pharmacokinetics and drug metabolism studies
- Clinical trials for neurodegenerative diseases
- Vaccine efficacy trials
- Patient-reported outcomes in clinical research
- Health disparities and clinical trial participation
Emerging Technologies and Innovations
- Single-cell omics technologies
- 3D bioprinting for tissue engineering
- CRISPR-based diagnostics
- Artificial intelligence applications in life sciences
- Organs-on-chip for drug screening
- Wearable biosensors for health monitoring
- Nanomedicine for targeted drug delivery
- Optogenetics for neuronal manipulation
- Quantum biology and biological systems
- Augmented reality in medical education
Ethical, Legal, and Social Implications (ELSI) in Life Sciences
- Privacy concerns in genomic research
- Ethical considerations in gene editing technologies
- Access to healthcare and genetic testing
- Intellectual property rights in biotechnology
- Informed consent in clinical trials
- Animal welfare in research
- Equity in environmental decision-making
- Data sharing and reproducibility in science
- Dual-use research and biosecurity
- Cultural perspectives on biomedicine and genetics
Public Health and Epidemiology
- Disease surveillance and outbreak investigation
- Global health disparities and access to healthcare
- Environmental factors in disease transmission
- Health impacts of climate change
- Social determinants of health
- Infectious disease modeling and forecasting
- Vaccination strategies and herd immunity
- Epidemiology of chronic diseases
- Mental health epidemiology
- Occupational health and safety
Plant Biology and Agriculture
- Crop domestication and evolution
- Plant-microbe interactions in agriculture
- Genetic engineering for crop improvement
- Plant hormone signaling pathways
- Abiotic stress tolerance mechanisms in plants
- Soil microbiology and nutrient cycling
- Agroecology and sustainable farming practices
- Plant secondary metabolites and natural products
- Plant developmental biology
- Plant epigenetics and environmental adaptation
Bioinformatics and Computational Biology
- Genome assembly and annotation algorithms
- Phylogenetic tree reconstruction methods
- Metagenomic data analysis pipelines
- Machine learning approaches for biomarker discovery
- Structural bioinformatics and protein modeling
- Systems biology and network analysis
- Transcriptomic data analysis tools
- Population genetics simulation software
- Evolutionary algorithms in bioinformatics
- Cloud computing in life sciences research
Toxicology and Environmental Health
- Mechanisms of chemical toxicity
- Risk assessment methodologies
- Environmental fate and transport of pollutants
- Endocrine disruptors and reproductive health
- Nanotoxicology and nanomaterial safety
- Biomonitoring of environmental contaminants
- Ecotoxicology and wildlife health
- Air pollution exposure and respiratory health
- Water quality and aquatic ecosystems
- Environmental justice and health disparities
Aquatic Biology and Oceanography
- Marine biodiversity conservation strategies
- Ocean acidification impacts on marine life
- Coral reef resilience and restoration
- Fisheries management and sustainable harvesting
- Deep-sea biodiversity and exploration
- Harmful algal blooms and ecosystem health
- Marine mammal conservation efforts
- Microplastics pollution in aquatic environments
- Ocean circulation and climate regulation
- Aquaculture and mariculture technologies
Social and Behavioral Sciences in Health
- Health behavior change interventions
- Social determinants of health disparities
- Health communication strategies
- Community-based participatory research
- Patient-centered care approaches
- Cultural competence in healthcare delivery
- Health literacy interventions
- Stigma reduction efforts in public health
- Health policy analysis and advocacy
- Digital health technologies for behavior monitoring
Bioethics and Biomedical Ethics
- Ethical considerations in human subjects research
- Research ethics in vulnerable populations
- Privacy and data protection in healthcare
- Professional integrity and scientific misconduct
- Ethical implications of genetic testing
- Access to healthcare and health equity
- End-of-life care and euthanasia debates
- Reproductive ethics and assisted reproduction
- Ethical challenges in emerging biotechnologies
Forensic Science and Criminalistics
- DNA fingerprinting techniques
- Forensic entomology and time of death estimation
- Trace evidence analysis methods
- Digital forensics in criminal investigations
- Ballistics and firearm identification
- Forensic anthropology and human identification
- Bloodstain pattern analysis
- Arson investigation techniques
- Forensic toxicology and drug analysis
- Forensic psychology and criminal profiling
Nutrition and Dietary Science
- Nutritional epidemiology studies
- Diet and chronic disease risk
- Functional foods and nutraceuticals
- Macronutrient metabolism pathways
- Micronutrient deficiencies and supplementation
- Gut microbiota and metabolic health
- Dietary interventions for weight management
- Food safety and risk assessment
- Sustainable diets and environmental impact
- Cultural influences on dietary habits
Entomology and Insect Biology
- Insect behavior and communication
- Insecticide resistance mechanisms
- Pollinator decline and conservation efforts
- Medical entomology and vector-borne diseases
- Invasive species management strategies
- Insect biodiversity in urban environments
- Agricultural pest management techniques
- Insect physiology and biochemistry
- Social insects and eusociality
- Insect symbiosis and microbial interactions
Zoology and Animal Biology
- Animal behavior and cognition
- Conservation genetics of endangered species
- Reproductive biology and breeding programs
- Wildlife forensics and illegal wildlife trade
- Comparative anatomy and evolutionary biology
- Animal welfare and ethics in research
- Physiological adaptations to extreme environments
- Zoological taxonomy and species discovery
- Animal communication and signaling
- Human-wildlife conflict mitigation strategies
Biochemistry and Molecular Biology
- Protein folding and misfolding diseases
- Enzyme kinetics and catalytic mechanisms
- Metabolic regulation in health and disease
- Signal transduction pathways
- DNA repair mechanisms and genome stability
- RNA biology and post-transcriptional regulation
- Lipid metabolism and membrane biophysics
- Molecular interactions in drug design
- Bioenergetics and cellular respiration
- Structural biology and X-ray crystallography
Cancer Biology and Oncology
- Tumor microenvironment and metastasis
- Cancer stem cells and therapy resistance
- Angiogenesis and tumor vasculature
- Immune checkpoint inhibitors in cancer therapy
- Liquid biopsy techniques for cancer detection
- Oncogenic signaling pathways
- Personalized medicine approaches in oncology
- Radiation therapy and tumor targeting strategies
- Cancer genomics and precision oncology
- Cancer prevention and lifestyle interventions
Developmental Biology and Embryology
- Embryonic stem cell differentiation
- Morphogen gradients and tissue patterning
- Developmental genetics and model organisms
- Regenerative potential in vertebrates and invertebrates
- Developmental plasticity and environmental cues
- Embryo implantation and pregnancy disorders
- Germ cell development and fertility preservation
- Cell fate determination in development
- Evolutionary developmental biology (evo-devo)
- Organogenesis and tissue morphogenesis
Pharmacology and Drug Discovery
- Drug-target interactions and pharmacokinetics
- High-throughput screening techniques
- Structure-activity relationship studies
- Drug repurposing strategies
- Natural product drug discovery
- Drug delivery systems and nanomedicine
- Pharmacovigilance and drug safety monitoring
- Pharmacoeconomics and healthcare outcomes
- Drug metabolism and drug-drug interactions
Stem Cell Research
- Induced pluripotent stem cells (iPSCs) technology
- Stem cell therapy applications in regenerative medicine
- Stem cell niche and microenvironment
- Stem cell banking and cryopreservation
- Stem cell-based disease modeling
What Are The 10 Examples of Life Science Research Paper Titles?
- Investigating the Role of Gut Microbiota in Neurological Disorders: Implications for Therapeutic Interventions.
- Genome-Wide Association Study Identifies Novel Genetic Markers for Cardiovascular Disease Risk.
- Understanding the Molecular Mechanisms of Cancer Metastasis: Insights from Cellular Signaling Pathways.
- The Impact of Climate Change on Plant-Pollinator Interactions: Implications for Biodiversity Conservation.
- Exploring the Potential of CRISPR/Cas9 Gene Editing Technology in Treating Genetic Disorders.
- Characterizing the Microbial Diversity of Extreme Environments: Insights from Deep-Sea Hydrothermal Vents.
- Assessment of Novel Drug Delivery Systems for Targeted Cancer Therapy: A Preclinical Study.
- Unraveling the Neurobiology of Addiction: Implications for Treatment Strategies.
- Investigating the Role of Epigenetics in Age-Related Diseases: From Mechanisms to Therapeutic Targets.
- Evaluating the Efficacy of Herbal Remedies in Traditional Medicine: A Systematic Review and Meta-Analysis.
Life science research is a journey of discovery, filled with wonder, excitement, and the occasional setback. Yet, through perseverance and ingenuity, researchers continue to push the boundaries of knowledge, unlocking the secrets of life itself. As we stand on the cusp of a new era of scientific discovery, one thing is clear: the future of life science research is brighter—and more promising—than ever before. I hope these life science research topics will help you to find the best topics for you.
Related Posts

Step by Step Guide on The Best Way to Finance Car

The Best Way on How to Get Fund For Business to Grow it Efficiently
Life Sciences | Graduate | Careers | Undergraduate
Aeronautics and astronautics, biological engineering, biophysics graduate certificate program, brain and cognitive sciences, chemical engineering, civil and environmental engineering (environmental microbiology), computational and systems biology, earth, atmospheric, and planetary sciences, hst - health, sciences and technology, materials science and engineering, mechanical engineering, microbiology program, nuclear science and engineering, sloan mba with a health care focus, sts- science, technology, and society, whoi joint program, writing and humanistic studies, life sciences at mit.
Many areas of research today have a Life Sciences focus. This is primarily due to the powerful tools of molecular biology, which form a common language and allow exciting and important interdisciplinary approaches. Experience in Life Sciences-based research opens multiple career paths.
This site collates the broad array of MIT graduate degree programs with a primary focus on biological questions, or that can include a Life Sciences focus. Applications for graduate study should be made through the appropriate program. Please explore this site, and our program offerings!
Find out more about:
Life at MIT and how to apply
School of Science
School of Engineering
Sloan School of Management
School of Humanities, Arts, and Social Sciences
Open Courseware
Printable List of Programs
List of Life Sciences Subjects at MIT
Graduate Life Sciences Programs at MIT
Office of Admissions
Mit open courseware.
Developed by the Dean's Office: MIT School of Science, in cooperation with MIT departments.
Copyright 2013 Massachusetts Institute of Technology. Site designed by Chrysos Designs .
Insights on Life Sciences

Generative AI in the pharmaceutical industry: Moving from hype to reality

Beyond the pandemic: The next chapter of innovation in vaccines

Early adoption of generative AI in commercial life sciences

What to expect from medtech in 2024

Here to stay: An attractive future for medical aesthetics

Accelerating clinical trials to improve biopharma R&D productivity

What early-stage investing reveals about biotech innovation
Digital diagnostics: A path forward for IVD
How artificial intelligence can power clinical development
Special collection.

Future of pharma operations

Coronavirus vaccines progress: What’s next?
McKinsey insights on cell and gene therapy
The rise of health technology
Want to learn more about how we help clients in life sciences, more insights, related practices.
Healthcare Systems & Services

Consumer Health


Public & Social Sector
Connect with our life sciences practice.

In the News

Wild megalopolis: Study shows unexpected pockets of biodiversity pepper Los Angeles

- Life Sciences undergraduates showcase scientific rigor and creativity at UCLA Undergraduate Research Week 2024

UCLA scientists develop new technology to position ‘off-the-shelf’ cancer immunotherapy for clinical use

Building on 62 years of research on climate change and marmots in the Rocky Mountains (PBS)
View All News
Dean’s Message
The importance of innovative science and education has never been more apparent, and UCLA Life Sciences is committed to addressing some of the most important questions we face today. I am honored to work with world-renowned faculty who are elucidating the intricate mechanisms of how living systems work…
Our groundbreaking research is leading to better diagnostics and treatments for disease; better strategies to protect our rapidly changing natural environment; a clearer understanding of mental health; and a deeper awareness of the role of science in society. It is my privilege to work alongside amazing instructors who provide a world-class education and research training to our students, with a keen eye toward inclusive teaching practices that foster the intellectual and personal growth of our diverse student body. Our current student body is one of the most racially and socio-economically diverse classes to enroll at UCLA, so the timing could not be better to think about how we approach the learning environment, through a lens of equity and inclusion. We are committed to investing in all of our students knowing they will be our next generation of leaders.
While we face unprecedented times, we have an incredible opportunity to rise to the moment and embrace the challenges before us, creatively and with a sense of purpose. We aim to be intentional about our scholarship, teaching, and scientific culture and in so doing provide leadership at UCLA and beyond. I invite you to explore for yourself what UCLA Life Sciences has to offer!

Tracy Johnson Dean, Life Sciences UCLA College
Quick Links
The Division of Life Sciences acknowledges the Gabrielino/Tongva peoples as the traditional land caretakers of Tovaangar (the Los Angeles basin and So. Channel Islands). As part of a California land grant institution, we pay our respects to the Honuukvetam (Ancestors), ‘Ahiihirom (Elders) and ‘Eyoohiinkem (our relatives/relations) past, present and emerging.
Interesting links
- “Dialogues on Inclusive Excellence in the Biosciences”
- “Divine Variations: How Christian Thought Became Racial Science” wins 2021 Iris Book Award
- $1.5 million CIRM grant funds UCLA research to advance stem cell-based technologies for treating intellectual disability syndromes (UCLA Broad Stem Cell Research Center)
- $6.2 million NIH grant to support UCLA study of how COVID-19 causes multiple organ failure
- 2018-19 Life Sciences Excellence Award Winners
- 2020 UCLA Life Sciences Excellence Awards
- 2021 Life Sciences Excellence Awards
- 2022 NSF Graduate Research Fellowship Program Awardees provide an inspiring glimpse into Life Sciences’ graduate student research
- 2022 UCLA Life Sciences’ Mautner Graduate Awardees
- 2022: Advancing Equity and Inclusion in UCLA Life Sciences
- 2024 Life Sciences Excellence Awards – Call for Nominations
- A ‘lost world’ in the Yucatan Peninsula reveals possible impacts of climate change on coastal plains
- A “Hot Ones”-style interview with Dean Johnson
- A decade after gene therapy, children born with deadly immune disorder remain healthy
- A digital nature tour of UCLA’s campus– highlighting our Mildred E. Mathias Botanical Garden
- A longer-lasting COVID vaccine? UCLA study points the way
- A True Bruin works to help fellow student-veterans in UCLA Life Sciences
- Affiliate Institutes & Centers
- American Indian Science and Engineering Society (AISES) at UCLA receives nationwide chapter award for outreach and community service
- Anatomy of a Vaccine (UCLA Magazine article)
- Announcing the new UCLA Rothman Family Institute for Food Studies
- APO Listings
- Available Academic Positions
- Basic tips for expanding your allyship
- Belonging at UCLA: U.S. Army infantryman to UCLA neuroscience graduate
- Big Data and Life Sciences
- Birds, climate change, and where we can make a difference- a conversation with Professor Morgan Tingley
- Black History in UCLA Life Sciences – Psychology’s first tenured Black professor recounts his early days at UCLA
- Celebrate Community: Asian Pacific Islander Heritage Month (May 2022)
- COVID and the need to address long-term grief
- COVID-19 and potential shifts in gender roles
- COVID-19 pandemic science and public health, made better through community partnership: 2022 Mautner Public Lecture with Joe Derisi, Ph.D.
- COVID-19 Vaccination: It Matters in Saving Black Lives (Recorded Town Hall Event)
- Curriculum Options for Biological Sciences Majors
- Dean Tracy Johnson interviewed on Univision show about women in science
- Departments
- Distinguished professor of ecology and evolutionary biology is honored with the UCLA Public Impact Research Award
- Diversity Committee
- Drug commonly used as antidepressant helps fight cancer in mice
- Ecology and Evolutionary Biology professor named 2019 Gold Shield Faculty Prize winner
- Ecology and Evolutionary Biology graduate student wins 2022 UCLA Grad Slam Final
- Embed iList
- Embryo models are not embryos, say leaders at the new UCLA Center for Reproductive Science, Health and Education
- Engaging in AAAS SEA Change: Q&A with Life Sciences Dean Tracy Johnson
- Facts, Myths, and Misinformation on COVID-19 – Protecting Black/Latinx Families – UCLA webinar January 13, 2022
- Faculty in UCLA Life Sciences listed among the world’s most influential researchers
- Faculty Resources
- Finding your Path in the Life Sciences • Fall 2022
- For UCLA-based startup, new muscular dystrophy treatment is a personal mission
- Frontpage (New)
- Gene therapy at UCLA gives man with sickle cell disease the chance for a better future
- Graduate Degrees
- Graduates & Post-Docs
- Homepage (New)
- Hope for patients with severe paralysis after spinal cord injury
- How exercise rejuvenates aging stem cells: a Q&A with Dr. Thomas Rando (Broad Stem Cell Research Center)
- How sourdough, seeds, shovels (and other basic survival needs) made a comeback in the pandemic
- In Memoriam: Kathryn Anderson, Pioneer of Developmental Biology and UCLA alum
- In memoriam: Allen Parducci, 97, influential cognitive psychologist and early pioneer of windsurfing
- In Memoriam: Distinguished Professor Bob Wayne, pioneer in evolutionary and conservation genetics
- In Memoriam: Seymour Feshbach, Professor Emeritus of Psychology
- Inaugural UCLA Jenessa Shapiro Memorial Award supports graduate student research on intraracial conversations
- Inclusion Research
- Inclusive Excellence
- Institute for Society and Genetics Professor Nanibaa’ Garrison
- Intricacies of L.A.’s urban ecosystem are the focus of a new UCLA podcast
- Life Sciences Core Curriculum
- Life Sciences Excellence Award Winners 2022
- Life Sciences Excellence Award Winners 2023
- Life Sciences Excellence Award Winners 2024
- Life Sciences professors named 2022 fellows of the American Association for the Advancement of Science
- Life Sciences Undergraduates – Study Abroad
- Life Sciences undergraduates publish research on pandemic inequities in the virtual classroom
- Majors & Minors
- Music’s Emotional Power Can Shape Memories—And Your Perception Of Time (Science Friday)
- New state grant funds UCLA research that will help guide interventions aimed at reducing COVID health risks for Black Californians
- Open Academic Positions
- Out of the Box
- Pangolin genome research aims to help protect pangolin species and reveals intriguing facts
- Planning Your Curriculum
- Professor Vickie Mays– stepping up to improve the health of underserved communities
- Q&A: Brandon Tsai, Triple Bruin and 2023 UC Grad Slam winner
- Research Programs
- Shrimp Parade: Why Walk on Land? UCLA Biologists Investigate (New York Times)
- Six UCLA Life Scientists on the 2020 list of the world’s most influential researchers
- Stellar Scientists & Mentor Professors
- Stem cell therapy promotes recovery from stroke and dementia in mice
- Stress eating? UCLA researcher provides insights and how you might train your brain to crave healthy foods.
- The Mystery of Monkeypox’s Global Spread (Wired)
- The U.S. is more racially diverse than ever. Will people of color unify politically?
- Tracy Johnson
- Two UCLA Life Sciences professors named inaugural HHMI Freeman Hrabowski Scholars
- UCLA awarded $10 million to improve liver transplantation
- UCLA Biologist named 2021 fellow of the California Academy of Sciences
- UCLA biologist receives Society of Vertebrate Paleontology’s highest award
- UCLA collaborative study finds nearly half of US prisons likely drawing from water contaminated with toxic PFAS
- UCLA distinguished speaker series engages L.A. County Supervisor in conversations about protecting Black mothers before, during and after pregnancy
- UCLA Graduate Programs in Bioscience’s Mentor Training for Faculty
- UCLA leads research to study female health across the animal kingdom–providing insights for a myriad of female health conditions
- UCLA Life Sciences faculty elected to the American Academy of Arts and Sciences
- UCLA Life Sciences Faculty Interviews on “The Science Show”
- UCLA Life Sciences New Student Welcome 2023
- UCLA Life Sciences professor named 2024 Fellow of the American Academy of Microbiology
- UCLA Life Sciences professors among BIOS Top Women in Academic Entrepreneurship
- UCLA Life Sciences professors lend their perspectives to the public event series, 10 Questions: If not now, when?
- UCLA Life Sciences’ undergraduates and mentors– making a difference through the Bunche Fellows Program
- UCLA Life Scientists aim to reduce greenhouse gases and create better tasting cultured meat
- UCLA Life Scientists awarded grants from CNSI Noble Family Innovation Fund to advance sustainability through nanoscience research
- UCLA molecular, cell and developmental biology transfer student receives prestigious 2022 Goldwater Scholarship
- UCLA predictive model identifies most vulnerable communities for COVID-19 public health measures
- UCLA Psychologist weighs in on why most diets don’t work
- UCLA Psychology Distinguished Professor Emerita, Shelley Taylor, receives 2023 National Medal of Science
- UCLA receives $5 million to establish new center to maximize neuroscience potential for public good
- UCLA research highlights the importance of reducing maternal stress to improve child health
- UCLA research paves the way for scaling the production of cultured meat
- UCLA research shows what a sense of belonging can offer to Latinx and African American college students
- UCLA research team receives $1 million grant to study long COVID
- UCLA researchers discover an unexpected regulator of heart repair
- UCLA researchers identify T cell receptors that could lead to new immunotherapies against prostate cancer
- UCLA SACNAS receives national award for promoting Justice, Equity, Diversity and Inclusion
- UCLA Science and Food x LA Times Food Bowl event: “People, Food, and Climate: Thinking Holistically About What We Eat” 9/28/22 (Recording)
- UCLA scientists create mouse research-model for COVID-19 virus and find multiple organ failures
- UCLA scientists make strides toward an ‘off-the-shelf’ immune cell therapy for cancer
- UCLA study finds inbreeding effects that could drive local extinction of Southern California mountain lions
- UCLA study suggests acne bacteria thrive when skin oil turns infection-fighting cells into accomplices
- UCLA survey finds most teens reject glamorized lifestyles in entertainment media
- UCLA undergraduates awarded for their podcast: “Effects of COVID-19 and Social Stigma on Califorina’s undocumented Latinx communities”
- UCLA-Charles R. Drew University partnership receives $11M grant to address inequities in HIV care and prevention
- UCLA-led study assesses effects of climate change and habitat loss on East African bird populations
- UCLA-led study develops a wearable sensor that could guide precision drug dosing
- UCLA-led team maps blood stem cell development – paving the way to better treatments for blood cancer and sickle cell disease
- Undergraduate Student Groups
- Undergraduates
- Undergraduates launch new pre-health program for UCLA’s military-connected students
- View: Event Card List No Image
- View: Featured News Slider
- View: Newsroom Cards
- Voting Information for Bruins
- Ways faculty can support DEI at UCLA
- When it comes to identifying new gene therapies, she’s in it for the long run (Broad Stem Cell Research Center story)
- Wildfires drive L.A.’s mountain lions to take deadly risks
- With $12 million in federal funding, UCLA to expand reach of its depression treatment
- Women’s History Month
- Featured News
- Life Science Scholars
- Uncategorized
- Undergraduate
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- October 2010
© 2024 Regents of the University of California
- Accessibility
- Report Misconduct
- Privacy & Terms of Use
Chemistry Early Path
** Students should meet with departmental Student Affairs Officers, to help pick the right courses and curricular pathway.**
Second Year
Fourth year.
* Only required for select Life Science majors.
Biology Exploration Path
* Only required for select Life Sciences majors.
Physics Early Path
*Physics does not require Physics 5B as a pre-requisite for Physics 5C
Biology First Path
* Only required for select Life Science Majors.
- Utility Menu
Undergraduate Science Education at Harvard
A world of exploration. a world of expertise..

Research FAQs
How do i get started.
Many Harvard undergraduates participate in life sciences research at one of Harvard’s campuses.
If you are a Harvard undergraduate interested in research, Undergraduate Science Research Advisor Kate Penner can help you navigate the process of finding a research group. She can help you:
- define your research interests
- navigate research group websites
- create and edit a science resume and cover letter
- identify and contact research groups
- apply for fellowships and funding
- integrate effectively into your research group & make the most of your research experiences
Before your meeting, think about what kinds of research interest you. What science courses did you enjoy the most? Did you read an article or hear a speaker discuss a topic that made you want to learn more?
How do I know what interests me if I have never done research before?
Even if you have not yet done independent research, you have been introduced to various topics in the life sciences. One strategy for choosing a research area is to recall particular class topics that captivated your interest and made you want to read more. Perhaps you completed a special project in high school or read a compelling science article. Maybe you even did research during high school. Because of the multitude of research opportunities available at Harvard, college is a great time for you to explore new research options and directions.
Begin by browsing the various department and research center web pages .
Invest time to read about faculty research ; you may find a project that grabs your attention in a research area that you weren’t aware of previously. In this broad-based search process, you also learn about the wide range of research projects at Harvard and its affiliated hospitals.
Once you identify a few labs whose research interests you (5 or 6 is usually sufficient), read 1-2 publications from each lab to learn about the group’s research focus, model systems, techniques, and overall goals. If you consider the amount of time that you will spend working in the lab or the field, whether it is full-time during the summer or part-time during the academic year, it makes sense to take the time to investigate a number of research options and identify the ones that will likely be a good fit for you and your interests and career goals.
For more detailed advice, please contact the Undergraduate Research Advisor, Kate Penner .
How do I contact and interview in the labs that I am interested in?
Once you have narrowed down your list of labs, send an introductory email inquiry directly to the faculty member who heads the research group (the Principal Investigator or P.I.). Tailor each email specifically for each lab; do not write a generic letter.
Opening few sentences: Start by introducing yourself and the purpose of your inquiry (i.e. you’d like to speak about summer research opportunities in their lab). Next, because you will have already done background reading, mention specific aspects of their research (citing the lab’s papers you have read) and state why they interest you . Your application will be stronger if you convey not only some knowledge of the lab’s scientific goals, but also a genuine interest in their research area and technical approaches.
Next paragraph: Tell them about yourself, what your goals are and why you want to do research with their group. Describe any previous research experience (as described below, attach your science resume). Previous experience may be helpful, but is not required for joining many research groups. Many undergraduates have not had much, if any, previous experience; professors are looking for students who are highly motivated to learn and dependable.
A brief closing: Give a timeline of your expected start date, how many hours per week you can devote during the academic term, and what your plans are for the summer.
Attach a copy of your science resume , which differs from a typical resume in its focus and concision. List the science courses you have taken and in which you are currently enrolled (if you are applying to labs outside of the Harvard FAS, list the course title in addition to the course number). Condense your high school information : list only the top 2-3 science experiences or accomplishments, and selected academic awards. A one-page science resume will convey key information and be easy to read.
Most faculty will respond to your email if it is clear that you are genuinely interested in their research and have not simply sent out a generic email. If you don’t receive a response with a week or ten days, you can follow up with an email asking if they have had a chance to consider your request. (Include you original correspondence at the end of your follow-up email.) Often faculty are traveling and don't have regular access to email, so you may have to be patient. It's also helpful to be aware of busy times of year (such as weeks at the start of the academic term for faculty who also teach).
If you get a response inviting you to an interview, make sure that you have a broad understanding of the major areas of the faculty's research program. Also be sure to read 1-2 published papers from the lab so you can ask specific questions about their research. If there are other undergraduates working in the lab already, you can contact them and ask about their experiences. For more detailed advise on interview preparation and making decision on which lab to choose from all your offers please contact the Undergraduate Research Advisor .
Each fall Science Education Office brings together Harvard scientists from various departments, institutes and hospitals together for the Harvard Undergraduate Research Opportunities in Science (HUROS) fair. HUROS is a great way to meet dozens of scientists in one day and explore multiple research areas before narrowing down your interest. Please register for the event at HUROS page of this site.
Do I have to stay in the same lab all 4 years or can I try different labs?
The short answer is no, you are not required to remain in the same lab for your entire undergraduate career. There are many reasons for changing a lab:
- your academic interests or concentration may have changed and thus the lab project is no longer appropriate
- you would like to study abroad (note that there is no additional cost in tuition for the term-time study abroad and Harvard has many fellowships for summer study abroad programs)
- your mentor may have moved on and there is no one in the lab to direct your project (it is not unusual for a postdoctoral fellow who is co-mentoring student to move as they secure a faculty position elsewhere)
- the project may not be working and the lab hasn’t offered an alternative
- or there may be personal reasons for leaving. It is acceptable to move on
If you do encounter difficulties, but you strongly prefer to remain in the lab, get help. Talk to your PI or mentor, or reach out to Undergraduate Research Advisor for advice. The PI may not be aware of the problem and bringing it to their attention may be all that is necessary to resolve it.
For students who are satisfied with their research experience, remaining in one lab for the duration of their undergraduate careers can have significant benefits. Students who spend two or three years in the same lab often find that they have become fully integrated members of the research group. In addition, the continuity of spending several years in one lab group often allows students to develop a high level of technical expertise that permits them to work on more sophisticated projects and perhaps produce more significant results.
Students may volunteer, receive a course credit or apply for Harvard Research Fellowships . Students who are on Financial Aid Work-Study may apply it towards their research stipend. Please contact Undergraduate Research Advisor for advise on your specific situation.
Student Responsibilities in the Lab: Lab citizenship and effort
Accepting an undergraduate into a research group and providing training for them is a very resource-intensive proposition for a lab, both in terms of the time commitment required from the lab mentors as well as the cost of laboratory supplies and reagents. It is incumbent upon students to recognize and respect this investment.
- One way for you to acknowledge the lab’s investment is to show that you appreciate the time that your mentors set aside from their own experiments to teach you. For example, try to be meticulous about letting your mentor know well in advance when you are unable to come to the lab as scheduled.
- On the other hand, showing up in the lab at a time that is not on your regular schedule and expecting that your mentor will be available to work with you is unrealistic because they may be in the middle of an experiment that cannot be interrupted for several hours.
- In addition to adhering to your lab schedule, show you respect the time that your mentor is devoting to you by putting forth a sincere effort when you are in the lab. This includes turning off your phone, ignoring text messages, avoiding surfing the web and chatting with your friends in the lab etc. You will derive more benefit from a good relationship with your lab both in terms of your achievements in research and future interactions with the PI if you demonstrate a sincere commitment to them. We have heard reports from some PIs who were unhappy with their undergraduates because they did not appear to appreciate the time that their mentors spent working with them.
- There will be “crunch” times, maybe even whole weeks, when you will be unable to work in the lab as many hours as you normally would because of midterms, finals, paper deadlines, illness or school vacations. This is fine and not unusual for students, but remember to let your mentor know in advance when you anticipate absences. Disappearing from the lab for days without communicating with your mentor is not acceptable. Your lab mentor and PI are much more likely to be understanding about schedule changes if you keep the lines of communication open but they may be less charitable if you simply disappear for days or weeks at a time. From our conversations with students, we have learned that maintaining good communication and a strong relationship with the lab mentor and/or PI correlates well with an undergraduate’s satisfaction and success in the laboratory.
- Perhaps the best way for you to demonstrate your appreciation of the lab’s commitment is to approach your project with genuine interest and intellectual curiosity. Regardless of how limited your time in the lab may be, especially for freshmen and sophomores, it is crucial to convey a sincere sense of engagement with your project and the lab’s research goals. You want to avoid giving the impression that you are there merely to fulfill a degree requirement or as prerequisite for a post-graduate program.
Funding Support
Students conducting research during the fall or spring terms typically either volunteer or earn course credit. For term time financial support, students may also apply for funding through the Harvard College Research Program (HCRP). Students may not simultaneously receive funding and also earn academic credit for a research project.
There are a number of fellowships available for Harvard undergraduates to support summer research projects. Students can also view numerous undergraduate programs and scholarships, including summer research, portable scholarships, and short-term opportunities on the Pathways to Science website .
For specific advise on fellowship selection and tips for writing research proposal please contact Undergraduate Research Advisor .
Research Blog Posts
- Undergraduate Researcher Profile: Jasmine Kung
- Undergraduate Researcher Profile: Tanisha Martheswaran
- Undergraduate Researcher Profile: Esther Yu
- Undergraduate Researcher Profile: Ellen Zhang
- Undergraduate Researcher Profile: Indu Prakash
- Harvard-affiliated Labs
- Research Opportunities and Funding
- Transportation for Researchers
- Undergraduate Research Opportunities (HUROS) Fair
- Undergraduate Research Spotlight
- Resume Template & Proposal Tips
- Lab Citizenship
- Research Ethics and Lab Safety
- Conference Presentation Grants
- Research Advising - Contact Us!
Post-bac Jobs & Resources
Are you a graduating senior who wants to work in a lab for a few years before starting a graduate school or medical school? See Post-Bac Positions Listings and Post-Bac Resources .
Life Sciences

The Graduate School of Arts and Sciences (GSAS) at Harvard University provides exceptional opportunities for study across the depth and breadth of the life sciences through the Harvard Integrated Life Sciences (HILS) federation. The HILS federation comprises 14 Ph.D. programs of study across four Harvard faculties—Harvard Faculty of Arts and Sciences, Harvard T. H. Chan School of Public Health, Harvard Medical School, and Harvard School of Dental Medicine. HILS offers flexibility, including options to take courses, do laboratory rotations, and even choose a dissertation advisor from across the HILS federation, subject to specific program requirements and lab availability.

Since 1983, the Life Sciences Research Foundation (LSRF) has funded nearly 650 outstanding postdoctoral fellows in all areas of the life sciences, and raised more than $60 million from generous industries, foundations and individuals to support this effort.
We believe that discoveries and application of innovations in biology for the public’s good depends upon the training and support of the highest quality young scientists. Every year our selection committee identifies the top 5% of applicants from an international pool of nearly 900 postdoctoral applicants. Once chosen, the LSRF solicits individuals, companies and foundations for the funds to support these young scientists at a critical juncture of their training for the three-year award period. The number of awards depends entirely on our fundraising efforts that year. LSRF does not have an endowment or existing pool of funds. Each year we seek support for our current group of finalists. We were able to fund 27 of 55 finalists in 2018, 27 of 58 finalists in 2019 and 22 of 60 finalists in 2020.
Brief History
In the late 1970s, biologists were founding and leading companies whose goal was to apply modern biological research technology to the solution of important problems in society. While academic and commercial branches of chemistry and engineering had a long history of collaboration, this had not been true of the biological sciences. In 1981 the founder of LSRF, Don Brown, sought to establish similar collaborations in biology, focusing on training the next generation of exceptional biologists. While for-profit enterprises of the life sciences were solicited as major sponsors for postdoctoral fellowships in the early years, sponsorship has evolved with the help of our scientific advisory board to include small and large not-for-profit foundations, and private individuals .
Because all scientists (Board, Peer Review Committee, LSRF Officers) serve LSRF without compensation, administrative costs to sponsors are approximately 4%.
- Share full article
Advertisement
Supported by
Guest Essay
The Long-Overlooked Molecule That Will Define a Generation of Science

By Thomas Cech
Dr. Cech is a biochemist and the author of the forthcoming book “The Catalyst: RNA and the Quest to Unlock Life’s Deepest Secrets,” from which this essay is adapted.
From E=mc² to splitting the atom to the invention of the transistor, the first half of the 20th century was dominated by breakthroughs in physics.
Then, in the early 1950s, biology began to nudge physics out of the scientific spotlight — and when I say “biology,” what I really mean is DNA. The momentous discovery of the DNA double helix in 1953 more or less ushered in a new era in science that culminated in the Human Genome Project, completed in 2003, which decoded all of our DNA into a biological blueprint of humankind.
DNA has received an immense amount of attention. And while the double helix was certainly groundbreaking in its time, the current generation of scientific history will be defined by a different (and, until recently, lesser-known) molecule — one that I believe will play an even bigger role in furthering our understanding of human life: RNA.
You may remember learning about RNA (ribonucleic acid) back in your high school biology class as the messenger that carries information stored in DNA to instruct the formation of proteins. Such messenger RNA, mRNA for short, recently entered the mainstream conversation thanks to the role they played in the Covid-19 vaccines. But RNA is much more than a messenger, as critical as that function may be.
Other types of RNA, called “noncoding” RNAs, are a tiny biological powerhouse that can help to treat and cure deadly diseases, unlock the potential of the human genome and solve one of the most enduring mysteries of science: explaining the origins of all life on our planet.
Though it is a linchpin of every living thing on Earth, RNA was misunderstood and underappreciated for decades — often dismissed as nothing more than a biochemical backup singer, slaving away in obscurity in the shadows of the diva, DNA. I know that firsthand: I was slaving away in obscurity on its behalf.
In the early 1980s, when I was much younger and most of the promise of RNA was still unimagined, I set up my lab at the University of Colorado, Boulder. After two years of false leads and frustration, my research group discovered that the RNA we’d been studying had catalytic power. This means that the RNA could cut and join biochemical bonds all by itself — the sort of activity that had been thought to be the sole purview of protein enzymes. This gave us a tantalizing glimpse at our deepest origins: If RNA could both hold information and orchestrate the assembly of molecules, it was very likely that the first living things to spring out of the primordial ooze were RNA-based organisms.
That breakthrough at my lab — along with independent observations of RNA catalysis by Sidney Altman at Yale — was recognized with a Nobel Prize in 1989. The attention generated by the prize helped lead to an efflorescence of research that continued to expand our idea of what RNA could do.
In recent years, our understanding of RNA has begun to advance even more rapidly. Since 2000, RNA-related breakthroughs have led to 11 Nobel Prizes. In the same period, the number of scientific journal articles and patents generated annually by RNA research has quadrupled. There are more than 400 RNA-based drugs in development, beyond the ones that are already in use. And in 2022 alone, more than $1 billion in private equity funds was invested in biotechnology start-ups to explore frontiers in RNA research.
What’s driving the RNA age is this molecule’s dazzling versatility. Yes, RNA can store genetic information, just like DNA. As a case in point, many of the viruses (from influenza to Ebola to SARS-CoV-2) that plague us don’t bother with DNA at all; their genes are made of RNA, which suits them perfectly well. But storing information is only the first chapter in RNA’s playbook.
Unlike DNA, RNA plays numerous active roles in living cells. It acts as an enzyme, splicing and dicing other RNA molecules or assembling proteins — the stuff of which all life is built — from amino acid building blocks. It keeps stem cells active and forestalls aging by building out the DNA at the ends of our chromosomes.
RNA discoveries have led to new therapies, such as the use of antisense RNA to help treat children afflicted with the devastating disease spinal muscular atrophy. The mRNA vaccines, which saved millions of lives during the Covid pandemic, are being reformulated to attack other diseases, including some cancers . RNA research may also be helping us rewrite the future; the genetic scissors that give CRISPR its breathtaking power to edit genes are guided to their sites of action by RNAs.
Although most scientists now agree on RNA's bright promise, we are still only beginning to unlock its potential. Consider, for instance, that some 75 percent of the human genome consists of dark matter that is copied into RNAs of unknown function. While some researchers have dismissed this dark matter as junk or noise, I expect it will be the source of even more exciting breakthroughs.
We don’t know yet how many of these possibilities will prove true. But if the past 40 years of research have taught me anything, it is never to underestimate this little molecule. The age of RNA is just getting started.
Thomas Cech is a biochemist at the University of Colorado, Boulder; a recipient of the Nobel Prize in Chemistry in 1989 for his work with RNA; and the author of “The Catalyst: RNA and the Quest to Unlock Life’s Deepest Secrets,” from which this essay is adapted.
The Times is committed to publishing a diversity of letters to the editor. We’d like to hear what you think about this or any of our articles. Here are some tips . And here’s our email: [email protected] .
Follow the New York Times Opinion section on Facebook , Instagram , TikTok , WhatsApp , X and Threads .
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
Collection 15 July 2019
Top 50: Life and Biological Sciences
We are pleased to share with you the 50 most read Nature Communications life and biological sciences articles* published in 2018. Featuring authors from around the world, these papers highlight valuable research from an international community.
Browse all Top 50 subject area collections here .
*Based on data from altmetric.com , covering January-December 2018

Embryos and embryonic stem cells from the white rhinoceros
The Southern (SWR) and Northern (NWR) are two subspecies of the White Rhinoceros with the NWR being almost extinct. Here, using assisted reproduction technology, the authors produce and cryopreserve SWR purebred and NWR-SWR hybrid embryos developed to the blastocyst stage, and also generate embryonic stem cell lines, in an attempt to save genes of the NWR.
- Thomas B. Hildebrandt
- Robert Hermes
- Cesare Galli
Scutoids are a geometrical solution to three-dimensional packing of epithelia
Cell arrangement in the plane of epithelia is well studied, but its three-dimensional packing is largely unknown. Here the authors model curved epithelia and predict that cells adopt a geometrical shape they call “scutoid”, resulting in different apical and basal neighbours, and confirm the presence of scutoids in curved tissues.
- Pedro Gómez-Gálvez
- Pablo Vicente-Munuera
- Luis M. Escudero

Similar neural responses predict friendship
Though we are often friends with people similar to ourselves, it is unclear if neural responses to perceptual stimuli are also similar. Here, authors show that the similarity of neural responses evoked by a range of videos was highest for close friends and decreased with increasing social distance.
- Carolyn Parkinson
- Adam M. Kleinbaum
- Thalia Wheatley
Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function
Cognitive function is associated with health and important life outcomes. Here, the authors perform a genome-wide association study for general cognitive function in 300,486 individuals and identify genetic loci that implicate neural and cell developmental pathways in this trait.
- Gail Davies
- Ian J. Deary
Wing bone geometry reveals active flight in Archaeopteryx
Archaeopteryx had a mix of traits seen in non-flying dinosaurs and flying birds, leading to debate on whether it had powered flight. Here, Voeten et al. compare wing bone architecture from Archaeopteryx and both flying and non-flying archosaurs, supporting that Archaeopteryx had powered flight but with a different stroke than that of modern birds.
- Dennis F. A. E. Voeten
- Sophie Sanchez
Hunting regulation favors slow life histories in a large carnivore
Hunting and harvesting are generally expected to select for faster life histories in the exploited species. Here, the authors analyse data from a hunted population of brown bears in Sweden and show that regulations protecting females with dependent young lead hunting to favor prolonged maternal care.
- Joanie Van de Walle
- Gabriel Pigeon
- Fanie Pelletier
Closed-loop stimulation of temporal cortex rescues functional networks and improves memory
Memory lapses can occur due to ineffective encoding, but it is unclear if targeted brain stimulation can improve memory performance. Here, authors use a closed-loop system to decode and stimulate periods of ineffective encoding, showing that stimulation of lateral temporal cortex can enhance memory.
- Youssef Ezzyat
- Paul A. Wanda
- Michael J. Kahana
Genome-wide study of hair colour in UK Biobank explains most of the SNP heritability
Natural hair colour in Europeans is a complex genetic trait. Here, the authors carry out a genome-wide association study using UK BioBank data, suggesting that in combination with pigmentation genes, variants with roles in hair texture and growth can affect hair colouration or our perception of it.
- Michael D. Morgan
- Erola Pairo-Castineira
- Ian J. Jackson
Diffusion markers of dendritic density and arborization in gray matter predict differences in intelligence
Previous studies suggest that individual differences in intelligence correlate with circuit complexity and dendritic arborization in the brain. Here the authors use NODDI, a diffusion MRI technique, to confirm that neurite density and arborization are inversely related to measures of intelligence.
- Christoph Fraenz
- Rex E. Jung
Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer
Production of aromatic monoterpene molecules in hop flowers is affected by genetic, environmental, and processing factors. Here, the authors engineer brewer’s yeast for the production of linalool and geraniol, and show pilot-scale beer produced by engineered strains reconstitutes some qualities of hop flavor.
- Charles M. Denby
- Rachel A. Li
- Jay D. Keasling

A bony-crested Jurassic dinosaur with evidence of iridescent plumage highlights complexity in early paravian evolution
A number of paravian dinosaurs have been described from the Jurassic Yanliao biota, but these have tended to be morphologically similar to Archaeopteryx . Here, Hu. describe the new paravian dinosaur, Caihong juji gen. et sp. nov., which possesses a suite of unusual skeletal and feather characteristics.
- Julia A. Clarke
Single-dose testosterone administration increases men’s preference for status goods
Testosterone is believed to be involved in social rank-related behavior. Here, the authors show that one dose of testosterone increases men’s preference for “high status” goods and brands, suggesting a role for testosterone in modern consumer behavior in men.
- H. Plassmann
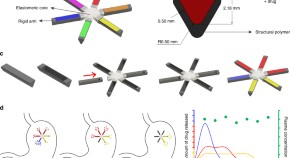
Development of an oral once-weekly drug delivery system for HIV antiretroviral therapy
Poor adherence to daily antiretrovirals can significantly affect treatment efficacy, but oral long-acting antiretrovirals are currently lacking. Here, the authors develop a once-weekly oral dosage form for anti-HIV drugs, assess its pharmacokinetics in pigs, and model its impact on viral resistance and disease epidemics.
- Ameya R. Kirtane
- Omar Abouzid
- Giovanni Traverso
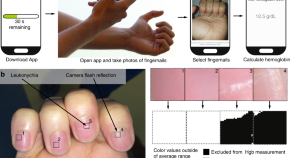
Smartphone app for non-invasive detection of anemia using only patient-sourced photos
Anemia has a global prevalence of over 2 billion people and is diagnosed via blood-based laboratory test. Here the authors describe a smartphone app that can estimate hemoglobin levels and detect anemia by analyzing pictures of fingernail beds taken with a smartphone and without the need of any external equipment.
- Robert G. Mannino
- David R. Myers
- Wilbur A. Lam
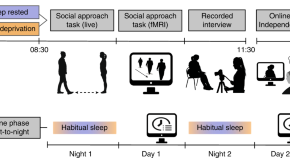
Sleep loss causes social withdrawal and loneliness
Loneliness markedly increases mortality and morbidity, yet the factors triggering loneliness remain largely unknown. This study shows that sleep loss leads to a neurobehavioral phenotype of human social separation and loneliness, one that is transmittable to non-sleep-deprived individuals.
- Eti Ben Simon
- Matthew P. Walker
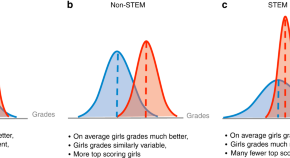
Gender differences in individual variation in academic grades fail to fit expected patterns for STEM
Men are over-represented in the STEM (science, technology, engineering and mathematics) workforce even though girls outperform boys in these subjects at school. Here, the authors cast doubt on one leading explanation for this paradox, the ‘variability hypothesis’.
- R. E. O’Dea
- S. Nakagawa
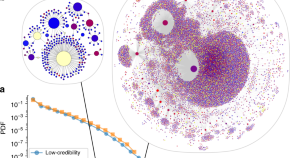
The spread of low-credibility content by social bots
Online misinformation is a threat to a well-informed electorate and undermines democracy. Here, the authors analyse the spread of articles on Twitter, find that bots play a major role in the spread of low-credibility content and suggest control measures for limiting the spread of misinformation.
- Chengcheng Shao
- Giovanni Luca Ciampaglia
- Filippo Menczer
Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere
Giant viruses are the largest viruses of the known virosphere and their genetic analysis can provide insights into virus evolution. Here, the authors discover Tupanvirus, a unique giant virus that has an unusually long tail and contains the largest translational apparatus of the known virosphere.
- Jônatas Abrahão
- Lorena Silva
- Bernard La Scola
Genome-wide meta-analysis implicates mediators of hair follicle development and morphogenesis in risk for severe acne
Acne vulgaris is a chronic inflammation of the skin, the genetic basis of which is incompletely understood. Here, Petridis et al. perform GWAS and meta-analysis for acne in 26,722 individuals and identify 12 novel risk loci that implicate structure and maintenance of the skin in severe acne risk.
- Christos Petridis
- Alexander A. Navarini
- Michael A. Simpson
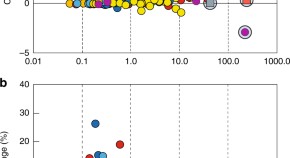
Patterns of island change and persistence offer alternate adaptation pathways for atoll nations
Inundation and erosion could make many atoll islands uninhabitable over the next century. Here the authors present an analysis of change in the atoll nation of Tuvalu that shows a 2.9% increase in land area over the past four decades, with 74% of islands increasing in size, despite rising sea levels.
- Paul S. Kench
- Murray R. Ford
- Susan D. Owen
Blood–brain barrier opening in Alzheimer’s disease using MR-guided focused ultrasound
Magnetic resonance-guided focused ultrasound with injected microbubbles has been used to temporarily open the blood–brain barrier (BBB) in animal models of Alzheimer's disease (AD). Here, the authors use this technology to non-invasively open the BBB in 5 patients with mild-to-moderate AD in a phase I trial, and show that the procedure is safe.
- Nir Lipsman
- Sandra E. Black
Cultural hitchhiking and competition between patrilineal kin groups explain the post-Neolithic Y-chromosome bottleneck
A population bottleneck 5000-7000 years ago in human males, but not females, has been inferred across several African, European and Asian populations. Here, Zeng and colleagues synthesize theory and data to suggest that competition among patrilineal kin groups produced the bottleneck pattern.
- Tian Chen Zeng
- Marcus W. Feldman
Caffeine-inducible gene switches controlling experimental diabetes
Control of transgene expression should ideally be easy and with minimal side effects. Here the authors present a synthetic biology-based approach in which the caffeine in coffee regulates a genetic circuit controlling glucagon-like peptide 1 expression in diabetic mice.
- Daniel Bojar
- Leo Scheller
- Martin Fussenegger
Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen
Microbes in the cow rumen are crucial for the breakdown of plant material. Here, Stewart et al. assemble over 900 bacterial and archaeal genomes from the cow rumen microbiome, revealing new species and genes encoding enzymes with potential roles in carbohydrate metabolism.
- Robert D. Stewart
- Marc D. Auffret
- Mick Watson
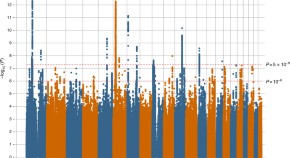
Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways
The UK Biobank provides data for three depression-related phenotypes. Here, Howard et al. perform a genome-association study for broad depression, probable major depressive disorder (MDD) and hospital record-coded MDD in up to 322,580 UK Biobank participants which highlights excitatory synaptic pathways.
- David M. Howard
- Mark J. Adams
- Andrew M. McIntosh
Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes
Interactions with other microbes may inhibit or facilitate the dispersal of bacteria. Here, Zhang et al. use cheese rind microbiomes as a model to show that physical networks created by filamentous fungi can affect the dispersal of motile bacteria and thus shape the diversity of microbial communities.
- Yuanchen Zhang
- Erik K. Kastman
- Benjamin E. Wolfe
Epidemiology is a science of high importance
Epidemiology dates back to the Age of Pericles in 5th Century B.C., but its standing as a ‘true’ science in 21st century is often questioned. This is unexpected, given that epidemiology directly impacts lives and our reliance on it will only increase in a changing world.
A low-gluten diet induces changes in the intestinal microbiome of healthy Danish adults
Gluten-free diets are increasingly common in the general population. Here, the authors report the results of a randomised cross-over trial involving middle-aged, healthy Danish adults, showing evidence that a low-gluten diet leads to gut microbiome changes, possibly due to variations in dietary fibres.
- Lea B. S. Hansen
- Henrik M. Roager
- Oluf Pedersen
In utero nanoparticle delivery for site-specific genome editing
The correction of genetic defects in utero could allow for improved outcomes of gene therapy. Here, the authors demonstrate safe delivery of nanoparticles to fetal mouse tissues, and show that nanoparticles containing peptide nucleic acids to edit the beta-globin gene are effective in a mouse model of beta-thalassemia.
- Adele S. Ricciardi
- Raman Bahal
- W. Mark Saltzman
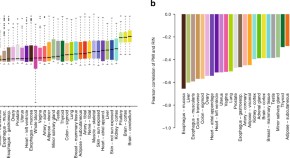
The effects of death and post-mortem cold ischemia on human tissue transcriptomes
RNA levels in post-mortem tissue can differ greatly from those before death. Studying the effect of post-mortem interval on the transcriptome in 36 human tissues, Ferreira et al. find that the response to death is largely tissue-specific and develop a model to predict time since death based on RNA data.
- Pedro G. Ferreira
- Manuel Muñoz-Aguirre
- Roderic Guigó
Gimap5-dependent inactivation of GSK3β is required for CD4 + T cell homeostasis and prevention of immune pathology
Loss of function GIMAP5 mutation is associated with lymphopenia, but how it mediates T cell homeostasis is unclear. Here the authors study Gimap5 −/− mice and a patient with GIMAP5 deficiency to show how this GTPAse negatively regulates GSK3β activity to prevent DNA damage and cell death.
- Andrew R. Patterson
- Mehari Endale
- Kasper Hoebe
3D virtual reconstruction of the Kebara 2 Neandertal thorax
How different Neandertal morphology was from that of modern humans has been a subject of long debate. Here, the authors develop a 3D virtual reconstruction of the thorax of an adult male Neandertal, showing similar size to modern humans, yet with greater respiratory capacity due to its different shape.
- Asier Gómez-Olivencia
- Alon Barash
Exercise induces new cardiomyocyte generation in the adult mammalian heart
The adult mammalian heart has a limited cardiomyogenic capacity. Here the authors show that intensive exercise leads to a 4.6-fold increase in murine cardiomyocyte proliferation requiring the expression of miR-222, and that exercise induces an extended cardiomyogenic response in the murine heart after infarction.
- Carolin Lerchenmüller
- Anthony Rosenzweig
Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses
Relatively well conserved domains of influenza A virus (IAV) proteins are potential candidates for the development of a universal IAV vaccine. Here, Deng et al . combine two such conserved antigens (M2e and HA stalk) in a double-layered protein nanoparticle and show that it protects against divergent IAVs in mice.
- Teena Mohan
- Bao-Zhong Wang
Intellectual synthesis in mentorship determines success in academic careers
While successful mentors tend to train successful students in academic career, it’s unclear how mentorship determines chances of a success in a trainee. Here, Liénard and colleagues analyze approximately 20 K mentor/trainee relationships in life sciences, and find that success of trainees is associated with an intellectual synthesis between their mentors’ research.
- Jean F. Liénard
- Titipat Achakulvisut
- Stephen V. David
Effective weight control via an implanted self-powered vagus nerve stimulation device
Developing new technologies for the neuromodulation of the vagus nerve can enable therapeutic strategies for body weight control in obese patients. Here, the authors present a battery-free self-powered implantable vagus nerve stimulation system that electrically responds to stomach movement.
- Xudong Wang
Olfactory receptor OR2AT4 regulates human hair growth
Increasing evidence suggest that olfactory receptors can carry additional functions besides olfaction. Here, Chéret et al. show that stimulation of the olfactory receptor ORT2A4 by the odorant Sandalore ® stimulates growth of human scalp hair follicles ex vivo, suggesting the use of ORT2A4-targeting odorants as hair growth-promoting agents.
- Jérémy Chéret
- Marta Bertolini

A diminutive perinate European Enantiornithes reveals an asynchronous ossification pattern in early birds
Fossil juvenile Mesozoic birds are exceedingly rare and can provide important insight into the early evolution of avian development. Here, Knoll et al. describe one of the smallest known Mesozoic avians, which indicates a clade-wide asynchronous pattern of osteogenesis and great variation in basal bird hatchling size and skeletal maturation tempo.
- Fabien Knoll
- Luis M. Chiappe
- Jose Luis Sanz
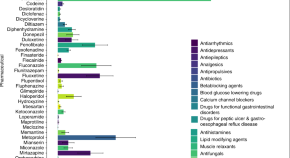
A diverse suite of pharmaceuticals contaminates stream and riparian food webs
Pharmaceuticals are widespread contaminants in surface waters. Here, Richmond and colleagues show that dozens of pharmaceuticals accumulate in food chains of streams, including in predators in adjacent terrestrial ecosystems.
- Erinn K. Richmond
- Emma J. Rosi
- Michael R. Grace
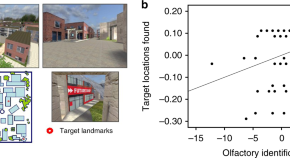
An intrinsic association between olfactory identification and spatial memory in humans
Olfaction, the sense of smell, may have originally evolved to aid navigation in space, but there is no direct evidence of a link between olfaction and navigation in humans. Here the authors show that olfaction and spatial memory abilities are correlated and rely on similar brain regions in humans.
- Louisa Dahmani
- Raihaan M. Patel
- Véronique D. Bohbot
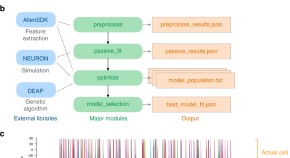
Systematic generation of biophysically detailed models for diverse cortical neuron types
Neocortical circuits exhibit diverse cell types that can be difficult to build into computational models. Here the authors employ a genetic algorithm-based parameter optimization to generate multi-compartment Hodgkin-Huxley models for diverse cell types in the Allen Cell Types Database.
- Nathan W. Gouwens
- Anton Arkhipov
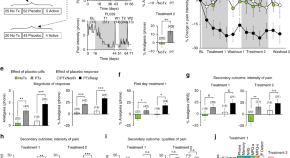
Brain and psychological determinants of placebo pill response in chronic pain patients
People vary in the extent to which they feel better after taking an inert, placebo, treatment, but the basis for individual placebo response is unclear. Here, the authors show how brain structural and functional variables, as well as personality traits, predict placebo response in those with chronic back pain.
- Etienne Vachon-Presseau
- Sara E. Berger
- A. Vania Apkarian
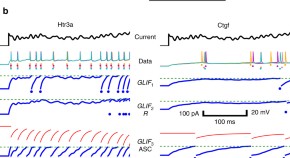
Generalized leaky integrate-and-fire models classify multiple neuron types
Simplified neuron models, such as generalized leaky integrate-and-fire (GLIF) models, are extensively used in network modeling. Here the authors systematically generate and compare GLIF models of varying complexity for their ability to classify cell types in the Allen Cell Types Database and faithfully reproduce spike trains.
- Corinne Teeter
- Ramakrishnan Iyer
- Stefan Mihalas
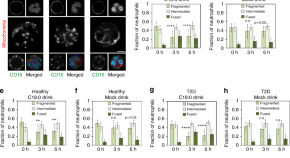
Dietary stearic acid regulates mitochondria in vivo in humans
Dietary fatty acids have different effects on human health. Here, the authors show that ingestion of the fatty acid C18:0, but not of C16:0, rapidly leads to fusion of mitochondria and fatty acid oxidation in humans, possibly explaining the health benefits of C18:0.
- Deniz Senyilmaz-Tiebe
- Daniel H. Pfaff
- Aurelio A. Teleman
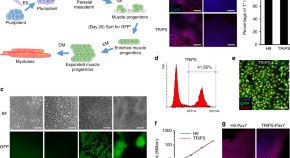
Engineering human pluripotent stem cells into a functional skeletal muscle tissue
The generation of functional skeletal muscle tissue from human pluripotent stem cells has not been reported. Here, the authors describe engineering of contractile skeletal muscle bundles in culture, which become vascularized and maintain functionality when transplanted into mice.
- Lingjun Rao
- Nenad Bursac
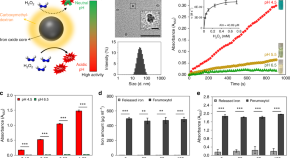
Topical ferumoxytol nanoparticles disrupt biofilms and prevent tooth decay in vivo via intrinsic catalytic activity
Ferumoxytol is a nanoparticle formulation approved for systemic use to treat iron deficiency. Liu et al. show that topical use of ferumoxytol, in combination with low concentrations of H 2 O 2 , disrupts intractable oral biofilms and prevents tooth decay in vitro and in an animal model.
- Pratap C. Naha
Honey bee Royalactin unlocks conserved pluripotency pathway in mammals
Royal jelly is the queen-maker for the honey bee that also has effects on longevity, fertility, and regeneration in mammals. Here the authors provide evidence that its major protein component Royalactin, and the mammalian structural analog Regina, maintain pluripotency in mouse ESCs by activating a ground-state pluripotency-like gene network.
- Derrick C. Wan
- Stefanie L. Morgan
- Kevin C. Wang
Deoxyribose and deoxysugar derivatives from photoprocessed astrophysical ice analogues and comparison to meteorites
Sugars are known to form from the UV photoprocessing of ices under astrophysical conditions. Here, the authors report the detection of deoxyribose, the sugar of DNA, and other deoxysugars from the UV photoprocessing of H 2 O:CH 3 OH ice mixtures, which are compared with materials from carbonaceous meteorites.
- Michel Nuevo
- George Cooper
- Scott A. Sandford
Genomic evidence of speciation reversal in ravens
Speciation reversal is known mainly from recently diverged lineages that have come into secondary contact following anthropogenic disturbance. Here, Kearns et al. use genomic and phylogenomic analyses to show that the Common Raven ( Corvus corax ) was formed by the ancient fusion of two non-sister lineages of ravens.
- Anna M. Kearns
- Marco Restani
- Kevin E. Omland
Analysis of 3800-year-old Yersinia pestis genomes suggests Bronze Age origin for bubonic plague
Yersinia pestis has caused infections (plague) in humans since the Early Bronze Age (5000 years ago). Here, Spyrou et al. reconstruct Y. pestis genomes from Late Bronze Age individuals, and find genomic evidence compatible with flea-mediated transmission causing bubonic plague.
- Maria A. Spyrou
- Rezeda I. Tukhbatova
- Johannes Krause
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Apply to UMaine
UMaine News

Graduate students sweep state competition for life science research
Three University of Maine graduate students placed as finalists in the 6th annual BioME Student Showcase for life science research. The annual event supports innovation and the commercialization of student ideas in Maine, and allows students the opportunity to network with professionals or connect with entrepreneurial resources. Ten UMaine undergraduate and graduate students were selected to participate in the competition, held April 27 at the University of Southern Maine McGoldrick Center in Portland.
Twenty-eight participated in the showcase across three different categories split by enrollment in high school or in a college undergraduate or graduate program. In order to be considered for the showcase, each student was required to submit an application.
Among college graduates, Sarah Holbrook placed first for her research titled “A Novel Category of Neuromuscular Disease: A Quest for a Cure”; Lola Holcomb finished second with research titled “ Developing ‘Microbiome Medicine’ with Broccoli and Bacteria”; and Samantha Costa placed third wit h “Increased PD-L1 expression in obese mice leads to bone marrow immunosuppression and increases the susceptibility for bone marrow metastases.” All three are UMaine students.
The graduate-level competition is called a “Fast Pitch” and was open to any graduate student enrolled in a Maine academic institution. Participants were required to share their research in a 5 minute pitch to judges with no more than five presentation slides, with the focus angled toward how their research could be commercialized. In-depth Q&A sessions followed their pitches and allowed them to demonstrate their knowledge of the subject. First to third place awards ranged from $2,500 to $700.
- UMaine Today Magazine
- Submit news
Origins of 'Welsh dragons' finally exposed by experts
A large fossil discovery has helped shed light on the history of dinosaurs in Wales.
Until recently, the land of the dragon didn't have any dinosaurs. However, in the last ten years, several dinosaurs have been reported, but their life conditions were not well known. In a new study by a team from the University of Bristol and published in Proceedings of the Geologists' Association , important details have been revealed for the first time.
They found that early Welsh dinosaurs from over 200 million year ago lived on a tropical lowland beside the sea. Dinosaur trackways are known from Barry and other sites nearby, showing that dinosaurs had walked across the warm lowlands.
The discovery was made at Lavernock Point, close to Cardiff and Penarth, where the cliffs of dark-coloured shales and limestones document ancient shallow seas. At several levels, there are accumulations of bones, including the remains of fish, sharks, marine reptiles and occasionally, dinosaurs.
Former student of the Bristol MSc in Palaeobiology Owain Evans led the study. He explained: "The bone bed paints the picture of a tropical archipelago, which was subjected to frequent storms, that washed material from around the surrounding area, both in land and out at sea, into a tidal zone. This means that from just one fossil horizon, we can reconstruct a complex ecological system, with a diverse array of marine reptiles like ichthyosaurs, plesiosaurs and placodonts in the water, and dinosaurs on land.
"I had visited the coast at Penarth all my life, growing up in Cardiff, but never noticed the fossils. Then, the more I read, the more amazing it became. Local geologists had been collecting bones since the 1870s, and most of these are in the National Museum of Wales in Cardiff."
Cindy Howells, Curator of Palaeontology at the National Museum of Wales, adds: "The collections from Lavernock go all the way back to the 19 th century, with many sections of the bone bed being collected over the years. The presence of dinosaur fossils at the site ensure that it remains one of the most significant localities for palaeontology in Wales."
Two discoveries made by the team while conducting fieldwork at Lavernock were the fossilized remains of a placodont osteoderm, and a single coelacanth gular bone. Supervisor Dr Chris Duffin said: "The remains of coelacanths and placodonts are relatively rare in the UK, which makes these finds even more remarkable. These two fossils alone help build a broader picture of what the Rhaetian in the UK would have looked like."
Professor Michael Benton from Bristol's School of Earth Sciences, another project supervisor, adds: "The volume of dinosaur remains found at Lavernock is extremely exciting, and is a chance to study a complex, and often mysterious period in their evolutionary history. We have identified the remains of a large Plateosaurus like animal, along with several bones which likely belonged to a predatory theropod."
A significant section of the paper is dedicated to the abundant microfossils found at the site, which include fish teeth, scales and bone fragments. By examining thousands of specimens, the team were able to identify the key species in the shallow seas and work out the relative importance of each.
The origins of the Welsh dragons have been pinned down at last.
- Paleontology
- Ancient DNA
- Tyrannosaurus Rex
- Origin of Life
- Anthropology
- Archaeopteryx
- Ichthyosaur
- Feathered dinosaurs
- Stegosaurus
- Paralititan
- Brachiosaurus
Story Source:
Materials provided by University of Bristol . Note: Content may be edited for style and length.
Related Multimedia :
- Artist’s depiction of a British Archipelago during the Triassic
Journal Reference :
- Owain Evans, Christopher J. Duffin, Claudia Hildebrandt, Michael J. Benton. Microvertebrates from the basal Rhaetian Bone Bed (Late Triassic) at Lavernock, South Wales . Proceedings of the Geologists' Association , 2024; DOI: 10.1016/j.pgeola.2024.05.001
Cite This Page :
Explore More
- Resting Brain: Neurons Rehearse for Future
- Observing Single Molecules
- A Greener, More Effective Way to Kill Termites
- One Bright Spot Among Melting Glaciers
- Martian Meteorites Inform Red Planet's Structure
- Volcanic Events On Jupiter's Moon Io: High Res
- What Negative Adjectives Mean to Your Brain
- 'Living Bioelectronics' Can Sense and Heal Skin
- Extinct Saber-Toothed Cat On Texas Coast
- Some Black Holes Survive in Globular Clusters
Trending Topics
Strange & offbeat.
UTA researcher recognized for data science leadership
Thursday, May 30, 2024 • Katherine Egan Bennett : contact

Xinlei “Sherry” Wang, director for research in the Division of Data Science at The University of Texas at Arlington, has been named a 2024 fellow of the American Statistical Association (ASA) .
Wang, the Jenkins Garret Endowed Professor of Statistics and Data Science, was selected for her outstanding contributions in developing and applying statistical and computational methods to complex data challenges, for excellence in mentoring doctoral students and junior researchers and for exceptional leadership of data science programs.
Founded in 1839, ASA is the world’s largest community of statisticians and the second oldest continuously operating professional association in the country. ASA fellows have been designated for nearly 100 years, and only up to one-third of 1% of the total association membership can be elected each year.
“I am honored to be elected an ASA fellow and recognized by my profession,” Wang said. “I'm truly grateful for the support I've received from my department, college and University since I joined UTA, which undoubtedly contributed to this achievement.”
Wang came to UTA in January 2023 as director of the College of Science’s newly created Center for Data Science Research and Education. She and her colleagues launched UTA’s first graduate degree in data science in fall 2023.
In January 2024, College of Science Dean Morteza Khaledi appointed Wang as director for research of the new Division of Data Science, which serves as a hub to organize, provide infrastructure support and facilitate growth in instructional and research programs involving data science.
“Dr. Wang is very deserving of this honor of being named an ASA fellow,” Dr. Khaledi said. “This recognition is a testament to her outstanding scholarship. She has played a major role in building and advancing our data science program in the College of Science, and we’re very fortunate to have her here at UTA.”
Wang received a Bachelor of Science in automatic control from the University of Science and Technology of China in 1997, a master’s in statistics from the University of Pittsburgh in 1998 and a doctorate in decision science and statistics from UT Austin in 2002. Her research has been funded by major grants from the National Science Foundation, National Institutes of Health, and the Cancer Prevention and Research Institute of Texas.
She has authored or co-authored more than 90 papers in refereed publications and made more than 75 presentations at conferences and seminars around the world. She has directed Ph.D. dissertations for 19 students, with seven more in progress.
Wang has been an ASA member since 1999 and has served the ASA in various capacities, including as vice president (2009-10) and president (2010-11) of the North Texas Chapter. She will be formally recognized as a 2024 fellow at an awards ceremony at the 2024 Joint Statistical Meetings in Portland, Oregon, in August.
- By Greg Pederson , College of Science

Life-changing impact
Student research shows the positive effects of gender-affirming voice therapy on trans individuals.
Have you ever listened to a recording of yourself and thought that it didn’t sound like you? Or maybe you didn’t like the way your voice sounded in the recording?
Mary Wilkens uses this simple analogy to help others understand the complexities of vocal dysphoria — the feeling that one’s voice doesn’t align with their gender identity. She said vocal dysphoria can be mentally exhausting for a transgender person as they transition from one gender identity to another and potentially cause feelings of unhappiness, stress, anxiety and depression.
“I’ve always felt that this is a really important thing that isn’t talked about during a lot of people’s transitions,” Wilkens said. “Vocal dysphoria is really huge, and people struggle to understand what it is and how it would feel.”
As a member of the LGBTQ+ community and a third-year speech language hearing sciences student at the University of Cincinnati, Wilkens is motivated to serve her community by advocating for the importance of gender-affirming voice therapy.
In January, she was accepted to present a case study from her research on the subject at the Voice Foundation’s 53rd Annual Symposium, which will take place in Philadelphia at the end of May. In January, Wilkens also received a $700 Undergraduate Research Fellowship Award from UC to help offset conference presentation–related costs.
How she came to research
Third-year speech language hearing sciences student Mary Wilkens
While she’s always been interested in gender-affirming voice therapy and anything related to voice studies, Wilkens said research was “actually a surprise interest,” one that stemmed from a conversation with a professor during her first semester at UC.
In a success in allied health class, Wilkens and her classmates were tasked with sharing what interested them in the wide field of speech pathology. After sharing her passion for voice studies, her professor Amy Hobek, PhD, connected her with Victoria McKenna, PhD, an assistant professor and director of UC’s Voice and Swallow Mechanics (VSM) Lab — a research lab that aims to increase access to clinical tools and improve the efficiency of diagnosis and treatment for voice and swallowing disorders.
The faculty encouraged Wilkens to apply for UC’s UPRISE (Undergraduates Pursuing Research in Science and Engineering) program, which is focused on increasing the pipeline of individuals from underserved communities into the STEMM fields. Wilkens was accepted and participated in a paid 12-week research opportunity in the VSM Lab the following summer, with McKenna serving as her mentor.
“I knew right away that Mary had an aptitude for voice research,” McKenna said. “The UPRISE program provided us an avenue to explore her different interests in voice science, and now we are working together on treatment outcomes for gender-affirming voice services.”
Though the UPRISE program was only a 12-week experience, Wilkens never stopped working with McKenna in the VSM Lab. Throughout her time in the lab, Wilkens has enjoyed working on a variety of research projects, but her favorite — the one that will inform her poster presentation at the Voice Foundation’s symposium in May — has been the Gender Diversity Project.
Her hands-on experience
When transgender individuals transition from one gender identity to another, they may seek gender-affirming medical care, including voice therapy or voice surgery to align their voice with their gender identity. The Gender Diversity Project was created to explore how specific speech and voice characteristics influence gender-perception and how socioeconomic and psychosocial factors influence access to transgender voice care and vocal outcomes.
“This project is really awesome because we’re looking throughout a person’s voice therapy process to see how their voice changes both perceptually and physically and proving that therapy is effective,” Wilkens said. “We’re proving that there is significant change and that the therapy is important and has a positive effect on their life.”
Wilkens joined the Gender Diversity Project in its early stages when the team was focused on spreading awareness of the project and surveying transgender people locally and throughout the United States about their experiences with voice therapy.
As the project gained participants — individuals undergoing gender-affirming voice therapy and surgery at UC Voice and Airway Center who were referred to the VSM Lab by a clinician — Wilkens began helping collect evidence to prove that the voice intervention the participants were receiving was effective. This involved taking baseline measurements of how an individual’s pitch had changed as a result of their therapy.
Wilkens’s role in this process was to help record an individual's larynx (or voice box) while McKenna used a scope (a high-speed camera) to assess the larynx’s anatomy. Wilkens also helped conduct questionnaires that measured the participants’ quality of life before and after receiving voice therapy, which helped further prove the positive impact the therapy had on their lives.
Wilkens’ upcoming national presentation will highlight a comprehensive case study on a participant who received vocal feminization surgery but developed a large granuloma (a growth on the vocal cord) during the healing process. Wilkens presented her preliminary findings of how the granuloma affected the participant’s healing and subsequent voice therapy at the VSM Lab’s biannual symposium in December and will present additional findings at the Voice Foundation’s symposium in May.
Mary Wilkens presents on her research.
“Mary has collected physiological, acoustical and quality-of-life outcomes over one whole year since our patient received gender-affirming voice surgery,” McKenna added about the case study. “Mary’s dedication to the project has been so important to its success. Very few studies are able to follow patients that long, and we are getting really important data on how transition impacts the lives of our patients long-term.”
Beyond the symposium, Wilkens looks forward to continuing researching transgender voice and working on her fourth-year capstone project in the VSM Lab. And while she still has graduation, graduate school and other challenges to tackle before entering the field as a full-time clinician, Wilkens has her sights set on working in a medical setting.
“I really enjoy medical work, and I love that they get to work with wide varieties of populations,” she said, “and if I’m in a clinical setting, I can continue my research, and I really love being able to research.”
Featured image at top: Mary Wilkens
Make an impact
Your research can make an impact in the everyday lives of others. Do you have a passion for healing? Apply to one of our highly ranked programs and ask us how you can make a difference through clinical and research opportunities here in the College of Allied Health Sciences.
Katie Coburn
- College of Allied Health Sciences
- Student Experience
- Next Lives Here
- Equity and Inclusion
- Experience-based Learning
Related Stories
A uc first: enrollment tops 48,000.
August 18, 2022
The University of Cincinnati anticipates record enrollment as classes begin Monday, Aug. 22, with a projected 48,300 students — a 3% increase over last year. It will mark nearly a decade of continuous growth of a student body that increasingly reflects the university’s core values.
May 31, 2024
Third-year speech language hearing sciences student Mary Wilkens is helping prove the positive effects that gender-affirming voice therapy has on transgender individuals’ lives.
President Pinto's 2021 year-in-review message
December 17, 2021
University of Cincinnati President Neville G. Pinto looks back on a historic year that brought students, faculty, staff and the community back together like never before.

IMAGES
COMMENTS
Top 50 Life and Biological Sciences Articles. We are pleased to share with you the 50 most read Nature Communications articles* in life and biological sciences published in 2019. Featuring authors ...
Research in the life sciences is a substantial national enterprise in which the United States invested $2,264 million in fiscal year 1967*; of this, 30 percent was provided by industry, 4.1 percent by foundations and other private granting agencies, 1.2 percent by academic institutions from their own resources, 0.3 percent by local and state ...
Life Science News. Updated daily with science research articles in all the life sciences.
Biological sciences encompasses all the divisions of natural sciences examining various aspects of vital processes. The concept includes anatomy, physiology, cell biology, biochemistry and ...
Browse the 50 most downloaded Nature Communications articles across life and biological sciences published in 2020. ... sample is an important clinical goal in current human microbiome research ...
Life Sciences is an international journal publishing articles that emphasize the molecular, cellular, and functional basis of therapy. The journal emphasizes the understanding of mechanism that is relevant to all aspects of human disease and translation to patients. ... Research in many fields is beginning to uncover mechanisms and potential ...
What Is The Life Sciences at Harvard University. Studying the life sciences will provide you with a foundation of scientific knowledge and ways of exploring the world. The life sciences pervade so many aspects of our lives - from health care, to the environment, to debates about stem cell research and genetic testing. While dramatic ...
The COVID-19 pandemic has mobilized the world scientific community in 2020, especially in the life sciences [ 1, 2 ]. In the first three months after the pandemic, the number of scientific papers about COVID-19 was fivefold the number of articles on H1N1 swine influenza [ 3 ]. Similarly, the number of clinical trials related to COVID-19 ...
In 1981 the founder of the Life Sciences Research Foundation, Donald D. Brown, sought to establish similar collaborations in biology, focusing on training the next generation of exceptional biologists. While for-profit enterprises of the life sciences were solicited as major sponsors for postdoctoral fellowships in the early years, sponsorship ...
Life Sciences. We're working on groundbreaking research aiming to revolutionize the field of life sciences. We're solving some of the most important issues humanity faces with artificial intelligence, developing novel and unconventional computing structures, as well as mathematical and computational modeling.
The simplest way to define life sciences is the study of living organisms and life processes. At NCBiotech, we see it as science involving cells and their components, products and processes. Biology, medicine and agriculture are the most obvious examples of the discipline. However, in recent years, a convergence is underway that requires a ...
Research in life science involves the systematic investigation and study of living organisms, their interactions, and their environments. It encompasses a wide range of disciplines, including biology, genetics, ecology, microbiology, neuroscience, and more. Life science research aims to expand our understanding of the fundamental principles ...
Life Sciences at MIT. for Graduate Students. Many areas of research today have a Life Sciences focus. This is primarily due to the powerful tools of molecular biology, which form a common language and allow exciting and important interdisciplinary approaches. Experience in Life Sciences-based research opens multiple career paths.
The rise of health technology. Health technology continues to push the boundaries of how healthcare is delivered and has the power to create breakthroughs in our understanding of disease. This collection of articles and interviews explores the evolving role of health technology in the life-sciences sector and implications for key stakeholders.
The importance of innovative science and education has never been more apparent, and UCLA Life Sciences is committed to addressing some of the most important questions we face today. I am honored to work with world-renowned faculty who are elucidating the intricate mechanisms of how living systems work…. Our groundbreaking research is leading ...
Each fall Science Education Office brings together Harvard scientists from various departments, institutes and hospitals together for the Harvard Undergraduate Research Opportunities in Science (HUROS) fair. HUROS is a great way to meet dozens of scientists in one day and explore multiple research areas before narrowing down your interest.
Life sciences is a major contributor to Stanford University's high-quality research output, as tracked by the Nature Index. It is home to pioneering work in DNA synthesis, stem-cell isolation ...
The Graduate School of Arts and Sciences (GSAS) at Harvard University provides exceptional opportunities for study across the depth and breadth of the life sciences through the Harvard Integrated Life Sciences (HILS) federation. The HILS federation comprises 14 Ph.D. programs of study across four Harvard faculties—Harvard Faculty of Arts and ...
This list of life sciences comprises the branches of science that involve the scientific study of life - such as microorganisms, plants, and animals including human beings.This science is one of the two major branches of natural science, the other being physical science, which is concerned with non-living matter. Biology is the overall natural science that studies life, with the other life ...
Since 1983, the Life Sciences Research Foundation (LSRF) has funded nearly 650 outstanding postdoctoral fellows in all areas of the life sciences, and raised more than $60 million from generous industries, foundations and individuals to support this effort. We believe that discoveries and application of innovations in biology for the public's ...
Advances in life sciences research and technologies are transforming global health and have the potential to provide new and improved ways to support healthier populations worldwide. However, research and emerging technologies in fields such as engineering and information technology, can pose risks to public health if misused, either inadvertently or deliberately, to cause harm.
Life science research is a broad field that requires an array of instrumentation and supplies. Life science research laboratories require numerous consumables including chemicals, reagents, and plasticware. In addition to consumables, life science research labs also employ numerous types of instruments that vary depending on their specific ...
Dr. Cech is a biochemist and the author of the forthcoming book "The Catalyst: RNA and the Quest to Unlock Life's Deepest Secrets," from which this essay is adapted. From E=mc² to splitting ...
Top 50: Life and Biological Sciences. We are pleased to share with you the 50 most read Nature Communications life and biological sciences articles* published in 2018. Featuring authors from ...
Three University of Maine graduate students placed as finalists in the 6th annual BioME Student Showcase for life science research. The annual event supports innovation and the commercialization of student ideas in Maine, and allows students the opportunity to network with professionals or connect with entrepreneurial resources.
With time, I began to complement my love of details with an ability to evaluate the how and why of research tasks and the relevance of my work beyond the experiments, data, and analyses. I was still getting lost in the details, but I was now able to understand why. I could also articulate the story behind my research.
Professor Michael Benton from Bristol's School of Earth Sciences, another project supervisor, adds: "The volume of dinosaur remains found at Lavernock is extremely exciting, and is a chance to ...
Thursday, May 30, 2024 • Katherine Egan Bennett : contact Xinlei "Sherry" Wang, director for research in the Division of Data Science at The University of Texas at Arlington, has been named a 2024 fellow of the American Statistical Association (ASA).. Wang, the Jenkins Garret Endowed Professor of Statistics and Data Science, was selected for her outstanding contributions in developing ...
The faculty encouraged Wilkens to apply for UC's UPRISE (Undergraduates Pursuing Research in Science and Engineering) program, which is focused on increasing the pipeline of individuals from underserved communities into the STEMM fields. Wilkens was accepted and participated in a paid 12-week research opportunity in the VSM Lab the following ...