- Daily Crossword
- Word Puzzle
- Word Finder
- Word of the Day
- Synonym of the Day
- Word of the Year
- Language stories
- All featured
- Gender and sexuality
- All pop culture
- Grammar Coach ™
- Writing hub
- Grammar essentials
- Commonly confused
- All writing tips
- Pop culture
- Writing tips

Did you mean assign ?

Definition of 'assignment'

Video: pronunciation of assignment

assignment in British English
Assignment in american english, examples of 'assignment' in a sentence assignment, cobuild collocations assignment, trends of assignment.
View usage for: All Years Last 10 years Last 50 years Last 100 years Last 300 years
Browse alphabetically assignment
- assigned randomly
- assigned risk
- assimilability
- assimilable
- All ENGLISH words that begin with 'A'
Related terms of assignment
- seat assignment
- tough assignment
- writing assignment
- challenging assignment
- difficult assignment
- View more related words
Quick word challenge
Quiz Review
Score: 0 / 5

Wordle Helper

Scrabble Tools

Synonyms of assignment
- as in lesson
- as in appointment
- More from M-W
- To save this word, you'll need to log in. Log In
Thesaurus Definition of assignment
Synonyms & Similar Words
- responsibility
- undertaking
- requirement
- designation
- appointment
- authorization
- installment
- installation
- destination
- emplacement
- investiture
- singling (out)
Antonyms & Near Antonyms
- dethronement
Synonym Chooser
How does the noun assignment contrast with its synonyms?
Some common synonyms of assignment are chore , duty , job , stint , and task . While all these words mean "a piece of work to be done," assignment implies a definite limited task assigned by one in authority.
When is it sensible to use chore instead of assignment ?
While the synonyms chore and assignment are close in meaning, chore implies a minor routine activity necessary for maintaining a household or farm.
When is duty a more appropriate choice than assignment ?
Although the words duty and assignment have much in common, duty implies an obligation to perform or responsibility for performance.
When might job be a better fit than assignment ?
The synonyms job and assignment are sometimes interchangeable, but job applies to a piece of work voluntarily performed; it may sometimes suggest difficulty or importance.
When could stint be used to replace assignment ?
In some situations, the words stint and assignment are roughly equivalent. However, stint implies a carefully allotted or measured quantity of assigned work or service.
When can task be used instead of assignment ?
The meanings of task and assignment largely overlap; however, task implies work imposed by a person in authority or an employer or by circumstance.
Thesaurus Entries Near assignment
assignments
Cite this Entry
“Assignment.” Merriam-Webster.com Thesaurus , Merriam-Webster, https://www.merriam-webster.com/thesaurus/assignment. Accessed 17 Apr. 2024.
More from Merriam-Webster on assignment
Nglish: Translation of assignment for Spanish Speakers
Britannica English: Translation of assignment for Arabic Speakers
Subscribe to America's largest dictionary and get thousands more definitions and advanced search—ad free!

Can you solve 4 words at once?
Word of the day, circumlocution.
See Definitions and Examples »
Get Word of the Day daily email!
Popular in Grammar & Usage
Your vs. you're: how to use them correctly, every letter is silent, sometimes: a-z list of examples, more commonly mispronounced words, how to use em dashes (—), en dashes (–) , and hyphens (-), absent letters that are heard anyway, popular in wordplay, a great big list of bread words, the words of the week - apr. 12, 10 scrabble words without any vowels, 12 more bird names that sound like insults (and sometimes are), 9 superb owl words, games & quizzes.

Words and phrases
Personal account.
- Access or purchase personal subscriptions
- Get our newsletter
- Save searches
- Set display preferences
Institutional access
Sign in with library card
Sign in with username / password
Recommend to your librarian
Institutional account management
Sign in as administrator on Oxford Academic
assigned adjective
- Hide all quotations
What does the adjective assigned mean?
There are three meanings listed in OED's entry for the adjective assigned . See ‘Meaning & use’ for definitions, usage, and quotation evidence.
Entry status
OED is undergoing a continuous programme of revision to modernize and improve definitions. This entry has not yet been fully revised.
How common is the adjective assigned ?
How is the adjective assigned pronounced, british english, u.s. english, australian english, where does the adjective assigned come from.
Earliest known use
Middle English
The earliest known use of the adjective assigned is in the Middle English period (1150—1500).
OED's earliest evidence for assigned is from around 1374, in a translation by Geoffrey Chaucer, poet and administrator.
assigned is formed within English, by derivation.
Etymons: assign v. , ‑ed suffix 1 .
Nearby entries
- assify, v. 1804–
- assign, n.¹ 1601–41
- assign, n.² c1450–
- assign, v. 1297–
- assignability, n. 1884–
- assignable, adj. 1659–
- assignably, adv. 1674–
- assignat, n. 1790–
- assignate, adj. 1471
- assignation, n. a1400–
- assigned, adj. c1374–
- assignee, adj. & n. 1419–
- assigneeism, n. 1883–
- assigneeship, n. 1829–
- assigner, n. 1667–
- assigning, n. 1580–
- assignment, n. 1393–
- assignor, n. 1690–
- assilag, n. 1698–
- assimilability, n. a1834–
- assimilable, adj. & n. 1646–
Thank you for visiting Oxford English Dictionary
To continue reading, please sign in below or purchase a subscription. After purchasing, please sign in below to access the content.
Meaning & use
Pronunciation, compounds & derived words, entry history for assigned, adj..
assigned, adj. was first published in 1885; not yet revised.
assigned, adj. was last modified in September 2023.
Revision of the OED is a long-term project. Entries in oed.com which have not been revised may include:
- corrections and revisions to definitions, pronunciation, etymology, headwords, variant spellings, quotations, and dates;
- new senses, phrases, and quotations which have been added in subsequent print and online updates.
Revisions and additions of this kind were last incorporated into assigned, adj. in September 2023.
Earlier versions of this entry were published in:
OED First Edition (1885)
- Find out more
OED Second Edition (1989)
- View assigned, ppl. a. in OED Second Edition
Please submit your feedback for assigned, adj.
Please include your email address if you are happy to be contacted about your feedback. OUP will not use this email address for any other purpose.
Citation details
Factsheet for assigned, adj., browse entry.

Understanding Assignments
What this handout is about.
The first step in any successful college writing venture is reading the assignment. While this sounds like a simple task, it can be a tough one. This handout will help you unravel your assignment and begin to craft an effective response. Much of the following advice will involve translating typical assignment terms and practices into meaningful clues to the type of writing your instructor expects. See our short video for more tips.
Basic beginnings
Regardless of the assignment, department, or instructor, adopting these two habits will serve you well :
- Read the assignment carefully as soon as you receive it. Do not put this task off—reading the assignment at the beginning will save you time, stress, and problems later. An assignment can look pretty straightforward at first, particularly if the instructor has provided lots of information. That does not mean it will not take time and effort to complete; you may even have to learn a new skill to complete the assignment.
- Ask the instructor about anything you do not understand. Do not hesitate to approach your instructor. Instructors would prefer to set you straight before you hand the paper in. That’s also when you will find their feedback most useful.
Assignment formats
Many assignments follow a basic format. Assignments often begin with an overview of the topic, include a central verb or verbs that describe the task, and offer some additional suggestions, questions, or prompts to get you started.
An Overview of Some Kind
The instructor might set the stage with some general discussion of the subject of the assignment, introduce the topic, or remind you of something pertinent that you have discussed in class. For example:
“Throughout history, gerbils have played a key role in politics,” or “In the last few weeks of class, we have focused on the evening wear of the housefly …”
The Task of the Assignment
Pay attention; this part tells you what to do when you write the paper. Look for the key verb or verbs in the sentence. Words like analyze, summarize, or compare direct you to think about your topic in a certain way. Also pay attention to words such as how, what, when, where, and why; these words guide your attention toward specific information. (See the section in this handout titled “Key Terms” for more information.)
“Analyze the effect that gerbils had on the Russian Revolution”, or “Suggest an interpretation of housefly undergarments that differs from Darwin’s.”
Additional Material to Think about
Here you will find some questions to use as springboards as you begin to think about the topic. Instructors usually include these questions as suggestions rather than requirements. Do not feel compelled to answer every question unless the instructor asks you to do so. Pay attention to the order of the questions. Sometimes they suggest the thinking process your instructor imagines you will need to follow to begin thinking about the topic.
“You may wish to consider the differing views held by Communist gerbils vs. Monarchist gerbils, or Can there be such a thing as ‘the housefly garment industry’ or is it just a home-based craft?”
These are the instructor’s comments about writing expectations:
“Be concise”, “Write effectively”, or “Argue furiously.”
Technical Details
These instructions usually indicate format rules or guidelines.
“Your paper must be typed in Palatino font on gray paper and must not exceed 600 pages. It is due on the anniversary of Mao Tse-tung’s death.”
The assignment’s parts may not appear in exactly this order, and each part may be very long or really short. Nonetheless, being aware of this standard pattern can help you understand what your instructor wants you to do.
Interpreting the assignment
Ask yourself a few basic questions as you read and jot down the answers on the assignment sheet:
Why did your instructor ask you to do this particular task?
Who is your audience.
- What kind of evidence do you need to support your ideas?
What kind of writing style is acceptable?
- What are the absolute rules of the paper?
Try to look at the question from the point of view of the instructor. Recognize that your instructor has a reason for giving you this assignment and for giving it to you at a particular point in the semester. In every assignment, the instructor has a challenge for you. This challenge could be anything from demonstrating an ability to think clearly to demonstrating an ability to use the library. See the assignment not as a vague suggestion of what to do but as an opportunity to show that you can handle the course material as directed. Paper assignments give you more than a topic to discuss—they ask you to do something with the topic. Keep reminding yourself of that. Be careful to avoid the other extreme as well: do not read more into the assignment than what is there.
Of course, your instructor has given you an assignment so that he or she will be able to assess your understanding of the course material and give you an appropriate grade. But there is more to it than that. Your instructor has tried to design a learning experience of some kind. Your instructor wants you to think about something in a particular way for a particular reason. If you read the course description at the beginning of your syllabus, review the assigned readings, and consider the assignment itself, you may begin to see the plan, purpose, or approach to the subject matter that your instructor has created for you. If you still aren’t sure of the assignment’s goals, try asking the instructor. For help with this, see our handout on getting feedback .
Given your instructor’s efforts, it helps to answer the question: What is my purpose in completing this assignment? Is it to gather research from a variety of outside sources and present a coherent picture? Is it to take material I have been learning in class and apply it to a new situation? Is it to prove a point one way or another? Key words from the assignment can help you figure this out. Look for key terms in the form of active verbs that tell you what to do.
Key Terms: Finding Those Active Verbs
Here are some common key words and definitions to help you think about assignment terms:
Information words Ask you to demonstrate what you know about the subject, such as who, what, when, where, how, and why.
- define —give the subject’s meaning (according to someone or something). Sometimes you have to give more than one view on the subject’s meaning
- describe —provide details about the subject by answering question words (such as who, what, when, where, how, and why); you might also give details related to the five senses (what you see, hear, feel, taste, and smell)
- explain —give reasons why or examples of how something happened
- illustrate —give descriptive examples of the subject and show how each is connected with the subject
- summarize —briefly list the important ideas you learned about the subject
- trace —outline how something has changed or developed from an earlier time to its current form
- research —gather material from outside sources about the subject, often with the implication or requirement that you will analyze what you have found
Relation words Ask you to demonstrate how things are connected.
- compare —show how two or more things are similar (and, sometimes, different)
- contrast —show how two or more things are dissimilar
- apply—use details that you’ve been given to demonstrate how an idea, theory, or concept works in a particular situation
- cause —show how one event or series of events made something else happen
- relate —show or describe the connections between things
Interpretation words Ask you to defend ideas of your own about the subject. Do not see these words as requesting opinion alone (unless the assignment specifically says so), but as requiring opinion that is supported by concrete evidence. Remember examples, principles, definitions, or concepts from class or research and use them in your interpretation.
- assess —summarize your opinion of the subject and measure it against something
- prove, justify —give reasons or examples to demonstrate how or why something is the truth
- evaluate, respond —state your opinion of the subject as good, bad, or some combination of the two, with examples and reasons
- support —give reasons or evidence for something you believe (be sure to state clearly what it is that you believe)
- synthesize —put two or more things together that have not been put together in class or in your readings before; do not just summarize one and then the other and say that they are similar or different—you must provide a reason for putting them together that runs all the way through the paper
- analyze —determine how individual parts create or relate to the whole, figure out how something works, what it might mean, or why it is important
- argue —take a side and defend it with evidence against the other side
More Clues to Your Purpose As you read the assignment, think about what the teacher does in class:
- What kinds of textbooks or coursepack did your instructor choose for the course—ones that provide background information, explain theories or perspectives, or argue a point of view?
- In lecture, does your instructor ask your opinion, try to prove her point of view, or use keywords that show up again in the assignment?
- What kinds of assignments are typical in this discipline? Social science classes often expect more research. Humanities classes thrive on interpretation and analysis.
- How do the assignments, readings, and lectures work together in the course? Instructors spend time designing courses, sometimes even arguing with their peers about the most effective course materials. Figuring out the overall design to the course will help you understand what each assignment is meant to achieve.
Now, what about your reader? Most undergraduates think of their audience as the instructor. True, your instructor is a good person to keep in mind as you write. But for the purposes of a good paper, think of your audience as someone like your roommate: smart enough to understand a clear, logical argument, but not someone who already knows exactly what is going on in your particular paper. Remember, even if the instructor knows everything there is to know about your paper topic, he or she still has to read your paper and assess your understanding. In other words, teach the material to your reader.
Aiming a paper at your audience happens in two ways: you make decisions about the tone and the level of information you want to convey.
- Tone means the “voice” of your paper. Should you be chatty, formal, or objective? Usually you will find some happy medium—you do not want to alienate your reader by sounding condescending or superior, but you do not want to, um, like, totally wig on the man, you know? Eschew ostentatious erudition: some students think the way to sound academic is to use big words. Be careful—you can sound ridiculous, especially if you use the wrong big words.
- The level of information you use depends on who you think your audience is. If you imagine your audience as your instructor and she already knows everything you have to say, you may find yourself leaving out key information that can cause your argument to be unconvincing and illogical. But you do not have to explain every single word or issue. If you are telling your roommate what happened on your favorite science fiction TV show last night, you do not say, “First a dark-haired white man of average height, wearing a suit and carrying a flashlight, walked into the room. Then a purple alien with fifteen arms and at least three eyes turned around. Then the man smiled slightly. In the background, you could hear a clock ticking. The room was fairly dark and had at least two windows that I saw.” You also do not say, “This guy found some aliens. The end.” Find some balance of useful details that support your main point.
You’ll find a much more detailed discussion of these concepts in our handout on audience .
The Grim Truth
With a few exceptions (including some lab and ethnography reports), you are probably being asked to make an argument. You must convince your audience. It is easy to forget this aim when you are researching and writing; as you become involved in your subject matter, you may become enmeshed in the details and focus on learning or simply telling the information you have found. You need to do more than just repeat what you have read. Your writing should have a point, and you should be able to say it in a sentence. Sometimes instructors call this sentence a “thesis” or a “claim.”
So, if your instructor tells you to write about some aspect of oral hygiene, you do not want to just list: “First, you brush your teeth with a soft brush and some peanut butter. Then, you floss with unwaxed, bologna-flavored string. Finally, gargle with bourbon.” Instead, you could say, “Of all the oral cleaning methods, sandblasting removes the most plaque. Therefore it should be recommended by the American Dental Association.” Or, “From an aesthetic perspective, moldy teeth can be quite charming. However, their joys are short-lived.”
Convincing the reader of your argument is the goal of academic writing. It doesn’t have to say “argument” anywhere in the assignment for you to need one. Look at the assignment and think about what kind of argument you could make about it instead of just seeing it as a checklist of information you have to present. For help with understanding the role of argument in academic writing, see our handout on argument .
What kind of evidence do you need?
There are many kinds of evidence, and what type of evidence will work for your assignment can depend on several factors–the discipline, the parameters of the assignment, and your instructor’s preference. Should you use statistics? Historical examples? Do you need to conduct your own experiment? Can you rely on personal experience? See our handout on evidence for suggestions on how to use evidence appropriately.
Make sure you are clear about this part of the assignment, because your use of evidence will be crucial in writing a successful paper. You are not just learning how to argue; you are learning how to argue with specific types of materials and ideas. Ask your instructor what counts as acceptable evidence. You can also ask a librarian for help. No matter what kind of evidence you use, be sure to cite it correctly—see the UNC Libraries citation tutorial .
You cannot always tell from the assignment just what sort of writing style your instructor expects. The instructor may be really laid back in class but still expect you to sound formal in writing. Or the instructor may be fairly formal in class and ask you to write a reflection paper where you need to use “I” and speak from your own experience.
Try to avoid false associations of a particular field with a style (“art historians like wacky creativity,” or “political scientists are boring and just give facts”) and look instead to the types of readings you have been given in class. No one expects you to write like Plato—just use the readings as a guide for what is standard or preferable to your instructor. When in doubt, ask your instructor about the level of formality she or he expects.
No matter what field you are writing for or what facts you are including, if you do not write so that your reader can understand your main idea, you have wasted your time. So make clarity your main goal. For specific help with style, see our handout on style .
Technical details about the assignment
The technical information you are given in an assignment always seems like the easy part. This section can actually give you lots of little hints about approaching the task. Find out if elements such as page length and citation format (see the UNC Libraries citation tutorial ) are negotiable. Some professors do not have strong preferences as long as you are consistent and fully answer the assignment. Some professors are very specific and will deduct big points for deviations.
Usually, the page length tells you something important: The instructor thinks the size of the paper is appropriate to the assignment’s parameters. In plain English, your instructor is telling you how many pages it should take for you to answer the question as fully as you are expected to. So if an assignment is two pages long, you cannot pad your paper with examples or reword your main idea several times. Hit your one point early, defend it with the clearest example, and finish quickly. If an assignment is ten pages long, you can be more complex in your main points and examples—and if you can only produce five pages for that assignment, you need to see someone for help—as soon as possible.
Tricks that don’t work
Your instructors are not fooled when you:
- spend more time on the cover page than the essay —graphics, cool binders, and cute titles are no replacement for a well-written paper.
- use huge fonts, wide margins, or extra spacing to pad the page length —these tricks are immediately obvious to the eye. Most instructors use the same word processor you do. They know what’s possible. Such tactics are especially damning when the instructor has a stack of 60 papers to grade and yours is the only one that low-flying airplane pilots could read.
- use a paper from another class that covered “sort of similar” material . Again, the instructor has a particular task for you to fulfill in the assignment that usually relates to course material and lectures. Your other paper may not cover this material, and turning in the same paper for more than one course may constitute an Honor Code violation . Ask the instructor—it can’t hurt.
- get all wacky and “creative” before you answer the question . Showing that you are able to think beyond the boundaries of a simple assignment can be good, but you must do what the assignment calls for first. Again, check with your instructor. A humorous tone can be refreshing for someone grading a stack of papers, but it will not get you a good grade if you have not fulfilled the task.
Critical reading of assignments leads to skills in other types of reading and writing. If you get good at figuring out what the real goals of assignments are, you are going to be better at understanding the goals of all of your classes and fields of study.
You may reproduce it for non-commercial use if you use the entire handout and attribute the source: The Writing Center, University of North Carolina at Chapel Hill
Make a Gift
Apple Business Manager User Guide
- Intro to Apple Business Manager
- Program requirements
- Participate in beta features
- Get support
- Provide feedback
- Edit preferences
- Configure locations
- Link to new domains
- About domain conflicts
- Disconnect federation from a domain
- Intro to federated authentication
- Use federated authentication with Google Workspace
- Use federated authentication with Microsoft Entra ID
- Use federated authentication with your identity provider
- Change a user’s domain information
- Transfer Apple services when federating
- Sync user accounts from Google Workspace
- Sync user accounts from Microsoft Entra ID
- Sync user accounts from your identity provider
- Get notified about user name conflicts
- Resolve Apple ID conflicts
- Resolve Google Workspace sync conflicts
- Resolve Microsoft Entra ID OIDC sync conflicts
- Resolve identity provider OIDC or SCIM sync conflicts
- Use Managed Apple IDs
- Customize user access to certain apps and services
- Use Sign in with Apple
- Intro to users and user groups
- Manually add users
- Manage existing users
- Add user groups
- Create or reset user passwords
- Intro to roles and privileges
- View and assign roles
- Add Content Managers to locations
- Add or reset verification phone numbers
- Intro to buying content
- Review content payment and billing information
- Select and buy content
- Learn about Custom Apps
- Manage content tokens
- Transfer licenses
- Migrate content tokens
- Invite VPP purchasers
- Plan for migration to Apps and Books
- Device workflow
- Manage device suppliers
- View device information
- Get device order progress reports
- Add devices from Apple Configurator
- Intro to MDM servers
- Link to a third-party MDM server
- Edit a third-party MDM server configuration
- Delete a third-party MDM server
- Review device assignments
- Assign, reassign, or unassign devices
- Assign a device that was serviced or replaced
- Release devices
- Create Shared iPad passcodes
- Sign in to Shared iPad
- Shared iPad and Managed Apple IDs
- Sign federated users out of devices
- How to search
- View activity
- Read log files
- Keyboard shortcuts
- Document revision history
Intro to buying content in Apple Business Manager
The App Store features thousands of apps. Fortunately, Apple Business Manager gives your organization a simple way to acquire and manage these apps in the Apps and Books Store. Using device management, you can install and update apps remotely, even if the App Store is disabled on the device.
Note: Depending on your local tax requirements, you may be required to provide tax information when you initially set up your organization to buy content.
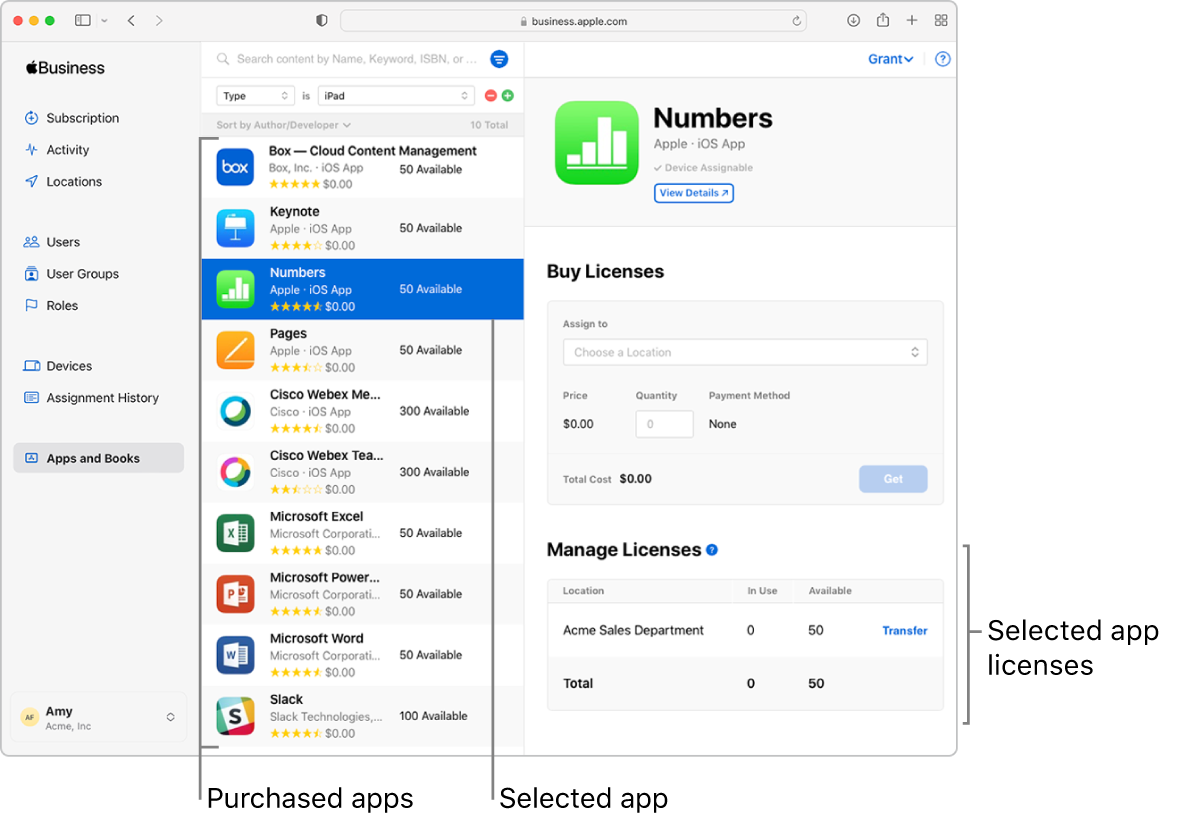
Distributing apps
Besides buying apps, you can also revoke and reassign apps to different devices and users. In this way, you always retain full ownership and control of apps you bought. You can assign the apps you bought through Apple Business Manager to any devices or users in any country or region where those apps are available from that location’s App Store. (The App Store Country or Region is determined by the address submitted when your organization signs up for Apple Business Manager. For example, if your organization signed up with an address in the United States, the App Store locale is set to United States.)
App distribution works best when the app is assigned before devices are configured or given to users. After the user receives the device and completes the Setup Assistant, Apple Business Manager can send the user an invitation by email or push notification. When the user accepts the invitation, the user has access to all of the apps—which are then remotely installed on the user’s devices.
Note: In-app purchases and subscriptions aren’t compatible with volume purchasing, Managed Apps, or Managed Apple IDs . Developers may offer a separate full-featured version of their apps (sometimes as a Custom App) for education or enterprise customers that need to deploy apps at scale.
Distribute Apple Books
Apple Books bought through Apple Business Manager can be distributed only to users, not devices. They can’t be revoked and reassigned. When books are assigned to users, those books follow the same country and region download restrictions as apps.
Note: Some Apple Books content isn’t available in certain countries or regions. To learn whether certain Apple Books content is available in your country or region, see the Apple Support article Availability of Apple programs and payment methods for education and business .
Recommendations
To simplify your deployment, it’s highly recommended that you assign apps to devices instead of users. With device-based app assignment, there’s no invitation process or requirement to use an Apple ID on the device. Books can’t be assigned to devices.
To alleviate bandwidth saturation during the initial mass deployment, consider distributing only the apps necessary for the first day of deployment, and then make additional apps available to users for download over time. If the device is supervised, apps are installed silently.
App and book assignment summary
The table below shows the types of apps or books, and the criteria for assigning them to devices and users.
- Open access
- Published: 15 April 2024
Demuxafy : improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods
- Drew Neavin 1 ,
- Anne Senabouth 1 ,
- Himanshi Arora 1 , 2 ,
- Jimmy Tsz Hang Lee 3 ,
- Aida Ripoll-Cladellas 4 ,
- sc-eQTLGen Consortium ,
- Lude Franke 5 ,
- Shyam Prabhakar 6 , 7 , 8 ,
- Chun Jimmie Ye 9 , 10 , 11 , 12 ,
- Davis J. McCarthy 13 , 14 ,
- Marta Melé 4 ,
- Martin Hemberg 15 &
- Joseph E. Powell ORCID: orcid.org/0000-0002-5070-4124 1 , 16
Genome Biology volume 25 , Article number: 94 ( 2024 ) Cite this article
42 Accesses
Metrics details
Recent innovations in single-cell RNA-sequencing (scRNA-seq) provide the technology to investigate biological questions at cellular resolution. Pooling cells from multiple individuals has become a common strategy, and droplets can subsequently be assigned to a specific individual by leveraging their inherent genetic differences. An implicit challenge with scRNA-seq is the occurrence of doublets—droplets containing two or more cells. We develop Demuxafy, a framework to enhance donor assignment and doublet removal through the consensus intersection of multiple demultiplexing and doublet detecting methods. Demuxafy significantly improves droplet assignment by separating singlets from doublets and classifying the correct individual.
Droplet-based single-cell RNA sequencing (scRNA-seq) technologies have provided the tools to profile tens of thousands of single-cell transcriptomes simultaneously [ 1 ]. With these technological advances, combining cells from multiple samples in a single capture is common, increasing the sample size while simultaneously reducing batch effects, cost, and time. In addition, following cell capture and sequencing, the droplets can be demultiplexed—each droplet accurately assigned to each individual in the pool [ 2 , 3 , 4 , 5 , 6 , 7 ].
Many scRNA-seq experiments now capture upwards of 20,000 droplets, resulting in ~16% (3,200) doublets [ 8 ]. Current demultiplexing methods can also identify doublets—droplets containing two or more cells—from different individuals (heterogenic doublets). These doublets can significantly alter scientific conclusions if they are not effectively removed. Therefore, it is essential to remove doublets from droplet-based single-cell captures.
However, demultiplexing methods cannot identify droplets containing multiple cells from the same individual (homogenic doublets) and, therefore, cannot identify all doublets in a single capture. If left in the dataset, those doublets could appear as transitional cells between two distinct cell types or a completely new cell type. Accordingly, additional methods have been developed to identify heterotypic doublets (droplets that contain two cells from different cell types) by comparing the transcriptional profile of each droplet to doublets simulated from the dataset [ 9 , 10 , 11 , 12 , 13 , 14 , 15 ]. It is important to recognise that demultiplexing methods achieve two functions—segregation of cells from different donors and separation of singlets from doublets—while doublet detecting methods solely classify singlets versus doublets.
Therefore, demultiplexing and transcription-based doublet detecting methods provide complementary information to improve doublet detection, providing a cleaner dataset and more robust scientific results. There are currently five genetic-based demultiplexing [ 2 , 3 , 4 , 5 , 6 , 7 , 16 ] and seven transcription-based doublet-detecting methods implemented in various languages [ 9 , 10 , 11 , 12 , 13 , 14 , 15 ]. Under different scenarios, each method is subject to varying performance and, in some instances, biases in their ability to accurately assign cells or detect doublets from certain conditions. The best combination of methods is currently unclear but will undoubtedly depend on the dataset and research question.
Therefore, we set out to identify the best combination of genetic-based demultiplexing and transcription-based doublet-detecting methods to remove doublets and partition singlets from different donors correctly. In addition, we have developed a software platform ( Demuxafy ) that performs these intersectional methods and provides additional commands to simplify the execution and interpretation of results for each method (Fig. 1 a).
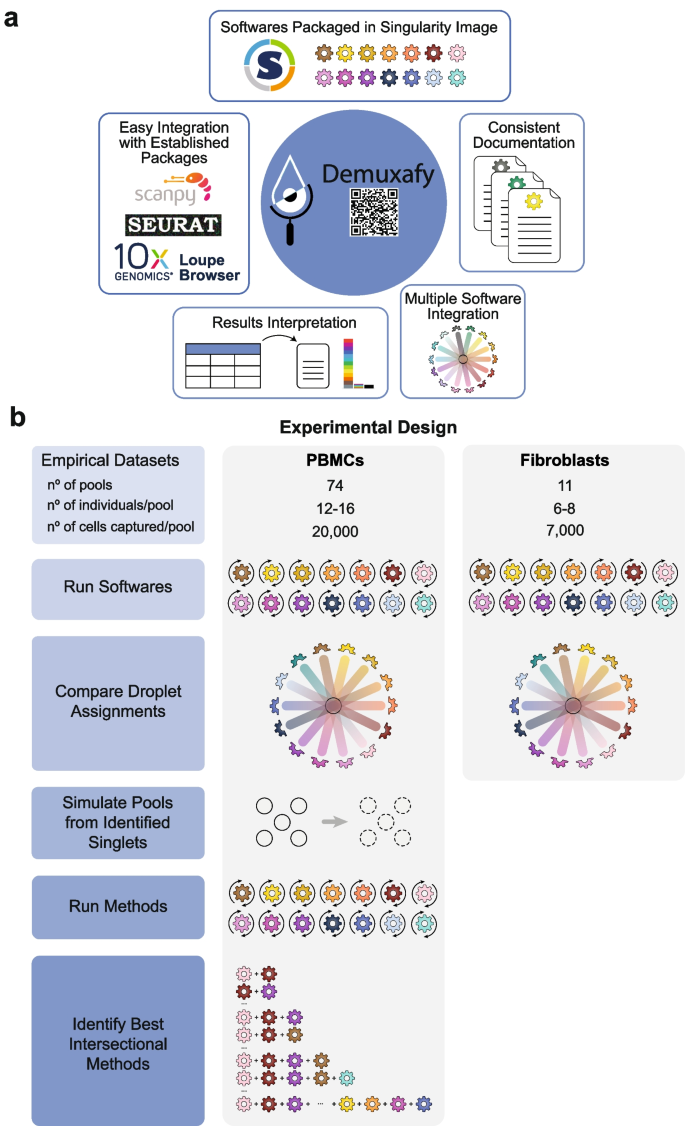
Study design and qualitative method classifications. a Demuxafy is a platform to perform demultiplexing and doublet detecting with consistent documentation. Demuxafy also provides wrapper scripts to quickly summarize the results from each method and assign clusters to each individual with reference genotypes when a reference-free demultiplexing method is used. Finally, Demuxafy provides a script to easily combine the results from multiple different methods into a single data frame and it provides a final assignment for each droplet based on the combination of multiple methods. In addition, Demuxafy provides summaries of the number of droplets classified as singlets or doublets by each method and a summary of the number of droplets assigned to each individual by each of the demultiplexing methods. b Two datasets are included in this analysis - a PBMC dataset and a fibroblast dataset. The PBMC dataset contains 74 pools that captured approximately 20,000 droplets each with 12-16 donor cells multiplexed per pool. The fibroblast dataset contains 11 pools of roughly 7,000 droplets per pool with sizes ranging from six to eight donors per pool. All pools were processed by all demultiplexing and doublet detecting methods and the droplet and donor classifications were compared between the methods and between the PBMCs and fibroblasts. Then the PBMC droplets that were classified as singlets by all methods were taken as ‘true singlets’ and used to generate new pools in silico. Those pools were then processed by each of the demultiplexing and doublet detecting methods and intersectional combinations of demultiplexing and doublet detecting methods were tested for different experimental designs
To compare the demultiplexing and doublet detecting methods, we utilised two large, multiplexed datasets—one that contained ~1.4 million peripheral blood mononuclear cells (PBMCs) from 1,034 donors [ 17 ] and one with ~94,000 fibroblasts from 81 donors [ 18 ]. We used the true singlets from the PBMC dataset to generate new in silico pools to assess the performance of each method and the multi-method intersectional combinations (Fig. 1 b).
Here, we compare 14 demultiplexing and doublet detecting methods with different methodological approaches, capabilities, and intersectional combinations. Seven of those are demultiplexing methods ( Demuxalot [ 6 ], Demuxlet [ 3 ], Dropulation [ 5 ], Freemuxlet [ 16 ], ScSplit [ 7 ], Souporcell [ 4 ], and Vireo [ 2 ]) which leverage the common genetic variation between individuals to identify cells that came from each individual and to identify heterogenic doublets. The seven remaining methods ( DoubletDecon [ 9 ], DoubletDetection [ 14 ], DoubletFinder [ 10 ], ScDblFinder [ 11 ], Scds [ 12 ], Scrublet [ 13 ], and Solo [ 15 ]) identify doublets based on their similarity to simulated doublets generated by adding the transcriptional profiles of two randomly selected droplets in the dataset. These methods assume that the proportion of real doublets in the dataset is low, so combining any two droplets will likely represent the combination of two singlets.
We identify critical differences in the performance of demultiplexing and doublet detecting methods to classify droplets correctly. In the case of the demultiplexing techniques, their performance depends on their ability to identify singlets from doublets and assign a singlet to the correct individual. For doublet detecting methods, the performance is based solely on their ability to differentiate a singlet from a doublet. We identify limitations in identifying specific doublet types and cell types by some methods. In addition, we compare the intersectional combinations of these methods for multiple experimental designs and demonstrate that intersectional approaches significantly outperform all individual techniques. Thus, the intersectional methods provide enhanced singlet classification and doublet removal—a critical but often under-valued step of droplet-based scRNA-seq processing. Our results demonstrate that intersectional combinations of demultiplexing and doublet detecting software provide significant advantages in droplet-based scRNA-seq preprocessing that can alter results and conclusions drawn from the data. Finally, to provide easy implementation of our intersectional approach, we provide Demuxafy ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/index.html ) a complete platform to perform demultiplexing and doublet detecting intersectional methods (Fig. 1 a).
Study design
To evaluate demultiplexing and doublet detecting methods, we developed an experimental design that applies the different techniques to empirical pools and pools generated in silico from the combination of true singlets—droplets identified as singlets by every method (Fig. 1 a). For the first phase of this study, we used two empirical multiplexed datasets—the peripheral blood mononuclear cell (PBMC) dataset containing ~1.4 million cells from 1034 donors and a fibroblast dataset of ~94,000 cells from 81 individuals (Additional file 1 : Table S1). We chose these two cell systems to assess the methods in heterogeneous (PBMC) and homogeneous (fibroblast) cell types.
Demultiplexing and doublet detecting methods perform similarly for heterogeneous and homogeneous cell types
We applied the demultiplexing methods ( Demuxalot , Demuxlet , Dropulation , Freemuxlet , ScSplit , Souporcell , and Vireo ) and doublet detecting methods ( DoubletDecon , DoubletDetection , DoubletFinder , ScDblFinder , Scds , Scrublet , and Solo ) to the two datasets and assessed the results from each method. We first compared the droplet assignments by identifying the number of singlets and doublets identified by a given method that were consistently annotated by all methods (Fig. 2 a–d). We also identified the percentage of droplets that were annotated consistently between pairs of methods (Additional file 2 : Fig S1). In the cases where two demultiplexing methods were compared to one another, both the droplet type (singlet or doublet) and the assignment of the droplet to an individual had to match to be considered in agreement. In all other comparisons (i.e. demultiplexing versus doublet detecting and doublet detecting versus doublet detecting), only the droplet type (singlet or doublet) was considered for agreement since doublet detecting methods cannot annotate donor assignment. We found that the two method types were more similar to other methods of the same type (i.e., demultiplexing versus demultiplexing and doublet detecting versus doublet detecting) than they were to methods from a different type (demultiplexing methods versus doublet detecting methods; Supplementary Fig 1). We found that the similarity of the demultiplexing and doublet detecting methods was consistent in the PBMC and fibroblast datasets (Pearson correlation R = 0.78, P -value < 2×10 −16 ; Fig S1a-c). In addition, demultiplexing methods were more similar than doublet detecting methods for both the PBMC and fibroblast datasets (Wilcoxon rank-sum test: P < 0.01; Fig. 2 a–b and Additional file 2 : Fig S1).
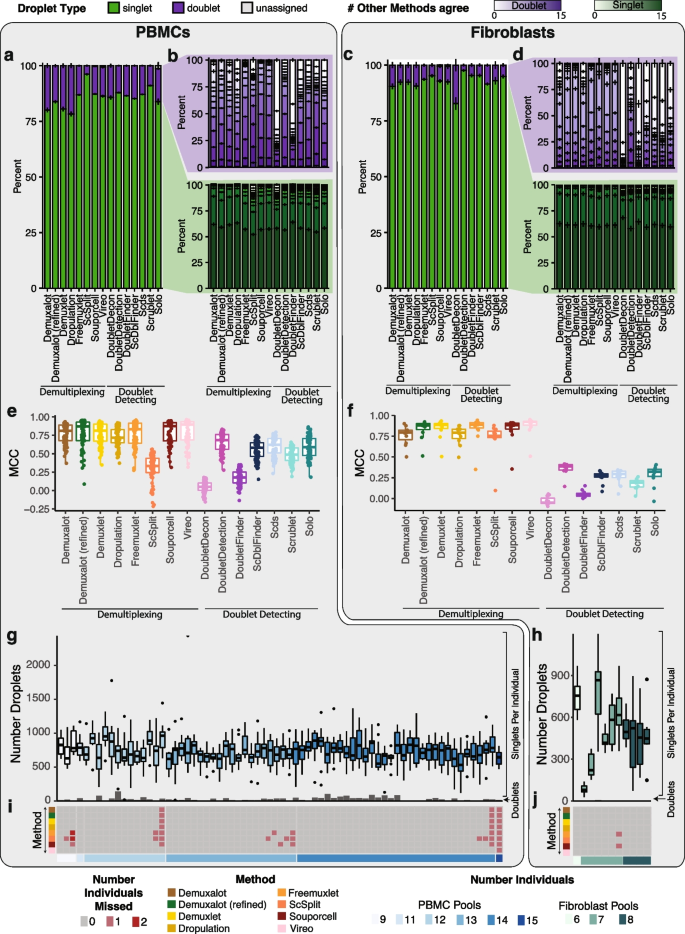
Demultiplexing and Doublet Detecting Method Performance Comparison. a The proportion of droplets classified as singlets and doublets by each method in the PBMCs. b The number of other methods that classified the singlets and doublets identified by each method in the PBMCs. c The proportion of droplets classified as singlets and doublets by each method in the fibroblasts. d The number of other methods that classified the singlets and doublets identified by each method in the fibroblasts. e - f The performance of each method when the majority classification of each droplet is considered the correct annotation in the PBMCs ( e ) and fibroblasts ( f ). g - h The number of droplets classified as singlets (box plots) and doublets (bar plots) by all methods in the PBMC ( g ) and fibroblast ( h ) pools. i - j The number of donors that were not identified by each method in each pool for PBMCs ( i ) and fibroblasts ( j ). PBMC: peripheral blood mononuclear cell. MCC: Matthew’s correlationcoefficient
The number of unique molecular identifiers (UMIs) and genes decreased in droplets that were classified as singlets by a larger number of methods while the mitochondrial percentage increased in both PBMCs and fibroblasts (Additional file 2 : Fig S2).
We next interrogated the performance of each method using the Matthew’s correlation coefficient (MCC) to calculate the consistency between Demuxify and true droplet classification. We identified consistent trends in the MCC scores for each method between the PBMCs (Fig. 2 e) and fibroblasts (Fig. 2 f). These data indicate that the methods behave similarly, relative to one another, for heterogeneous and homogeneous datasets.
Next, we sought to identify the droplets concordantly classified by all demultiplexing and doublet detecting methods in the PBMC and fibroblast datasets. On average, 732 singlets were identified for each individual by all the methods in the PBMC dataset. Likewise, 494 droplets were identified as singlets for each individual by all the methods in the fibroblast pools. However, the concordance of doublets identified by all methods was very low for both datasets (Fig. 2 e–f). Notably, the consistency of classifying a droplet as a doublet by all methods was relatively low (Fig. 2 b,d,g, and h). This suggests that doublet identification is not consistent between all the methods. Therefore, further investigation is required to identify the reasons for these inconsistencies between methods. It also suggests that combining multiple methods for doublet classification may be necessary for more complete doublet removal. Further, some methods could not identify all the individuals in each pool (Fig. 2 i–j). The non-concordance between different methods demonstrates the need to effectively test each method on a dataset where the droplet types are known.

Computational resources vary for demultiplexing and doublet detecting methods
We recorded each method’s computational resources for the PBMC pools, with ~20,000 cells captured per pool (Additional file 1 : Table S1). Of the demultiplexing methods, ScSplit took the most time (multiple days) and required the most steps, but Demuxalot , Demuxlet , and Freemuxlet used the most memory. Solo took the longest time (median 13 h) and most memory to run for the doublet detecting methods but is the only method built to be run directly from the command line, making it easy to implement (Additional file 2 : Fig S3).
Generate pools with known singlets and doublets
However, there is no gold standard to identify which droplets are singlets or doublets. Therefore, in the second phase of our experimental design (Fig. 1 b), we used the PBMC droplets classified as singlets by all methods to generate new pools in silico. We chose to use the PBMC dataset since our first analyses indicated that method performance is similar for homogeneous (fibroblast) and heterogeneous (PBMC) cell types (Fig. 2 and Additional file 2 : Fig S1) and because we had many more individuals available to generate in silico pools from the PBMC dataset (Additional file 1 : Table S1).
We generated 70 pools—10 each of pools that included 2, 4, 8, 16, 32, 64, or 128 individuals (Additional file 1 : Table S2). We assume a maximum 20% doublet rate as it is unlikely researchers would use a technology that has a higher doublet rate (Fig. 3 a).
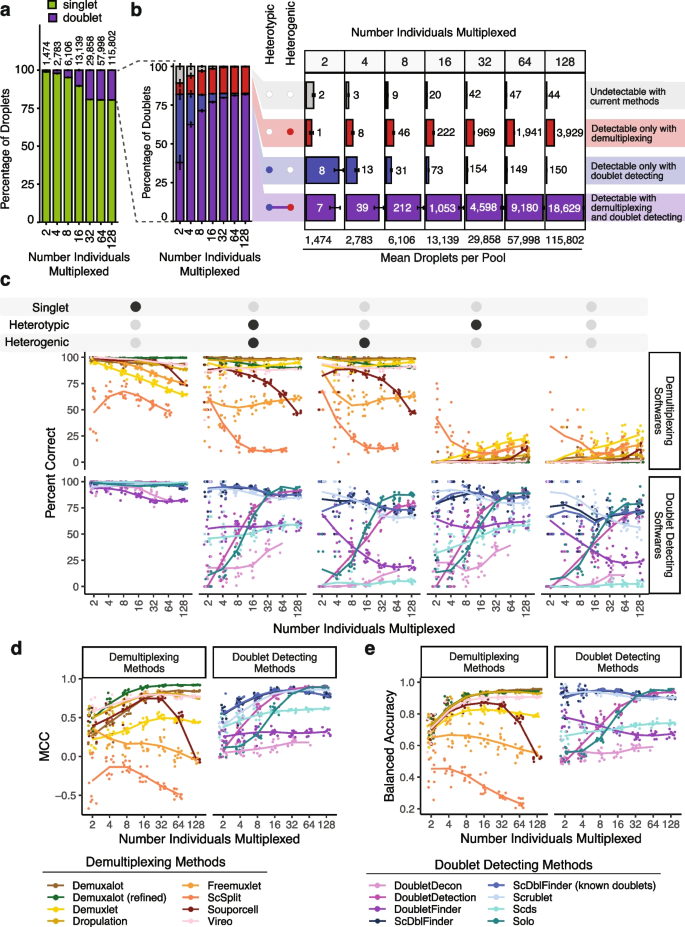
In silico Pool Doublet Annotation and Method Performance. a The percent of singlets and doublets in the in -silico pools - separated by the number of multiplexed individuals per pool. b The percentage and number of doublets that are heterogenic (detectable by demultiplexing methods), heterotypic (detectable by doublet detecting methods), both (detectable by either method category) and neither (not detectable with current methods) for each multiplexed pool size. c Percent of droplets that each of the demultiplexing and doublet detecting methods classified correctly for singlets and doublet subtypes for different multiplexed pool sizes. d Matthew’s Correlation Coefficient (MCC) for each of the methods for each of the multiplexed pool sizes. e Balanced accuracy for each of the methods for each of the multiplexed pool sizes
We used azimuth to classify the PBMC cell types for each droplet used to generate the in silico pools [ 19 ] (Additional file 2 : Fig S4). As these pools have been generated in silico using empirical singlets that have been well annotated, we next identified the proportion of doublets in each pool that were heterogenic, heterotypic, both, and neither. This approach demonstrates that a significant percentage of doublets are only detectable by doublet detecting methods (homogenic and heterotypic) for pools with 16 or fewer donors multiplexed (Fig. 3 b).
While the total number of doublets that would be missed if only using demultiplexing methods appears small for fewer multiplexed individuals (Fig. 3 b), it is important to recognise that this is partly a function of the ~732 singlet cells per individual used to generate these pools. Hence, the in silico pools with fewer individuals also have fewer cells. Therefore, to obtain numbers of doublets that are directly comparable to one another, we calculated the number of each doublet type that would be expected to be captured with 20,000 cells when 2, 4, 8, 16, or 32 individuals were multiplexed (Additional file 2 : Fig S5). These results demonstrate that many doublets would be falsely classified as singlets since they are homogenic when just using demultiplexing methods for a pool of 20,000 cells captured with a 16% doublet rate (Additional file 2 : Fig S5). However, as more individuals are multiplexed, the number of droplets that would not be detectable by demultiplexing methods (homogenic) decreases. This suggests that typical workflows that use only one demultiplexing method to remove doublets from pools that capture 20,000 droplets with 16 or fewer multiplexed individuals fail to adequately remove between 173 (16 multiplexed individuals) and 1,325 (2 multiplexed individuals) doublets that are homogenic and heterotypic which could be detected by doublet detecting methods (Additional file 2 : Fig S5). Therefore, a technique that uses both demultiplexing and doublet detecting methods in parallel will complement more complete doublet removal methods. Consequently, we next set out to identify the demultiplexing and doublet detecting methods that perform the best on their own and in concert with other methods.
Doublet and singlet droplet classification effectiveness varies for demultiplexing and doublet detecting methods
Demultiplexing methods fail to classify homogenic doublets.
We next investigated the percentage of the droplets that were correctly classified by each demultiplexing and doublet detecting method. In addition to the seven demultiplexing methods, we also included Demuxalot with the additional steps to refine the genotypes that can then be used for demultiplexing— Demuxalot (refined). Demultiplexing methods correctly classify a large portion of the singlets and heterogenic doublets (Fig. 3 c). This pattern is highly consistent across different cell types, with the notable exceptions being decreased correct classifications for erythrocytes and platelets when greater than 16 individuals are multiplexed (Additional file 2 : Fig S6).
However, Demuxalot consistently demonstrates the highest correct heterogenic doublet classification. Further, the percentage of the heterogenic doublets classified correctly by Souporcell decreases when large numbers of donors are multiplexed. ScSplit is not as effective as the other demultiplexing methods at classifying heterogenic doublets, partly due to the unique doublet classification method, which assumes that the doublets will generate a single cluster separate from the donors (Table 1 ). Importantly, the demultiplexing methods identify almost none of the homogenic doublets for any multiplexed pool size—demonstrating the need to include doublet detecting methods to supplement the demultiplexing method doublet detection.
Doublet detecting method classification performances vary greatly
In addition to assessing each of the methods with default settings, we also evaluated ScDblFinder with ‘known doublets’ provided. This method can take already known doublets and use them when detecting doublets. For these cases, we used the droplets that were classified as doublets by all the demultiplexing methods as ‘known doublets’.
Most of the methods classified a similarly high percentage of singlets correctly, with the exceptions of DoubletDecon and DoubletFinder for all pool sizes (Fig. 3 c). However, unlike the demultiplexing methods, there are explicit cell-type-specific biases for many of the doublet detecting methods (Additional file 2 : Fig S7). These differences are most notable for cell types with fewer cells (i.e. ASDC and cDC2) and proliferating cells (i.e. CD4 Proliferating, CD8 Proliferating, and NK Proliferating). Further, all of the softwares demonstrate high correct percentages for some cell types including CD4 Naïve and CD8 Naïve (Additional file 2 : Fig S7).
As expected, all doublet detecting methods identified heterotypic doublets more effectively than homotypic doublets (Fig. 3 c). However, ScDblFinder and Scrublet classified the most doublets correctly across all doublet types for pools containing 16 individuals or fewer. Solo was more effective at identifying doublets than Scds for pools containing more than 16 individuals. It is also important to note that it was not feasible to run DoubletDecon for the largest pools containing 128 multiplexed individuals and an average of 115,802 droplets (range: 113,594–119,126 droplets). ScDblFinder performed similarly when executed with and without known doublets (Pearson correlation P = 2.5 × 10 -40 ). This suggests that providing known doublets to ScDblFinder does not offer an added benefit.
Performances vary between demultiplexing and doublet detecting method and across the number of multiplexed individuals
We assessed the overall performance of each method with two metrics: the balanced accuracy and the MCC. We chose to use balanced accuracy since, with unbalanced group sizes, it is a better measure of performance than accuracy itself. Further, the MCC has been demonstrated as a more reliable statistical measure of performance since it considers all possible categories—true singlets (true positives), false singlets (false positives), true doublets (true negatives), and false doublets (false negatives). Therefore, a high score on the MCC scale indicates high performance in each metric. However, we provide additional performance metrics for each method (Additional file 1 : Table S3). For demultiplexing methods, both the droplet type (singlet or doublet) and the individual assignment were required to be considered a ‘true singlet’. In contrast, only the droplet type (singlet or doublet) was needed for doublet detection methods.
The MCC and balanced accuracy metrics are similar (Spearman’s ⍴ = 0.87; P < 2.2 × 10 -308 ). Further, the performance of Souporcell decreases for pools with more than 32 individuals multiplexed for both metrics (Student’s t -test for MCC: P < 1.1 × 10 -9 and balanced accuracy: P < 8.1 × 10 -11 ). Scds , ScDblFinder , and Scrublet are among the top-performing doublet detecting methods Fig. 3 d–e).
Overall, between 0.4 and 78.8% of droplets were incorrectly classified by the demultiplexing or doublet detecting methods depending on the technique and the multiplexed pool size (Additional file 2 : Fig S8). Demuxalot (refined) and DoubletDetection demonstrated the lowest percentage of incorrect droplets with about 1% wrong in the smaller pools (two multiplexed individuals) and about 3% incorrect in pools with at least 16 multiplexed individuals. Since some transitional states and cell types are present in low percentages in total cell populations (i.e. ASDCs at 0.02%), incorrect classification of droplets could alter scientific interpretations of the data, and it is, therefore, ideal for decreasing the number of erroneous assignments as much as possible.
False singlets and doublets demonstrate different metrics than correctly classified droplets
We next asked whether specific cell metrics might contribute to false singlet and doublet classifications for different methods. Therefore, we compared the number of genes, number of UMIs, mitochondrial percentage and ribosomal percentage of the false singlets and doublets to equal numbers of correctly classified cells for each demultiplexing and doublet detecting method.
The number of UMIs (Additional file 2 : Fig S9 and Additional file 1 : Table S4) and genes (Additional file 2 : Fig S10 and Additional file 1 : Table S5) demonstrated very similar distributions for all comparisons and all methods (Spearman ⍴ = 0.99, P < 2.2 × 10 -308 ). The number of UMIs and genes were consistently higher in false singlets and lower in false doublets for most demultiplexing methods except some smaller pool sizes (Additional file 2 : Fig S9a and Additional file 2 : Fig S10a; Additional file 1 : Table S4 and Additional file 1 : Table S5). The number of UMIs and genes was consistently higher in droplets falsely classified as singlets by the doublet detecting methods than the correctly identified droplets (Additional file 2 : Fig S9b and Additional file 2 : Fig S10b; Additional file 1 : Table S4 and Additional file 1 : Table S5). However, there was less consistency in the number of UMIs and genes detected in false singlets than correctly classified droplets between the different doublet detecting methods (Additional file 2 : Fig S9b and Additional file 2 : Fig S10b; Additional file 1 : Table S4 and Additional file 1 : Table S5).
The ribosomal percentage of the droplets falsely classified as singlets or doublets is similar to the correctly classified droplets for most methods—although they are statistically different for larger pool sizes (Additional file 2 : Fig S11a and Additional file 1 : Table S6). However, the false doublets classified by some demultiplexing methods ( Demuxalot , Demuxalot (refined), Demuxlet , ScSplit , Souporcell , and Vireo ) demonstrated higher ribosomal percentages. Some doublet detecting methods ( ScDblFinder , ScDblFinder with known doublets and Solo) demonstrated higher ribosomal percentages for the false doublets while other demonstrated lower ribosomal percentages ( DoubletDecon , DoubletDetection , and DoubletFinder ; Additional file 2 : Fig S11b and Additional file 1 : Table S6).
Like the ribosomal percentage, the mitochondrial percentage in false singlets is also relatively similar to correctly classified droplets for both demultiplexing (Additional file 2 : Fig S12a and Additional file 1 : Table S7) and doublet detecting methods (Additional file 2 : Fig S12b). The mitochondrial percentage for false doublets is statistically lower than the correctly classified droplets for a few larger pools for Freemuxlet , ScSplit , and Souporcell . The doublet detecting method Solo also demonstrates a small but significant decrease in mitochondrial percentage in the false doublets compared to the correctly annotated droplets. However, other doublet detecting methods including DoubletFinder and the larger pools of most other methods demonstrated a significant increase in mitochondrial percent in the false doublets compared to the correctly annotated droplets (Additional file 2 : Fig S12b).
Overall, these results demonstrate a strong relationship between the number of genes and UMIs and limited influence of ribosomal or mitochondrial percentage in a droplet and false classification, suggesting that the number of genes and UMIs can significantly bias singlet and doublet classification by demultiplexing and doublet detecting methods.
Ambient RNA, number of reads per cell, and uneven pooling impact method performance
To further quantify the variables that impact the performance of each method, we simulated four conditions that could occur with single-cell RNA-seq experiments: (1) decreased number of reads (reduced 50%), (2) increased ambient RNA (10%, 20% and 50%), (3) increased mitochondrial RNA (5%, 10% and 25%) and 4) uneven donor pooling from single donor spiking (0.5 or 0.75 proportion of pool from one donor). We chose these scenarios because they are common technical effects that can occur.
We observed a consistent decrease in the demultiplexing method performance when the number of reads were decreased by 50% but the degree of the effect varied for each method and was larger in pools containing more multiplexed donors (Additional file 2 : Fig S13a and Additional file 1 : Table S8). Decreasing the number of reads did not have a detectable impact on the performance of the doublet detecting methods.
Simulating additional ambient RNA (10%, 20%, or 50%) decreased the performance of all the demultiplexing methods (Additional file 2 : Fig S13b and Additional file 1 : Table S9) but some were unimpacted in pools that had 16 or fewer individuals multiplexed ( Souporcell and Vireo ). The performance of some of the doublet detecting methods were impacted by the ambient RNA but the performance of most methods did not decrease. Scrublet and ScDblFinder were the doublet detecting methods most impacted by ambient RNA but only in pools with at least 32 multiplexed donors (Additional file 2 : Fig S13b and Additional file 1 : Table S9).
Increased mitochondrial percent did not impact the performance of demultiplexing or doublet detecting methods (Additional file 2 : Fig S13c and Additional file 1 : Table S10).
We also tested whether experimental designs that pooling uneven proportions of donors would alter performance. We tested scenarios where either half the pool was composed of a single donor (0.5 spiked donor proportion) or where three quarters of the pool was composed of a single donor. This experimental design significantly reduced the demultiplexing method performance (Additional file 2 : Fig S13d and Additional file 1 : Table S11) with the smallest influence on Freemuxlet . The performance of most of the doublet detecting methods were unimpacted except for DoubletDetection that demonstrated significant decreases in performance in pools where at least 16 donors were multiplexed. Intriguingly, the performance of Solo increased with the spiked donor pools when the pools consisted of 16 donors or less.
Our results demonstrate significant differences in overall performance between different demultiplexing and doublet detecting methods. We further noticed some differences in the use of the methods. Therefore, we have accumulated these results and each method’s unique characteristics and benefits in a heatmap for visual interpretation (Fig. 4 ).
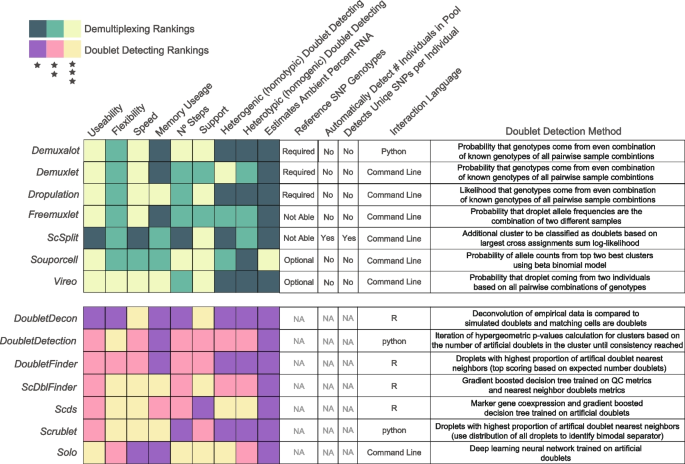
Assessment of each of the demultiplexing and doublet detecting methods. Assessments of a variety of metrics for each of the demultiplexing (top) and doublet detecting (bottom) methods
Framework for improving singlet classifications via method combinations
After identifying the demultiplexing and doublet detecting methods that performed well individually, we next sought to test whether using intersectional combinations of multiple methods would enhance droplet classifications and provide a software platform— Demuxafy —capable of supporting the execution of these intersectional combinations.
We recognise that different experimental designs will be required for each project. As such, we considered this when testing combinations of methods. We considered multiple experiment designs and two different intersectional methods: (1) more than half had to classify a droplet as a singlet to be called a singlet and (2) at least half of the methods had to classify a droplet as a singlet to be called a singlet. Significantly, these two intersectional methods only differ when an even number of methods are being considered. For combinations that include demultiplexing methods, the individual called by the majority of the methods is the individual used for that droplet. When ties occur, the individual is considered ‘unassigned’.
Combining multiple doublet detecting methods improve doublet removal for non-multiplexed experimental designs
For the non-multiplexed experimental design, we considered all possible method combinations (Additional file 1 : Table S12). We identified important differences depending on the number of droplets captured and have provided recommendations accordingly. We identified that DoubletFinder , Scrublet , ScDblFinder and Scds is the ideal combination for balanced droplet calling when less than 2,000 droplets are captured. Scds and ScDblFinder or Scrublet , Scds and ScDblFinder is the best combination when 2,000–10,000 droplets are captured. Scds , Scrublet, ScDblFinder and DoubletDetection is the best combination when 10,000–20,000 droplets are captured and Scrublet , Scds , DoubletDetection and ScDblFinder . It is important to note that even a slight increase in the MCC significantly impacts the number of true singlets and true doublets classified with the degree of benefit highly dependent on the original method performance. The combined method increases the MCC compared to individual doublet detecting methods on average by 0.11 and up to 0.33—a significant improvement in the MCC ( t -test FDR < 0.05 for 95% of comparisons). For all combinations, the intersectional droplet method requires more than half of the methods to consider the droplet a singlet to classify it as a singlet (Fig. 5 ).
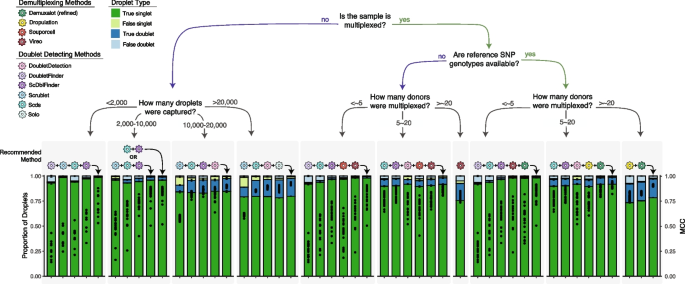
Recommended Method Combinations Dependent on Experimental Design. Method combinations are provided for different experimental designs, including those that are not multiplexed (left) and multiplexed (right), including experiments that have reference SNP genotypes available vs those that do not and finally, multiplexed experiments with different numbers of individuals multiplexed. The each bar represents either a single method (shown with the coloured icon above the bar) or a combination of methods (shown with the addition of the methods and an arrow indicating the bar). The proportion of true singlets, true doublets, false singlets and false doublets for each method or combination of methods is shown with the filled barplot and the MCC is shown with the black points overlaid on the barplot. MCC: Matthew’s Correlation Coefficient
Demuxafy performs better than Chord
Chord is an ensemble machine learning doublet detecting method that uses Scds and DoubletFinder to identify doublets. We compared Demuxafy using Scds and DoubletFinder to Chord and identified that Demuxafy outperformed Chord in pools that contained at least eight donors and was equivalent in pools that contained less than eight donors (Additional file 2 : Fig S14). This is because Chord classifies more droplets as false singlets and false doublets than Demuxafy . In addition, Chord failed to complete for two of the pools that contained 128 multiplexed donors.
Combining multiple demultiplexing and doublet detecting methods improve doublet removal for multiplexed experimental designs
For experiments where 16 or fewer individuals are multiplexed with reference SNP genotypes available, we considered all possible combinations between the demultiplexing and doublet detecting methods except ScDblFinder with known doublets due to its highly similar performance to ScDblFinder (Fig 3 ; Additional file 1 : Table S13). The best combinations are DoubletFinder , Scds , ScDblFinder , Vireo and Demuxalot (refined) (<~5 donors) and Scrublet , ScDblFinder , DoubletDetection , Dropulation and Demuxalot (refined) (Fig. 5 ). These intersectional methods increase the MCC compared to the individual methods ( t -test FDR < 0.05), generally resulting in increased true singlets and doublets compared to the individual methods. The improvement in MCC depends on every single method’s performance but, on average, increases by 0.22 and up to 0.71. For experiments where the reference SNP genotypes are unknown, the individuals multiplexed in the pool with 16 or fewer individuals multiplexed, DoubletFinder , ScDblFinder, Souporcell and Vireo (<~5 donors) and Scds , ScDblFinder , DoubletDetection , Souporcell and Vireo are the ideal methods (Fig. 5 ). These intersectional methods again significantly increase the MCC up to 0.87 compared to any of the individual techniques that could be used for this experimental design ( t -test FDR < 0.05 for 94.2% of comparisons). In both cases, singlets should only be called if more than half of the methods in the combination classify the droplet as a singlet.
Combining multiple demultiplexing methods improves doublet removal for large multiplexed experimental designs
For experiments that multiplex more than 16 individuals, we considered the combinations between all demultiplexing methods (Additional file 1 : Table S14) since only a small proportion of the doublets would be undetectable by demultiplexing methods (droplets that are homogenic; Fig 3 b). To balance doublet removal and maintain true singlets, we recommend the combination of Demuxalot (refined) and Dropulation . These method combinations significantly increase the MCC by, on average, 0.09 compared to all the individual methods ( t -test FDR < 0.05). This substantially increases true singlets and true doublets relative to the individual methods. If reference SNP genotypes are not available for the individuals multiplexed in the pools, Vireo performs the best (≥ 16 multiplexed individuals; Fig. 5 ). This is the only scenario in which executing a single method is advantageous to a combination of methods. This is likely due to the fact that most of the methods perform poorly for larger pool sizes (Fig. 3 c).
These results collectively demonstrate that, regardless of the experimental design, demultiplexing and doublet detecting approaches that intersect multiple methods significantly enhance droplet classification. This is consistent across different pool sizes and will improve singlet annotation.
Demuxafy improves doublet removal and improves usability
To make our intersectional approaches accessible to other researchers, we have developed Demuxafy ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/index.html ) - an easy-to-use software platform powered by Singularity. This platform provides the requirements and instructions to execute each demultiplexing and doublet detecting methods. In addition, Demuxafy provides wrapper scripts that simplify method execution and effectively summarise results. We also offer tools that help estimate expected numbers of doublets and provide method combination recommendations based on scRNA-seq pool characteristics. Demuxafy also combines the results from multiple different methods, provides classification combination summaries, and provides final integrated combination classifications based on the intersectional techniques selected by the user. The significant advantages of Demuxafy include a centralised location to execute each of these methods, simplified ways to combine methods with an intersectional approach, and summary tables and figures that enable practical interpretation of multiplexed datasets (Fig. 1 a).
Demultiplexing and doublet detecting methods have made large-scale scRNA-seq experiments achievable. However, many demultiplexing and doublet detecting methods have been developed in the recent past, and it is unclear how their performances compare. Further, the demultiplexing techniques best detect heterogenic doublets while doublet detecting methods identify heterotypic doublets. Therefore, we hypothesised that demultiplexing and doublet detecting methods would be complementary and be more effective at removing doublets than demultiplexing methods alone.
Indeed, we demonstrated the benefit of utilising a combination of demultiplexing and doublet detecting methods. The optimal intersectional combination of methods depends on the experimental design and capture characteristics. Our results suggest super loaded captures—where a high percentage of doublets is expected—will benefit from multiplexing. Further, when many donors are multiplexed (>16), doublet detecting is not required as there are few doublets that are homogenic and heterotypic.
We have provided different method combination recommendations based on the experimental design. This decision is highly dependent on the research question.
Conclusions
Overall, our results provide researchers with important demultiplexing and doublet detecting performance assessments and combinatorial recommendations. Our software platform, Demuxafy ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/index.html ), provides a simple implementation of our methods in any research lab around the world, providing cleaner scRNA-seq datasets and enhancing interpretation of results.
PBMC scRNA-seq data
Blood samples were collected and processed as described previously [ 17 ]. Briefly, mononuclear cells were isolated from whole blood samples and stored in liquid nitrogen until thawed for scRNA-seq capture. Equal numbers of cells from 12 to 16 samples were multiplexed per pool and single-cell suspensions were super loaded on a Chromium Single Cell Chip A (10x Genomics) to capture 20,000 droplets per pool. Single-cell libraries were processed per manufacturer instructions and the 10× Genomics Cell Ranger Single Cell Software Suite (v 2.2.0) was used to process the data and map it to GRCh38. Cellbender v0.1.0 was used to identify empty droplets. Almost all droplets reported by Cell Ranger were identified to contain cells by Cellbender (mean: 99.97%). The quality control metrics of each pool are demonstrated in Additional file 2 : Fig S15.
PBMC DNA SNP genotyping
SNP genotype data were prepared as described previously [ 17 ]. Briefly, DNA was extracted from blood with the QIAamp Blood Mini kit and genotyped on the Illumina Infinium Global Screening Array. SNP genotypes were processed with Plink and GCTA before imputing on the Michigan Imputation Server using Eagle v2.3 for phasing and Minimac3 for imputation based on the Haplotype Reference Consortium panel (HRCr1.1). SNP genotypes were then lifted to hg38 and filtered for > 1% minor allele frequency (MAF) and an R 2 > 0.3.
Fibroblast scRNA-seq data
The fibroblast scRNA-seq data has been described previously [ 18 ]. Briefly, human skin punch biopsies from donors over the age of 18 were cultured in DMEM high glucose supplemented with 10% fetal bovine serum (FBS), L-glutamine, 100 U/mL penicillin and 100 μg/mL (Thermo Fisher Scientific, USA).
For scRNA-seq, viable cells were flow sorted and single cell suspensions were loaded onto a 10× Genomics Single Cell 3’ Chip and were processed per 10× instructions and the Cell Ranger Single Cell Software Suite from 10× Genomics was used to process the sequencing data into transcript count tables as previously described [ 18 ]. Cellbender v0.1.0 was used to identify empty droplets. Almost all droplets reported by Cell Ranger were identified to contain cells by Cellbender (mean: 99.65%). The quality control metrics of each pool are demonstrated in Additional file 2 : Fig S16.
Fibroblast DNA SNP genotyping
The DNA SNP genotyping for fibroblast samples has been described previously [ 18 ]. Briefly, DNA from each donor was genotyped on an Infinium HumanCore-24 v1.1 BeadChip (Illumina). GenomeStudioTM V2.0 (Illumina), Plink and GenomeStudio were used to process the SNP genotypes. Eagle V2.3.5 was used to phase the SNPs and it was imputed with the Michigan Imputation server using minimac3 and the 1000 genome phase 3 reference panel as described previously [ 18 ].
Demultiplexing methods
All the demultiplexing methods were built and run from a singularity image.
Demuxalot [ 6 ] is a genotype reference-based single cell demultiplexing method. Demualot v0.2.0 was used in python v3.8.5 to annotate droplets. The likelihoods, posterior probabilities and most likely donor for each droplet were estimated using the Demuxalot Demultiplexer.predict_posteriors function. We also used Demuxalot Demultiplexer.learn_genotypes function to refine the genotypes before estimating the likelihoods, posterior probabilities and likely donor of each droplet with the refined genotypes as well.
The Popscle v0.1-beta suite [ 16 ] for population genomics in single cell data was used for Demuxlet and Freemuxlet demultiplexing methods. The popscle dsc-pileup function was used to create a pileup of variant calls at known genomic locations from aligned sequence reads in each droplet with default arguments.
Demuxlet [ 3 ] is a SNP genotype reference-based single cell demultiplexing method. Demuxlet was run with a genotype error coefficient of 1 and genotype error offset rate of 0.05 and the other default parameters using the popscle demuxlet command from Popscle (v0.1-beta).
Freemuxlet [ 16 ] is a SNP genotype reference-free single cell demultiplexing method. Freemuxlet was run with default parameters including the number of samples included in the pool using the popscle freemuxlet command from Popscle (v0.1-beta).
Dropulation
Dropulation [ 5 ] is a SNP genotype reference-based single cell demultiplexing method that is part of the Drop-seq software. Dropulation from Drop-seq v2.5.1 was implemented for this manuscript. In addition, the method for calling singlets and doublets was provided by the Dropulation developer and implemented in a custom R script available on Github and Zenodo (see “Availability of data and materials”).
ScSplit v1.0.7 [ 7 ] was downloaded from the ScSplit github and the recommended steps for data filtering quality control prior to running ScSplit were followed. Briefly, reads that had read quality lower than 10, were unmapped, were secondary alignments, did not pass filters, were optical PCR duplicates or were duplicate reads were removed. The resulting bam file was then sorted and indexed followed by freebayes to identify single nucleotide variants (SNVs) in the dataset. The resulting SNVs were filtered for quality scores greater than 30 and for variants present in the reference SNP genotype vcf. The resulting filtered bam and vcf files were used as input for the s cSplit count command with default settings to count the number of reference and alternative alleles in each droplet. Next the allele matrices were used to demultiplex the pool and assign cells to different clusters using the scSplit run command including the number of individuals ( -n ) option and all other options set to default. Finally, the individual genotypes were predicted for each cluster using the scSplit genotype command with default parameters.
Souporcell [ 4 ] is a SNP genotype reference-free single cell demultiplexing method. The Souporcell v1.0 singularity image was downloaded via instructions from the gihtub page. The Souporcell pipeline was run using the souporcell_pipeline.py script with default options and the option to include known variant locations ( --common_variants ).
Vireo [ 2 ] is a single cell demultiplexing method that can be used with reference SNP genotypes or without them. For this assessment, Vireo was used with reference SNP genotypes. Per Vireo recommendations, we used model 1 of the cellSNP [ 20 ] version 0.3.2 to make a pileup of SNPs for each droplet with the recommended options using the genotyped reference genotype file as the list of common known SNP and filtered with SNP locations that were covered by at least 20 UMIs and had at least 10% minor allele frequency across all droplets. Vireo version 0.4.2 was then used to demultiplex using reference SNP genotypes and indicating the number of individuals in the pools.
Doublet detecting methods
All doublet detecting methods were built and run from a singularity image.
DoubletDecon
DoubletDecon [ 9 ] is a transcription-based deconvolution method for identifying doublets. DoubletDecon version 1.1.6 analysis was run in R version 3.6.3. SCTransform [ 21 ] from Seurat [ 22 ] version 3.2.2 was used to preprocess the scRNA-seq data and then the Improved_Seurat_Pre_Process function was used to process the SCTransformed scRNA-seq data. Clusters were identified using Seurat function FindClusters with resolution 0.2 and 30 principal components (PCs). Then the Main_Doublet_Decon function was used to deconvolute doublets from singlets for six different rhops—0.6, 0.7, 0.8, 0.9, 1.0 and 1.1. We used a range of rhop values since the doublet annotation by DoubletDecon is dependent on the rhop parameter which is selected by the user. The rhop that resulted in the closest number of doublets to the expected number of doublets was selected on a per-pool basis and used for all subsequent analysis. Expected number of doublets were estimated with the following equation:
where N is the number of droplets captured and D is the number of expected doublets.
DoubletDetection
DoubletDetection [ 14 ] is a transcription-based method for identifying doublets. DoubletDetection version 2.5.2 analysis was run in python version 3.6.8. Droplets without any UMIs were removed before analysis with DoubletDetection . Then the doubletdetection.BoostClassifier function was run with 50 iterations with use_phenograph set to False and standard_scaling set to True. The predicted number of doublets per iteration was visualised across all iterations and any pool that did not converge after 50 iterations, it was run again with increasing numbers of iterations until they reached convergence.
DoubletFinder
DoubletFinder [ 10 ] is a transcription-based doublet detecting method. DoubletFinder version 2.0.3 was implemented in R version 3.6.3. First, droplets that were more than 3 median absolute deviations (mad) away from the median for mitochondrial per cent, ribosomal per cent, number of UMIs or number of genes were removed per developer recommendations. Then the data was normalised with SCTransform followed by cluster identification using FindClusters with resolution 0.3 and 30 principal components (PCs). Then, pKs were selected by the pK that resulted in the largest BC MVN as recommended by DoubletFinder. The pK vs BC MVN relationship was visually inspected for each pool to ensure an effective BC MVN was selected for each pool. Finally, the homotypic doublet proportions were calculated and the number of expected doublets with the highest doublet proportion were classified as doublets per the following equation:
ScDblFinder
ScDblFinder [ 11 ] is a transcription-based method for detecting doublets from scRNA-seq data. ScDblFinder 1.3.25 was implemented in R version 4.0.3. ScDblFinder was implemented with two sets of options. The first included implementation with the expected doublet rate as calculated by:
where N is the number of droplets captured and R is the expected doublet rate. The second condition included the same expected number of doublets and included the doublets that had already been identified by all the demultiplexing methods.
Scds [ 12 ] is a transcription-based doublet detecting method. Scds version 1.1.2 analysis was completed in R version 3.6.3. Scds was implemented with the cxds function and bcds functions with default options followed by the cxds_bcds_hybrid with estNdbl set to TRUE so that doublets will be estimated based on the values from the cxds and bcds functions.
Scrublet [ 13 ] is a transcription-based doublet detecting method for single-cell RNA-seq data. Scrublet was implemented in python version 3.6.3. Scrublet was implemented per developer recommendations with at least 3 counts per droplet, 3 cells expressing a given gene, 30 PCs and a doublet rate based on the following equation:
where N is the number of droplets captured and R is the expected doublet rate. Four different minimum number of variable gene percentiles: 80, 85, 90 and 95. Then, the best variable gene percentile was selected based on the distribution of the simulated doublet scores and the location of the doublet threshold selection. In the case that the selected threshold does not fall between a bimodal distribution, those pools were run again with a manual threshold set.
Solo [ 15 ] is a transcription-based method for detecting doublets in scRNA-seq data. Solo was implemented with default parameters and an expected number of doublets based on the following equation:
where N is the number of droplets captured and D is the number of expected doublets. Solo was additionally implemented in a second run for each pool with the doublets that were identified by all the demultiplexing methods as known doublets to initialize the model.
In silico pool generation
Cells that were identified as singlets by all methods were used to simulate pools. Ten pools containing 2, 4, 8, 16, 32, 64 and 128 individuals were simulated assuming a maximum 20% doublet rate as it is unlikely researchers would use a technology that has a higher doublet rate. The donors for each simulated pool were randomly selected using a custom R script which is available on Github and Zenodo (see ‘Availability of data and materials’). A separate bam for the cell barcodes for each donor was generated using the filterbarcodes function from the sinto package (v0.8.4). Then, the GenerateSyntheticDoublets function provided by the Drop-seq [ 5 ] package was used to simulate new pools containing droplets with known singlets and doublets.
Twenty-one total pools—three pools from each of the different simulated pool sizes (2, 4, 8, 16, 32, 64 and 128 individuals) —were used to simulate different experimental scenarios that may be more challenging for demultiplexing and doublet detecting methods. These include simulating higher ambient RNA, higher mitochondrial percent, decreased read coverage and imbalanced donor proportions as described subsequently.
High ambient RNA simulations
Ambient RNA was simulated by changing the barcodes and UMIs on a random selection of reads for 10, 20 or 50% of the total UMIs. This was executed with a custom R script that is available in Github and Zenodo (see ‘Availability of data and materials’).
High mitochondrial percent simulations
High mitochondrial percent simulations were produced by replacing reads in 5, 10 or 25% of the randomly selected cells with mitochondrial reads. The number of reads to replace was derived from a normal distribution with an average of 30 and a standard deviation of 3. This was executed with a custom R script available in Github and Zenodo (see ‘Availability of data and materials’).
Imbalanced donor simulations
We simulated pools that contained uneven proportions of the donors in the pools to identify if some methods are better at demultiplexing pools containing uneven proportions of each donor in the pool. We simulated pools where 50, 75 or 95% of the pool contained cells from a single donor and the remainder of the pool was even proportions of the remaining donors in the pool. This was executed with a custom R script available in Github and Zenodo (see ‘Availability of data and materials’).
Decrease read coverage simulations
Decreased read coverage of pools was simulated by down-sampling the reads by two-thirds of the original coverage.
Classification annotation
Demultiplexing methods classifications were considered correct if the droplet annotation (singlet or doublet) and the individual annotation was correct. If the droplet type was correct but the individual annotation was incorrect (i.e. classified as a singlet but annotated as the wrong individual), then the droplet was incorrectly classified.
Doublet detecting methods were considered to have correct classifications if the droplet annotation matched the known droplet type.
All downstream analyses were completed in R version 4.0.2.
Availability of data and materials
All data used in this manuscript is publicly available. The PBMC data is available on GEO (Accession: GSE196830) [ 23 ] as originally described in [ 17 ]. The fibroblast data is available on ArrayExpress (Accession Number: E-MTAB-10060) [ 24 ] and as originally described in [ 18 ]. The code used for the analyses in this manuscript are provided on Github ( https://github.com/powellgenomicslab/Demuxafy_manuscript/tree/v4 ) and Zenodo ( https://zenodo.org/records/10813452 ) under an MIT Open Source License [ 25 , 26 ]. Demuxafy is provided as a package with source code available on Github ( https://github.com/drneavin/Demultiplexing_Doublet_Detecting_Docs ) and instructions on ReadTheDocs ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/ ) under an MIT Open Source License [ 27 ]. Demuxafy is also available on Zenodo with the link https://zenodo.org/records/10870989 [ 28 ].
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:1–12.
Article Google Scholar
Huang Y, McCarthy DJ, Stegle O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 2019;20:273.
Article PubMed PubMed Central Google Scholar
Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36:89–94.
Article CAS PubMed Google Scholar
Heaton H, Talman AM, Knights A, Imaz M, Gaffney DJ, Durbin R, et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat Methods. 2020;17:615–20.
Wells MF, Nemesh J, Ghosh S, Mitchell JM, Salick MR, Mello CJ, et al. Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages. Cell Stem Cell. 2023;30:312–332.e13.
Article CAS PubMed PubMed Central Google Scholar
Rogozhnikov A, Ramkumar P, Shah K, Bedi R, Kato S, Escola GS. Demuxalot: scaled up genetic demultiplexing for single-cell sequencing. bioRxiv. 2021;2021.05.22.443646.
Xu J, Falconer C, Nguyen Q, Crawford J, McKinnon BD, Mortlock S, et al. Genotype-free demultiplexing of pooled single-cell RNA-seq. Genome Biol. 2019;20:290.
What is the maximum number of cells that can be profiled?. Available from: https://kb.10xgenomics.com/hc/en-us/articles/360001378811-What-is-the-maximum-number-of-cells-that-can-be-profiled -
DePasquale EAK, Schnell DJ, Van Camp PJ, Valiente-Alandí Í, Blaxall BC, Grimes HL, et al. DoubletDecon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep. 2019;29:1718–1727.e8.
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337.e4.
Germain P-L, Lun A, Meixide CG, Macnair W, Robinson MD. Doublet identification in single-cell sequencing data. 2022;
Bais AS, Kostka D. Scds: Computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics. 2020;36:1150–8.
Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9.
Shor, Jonathan. DoubletDetection. Available from: https://github.com/JonathanShor/DoubletDetection .
Bernstein NJ, Fong NL, Lam I, Roy MA, Hendrickson DG, Kelley DR. Solo: doublet identification in single-cell RNA-Seq via semi-supervised deep learning. Cell Syst. 2020;11:95–101.e5.
popscle. Available from: https://github.com/statgen/popscle .
Yazar S, Alquicira-Hernandez J, Wing K, Senabouth A, Gordon MG, Andersen S, et al. Single-cell eQTL mapping identifies cell type–specific genetic control of autoimmune disease. Science. 2022;376:eabf3041.
Neavin D, Nguyen Q, Daniszewski MS, Liang HH, Chiu HS, Senabouth A, et al. Single cell eQTL analysis identifies cell type-specific genetic control of gene expression in fibroblasts and reprogrammed induced pluripotent stem cells. Genome Biol. 2021;1–19.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29.
Huang X, Huang Y. Cellsnp-lite: an efficient tool for genotyping single cells. bioRxiv. 2021;2020.12.31.424913.
Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. bioRxiv. 2019;576827.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21.
Powell JE. Single-cell eQTL mapping identifies cell type specific genetic control of autoimmune disease. Datasets. Gene Expression Omnibus. 2022. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196830 .
Nguyen Q, Powell JE. scRNA-seq in 79 fibroblast cell lines and 31 reprogrammed induced pluripotent stem cell lines for sceQTL analysis. Datasets. ArrayExpress. 2021. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-10060?query=E-MTAB-10060 .
Neavin DR. Demuxafy analyses. Github. 2024. https://github.com/powellgenomicslab/Demuxafy_manuscript/tree/v4 .
Neavin DR. Demuxafy analyses. Zenodo. 2024. https://zenodo.org/records/10813452 .
Neavin D. Demuxafy. Github. 2024. https://github.com/drneavin/Demultiplexing_Doublet_Detecting_Docs .
Neavin D. Demuxafy. Zenodo. 2024. https://zenodo.org/records/10870989 .
McCaughey T, Liang HH, Chen C, Fenwick E, Rees G, Wong RCB, et al. An interactive multimedia approach to improving informed consent for induced pluripotent stem cell research. Cell Stem Cell. 2016;18:307–8.
Download references
Authors’ Twitter handles
Twitter handles: @drneavin (Drew Neavin), @thjimmylee (Jimmy Tsz Hang Lee), @marta_mele_m (Marta Melé)
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 3 .
This work was funded by the National Health and Medical Research Council (NHMRC) Investigator grant (1175781), and funding from the Goodridge foundation. J.E.P is also supported by a fellowship from the Fok Foundation.
Author information
Authors and affiliations.
Garvan-Weizmann Centre for Cellular Genomics, Garvan Institute for Medical Research, Darlinghurst, NSW, Australia
Drew Neavin, Anne Senabouth, Himanshi Arora & Joseph E. Powell
Present address: Statewide Genomics at NSW Health Pathology, Sydney, NSW, Australia
Himanshi Arora
Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK
Jimmy Tsz Hang Lee
Life Sciences Department, Barcelona Supercomputing Center, Barcelona, Catalonia, Spain
Aida Ripoll-Cladellas & Marta Melé
Department of Genetics, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands
Lude Franke
Spatial and Single Cell Systems Domain, Genome Institute of Singapore (GIS), Agency for Science, Technology and Research (A*STAR), Singapore, Republic of Singapore
Shyam Prabhakar
Population and Global Health, Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Republic of Singapore
Cancer Science Institute of Singapore, National University of Singapore, Singapore, Republic of Singapore
Bakar Institute for Computational Health Sciences, University of California, San Francisco, CA, USA
Chun Jimmie Ye
Institute for Human Genetics, University of California, San Francisco, San Francisco, CA, USA
Division of Rheumatology, Department of Medicine, University of California, San Francisco, San Francisco, CA, USA
Chan Zuckerberg Biohub, San Francisco, CA, USA
Bioinformatics and Cellular Genomics, St Vincent’s Institute of Medical Research, Fitzroy, Australia
Davis J. McCarthy
Melbourne Integrative Genomics, School of BioSciences–School of Mathematics & Statistics, Faculty of Science, University of Melbourne, Melbourne, Australia
Present address: The Gene Lay Institute of Immunology and Inflammation, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA
Martin Hemberg
UNSW Cellular Genomics Futures Institute, University of New South Wales, Kensington, NSW, Australia
Joseph E. Powell
You can also search for this author in PubMed Google Scholar
sc-eQTLGen Consortium
Contributions.
DRN and JEP conceived the project idea and study design. JTHL, AR, LF, SP, CJY, DJM, MM and MH provided feedback on experimental design. DRN carried out analyses with support on coding from AS. JTHL and AR tested Demuxafy and provided feedback. DRN and JEP wrote the manuscript. All authors reviewed and provided feedback on the manuscript.
Corresponding authors
Correspondence to Drew Neavin or Joseph E. Powell .
Ethics declarations
Ethics approval and consent to participate.
Briefly, all work was approved by the Royal Hobart Hospital, the Hobart Eye Surgeons Clinic, Human Research Ethics Committees of the Royal Victorian Eye and Ear Hospital (11/1031), University of Melbourne (1545394) and University of Tasmania (H0014124) in accordance with the requirements of the National Health & Medical Research Council of Australia (NHMRC) and conformed with the Declaration of Helsinki [ 29 ].
Consent for publication
No personal data for any individual requiring consent for publication was included in this manuscript.
Competing interests
C.J.Y. is founder for and holds equity in DropPrint Genomics (now ImmunAI) and Survey Genomics, a Scientific Advisory Board member for and holds equity in Related Sciences and ImmunAI, a consultant for and holds equity in Maze Therapeutics, and a consultant for TReX Bio, HiBio, ImYoo, and Santa Ana. Additionally, C.J.Y is also newly an Innovation Investigator for the Arc Institute. C.J.Y. has received research support from Chan Zuckerberg Initiative, Chan Zuckerberg Biohub, Genentech, BioLegend, ScaleBio and Illumina.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: supplementary tables and legends., additional file 2: supplementary figures and legends., additional file 3..
Review history.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Neavin, D., Senabouth, A., Arora, H. et al. Demuxafy : improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods. Genome Biol 25 , 94 (2024). https://doi.org/10.1186/s13059-024-03224-8
Download citation
Received : 07 March 2023
Accepted : 25 March 2024
Published : 15 April 2024
DOI : https://doi.org/10.1186/s13059-024-03224-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Single-cell analysis
- Genetic demultiplexing
- Doublet detecting
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
How Draymond Green and Trayce Jackson-Davis have revived the Warriors' defense
Warriors finish regular season with win over Jazz (2:11)
GOLDEN STATE WARRIORS guard Chris Paul called it the best defensive play he had seen in his 19-year career.
With 1:30 left in the fourth quarter against the Dallas Mavericks on April 2 and the Warriors holding a 98-92 lead, Mavericks guard Luka Doncic feigned a dribble penetration from the top of the 3-point line before dishing the ball to guard Kyrie Irving . As Warriors forward Draymond Green roamed the paint, Irving sidestepped the defending Andrew Wiggins , took two steps toward Green and then passed to center Daniel Gafford near the baseline.
Just as quickly, Green changed his positioning and met Gafford at the rim for a spectacular block. Green ripped the rebound away from Irving, firing up the Chase Center crowd, with the Warriors then holding on for a 104-100 win.
"We're starting to understand that defense is going to be what we have to hang our hat on," Paul said after the game. "Then on offense, we of course have amazing shooters and scorers. But when we defend, it opens everything else up."
Draymond Green's big-time block on Daniel Gafford keeps the Warriors ahead late in the fourth quarter.
The team has fought through several on-court issues this season, including Green's suspensions and late-game struggles, Stephen Curry 's drop in offensive production since the All-Star break and Thompson finishing the regular season with his lowest scoring average since 2013-14.
But the victory against Dallas was part of a six-game winning streak fueled by an improved defense led by Green and the emergence of rookie center Trayce Jackson-Davis .
Golden State enters Tuesday's play-in tournament game against the Sacramento Kings with 10 wins in its final 12 regular season games. The Warriors also boast the league's seventh-best defense since the All-Star break and are fifth in defensive efficiency since Jackson-Davis entered the starting lineup March 27.
The Warriors' road back to the playoffs remains tough. They'll need to win back-to-back road games just to secure a first-round series against the top-seeded Oklahoma City Thunder .
But it was only three weeks ago when the Warriors were in danger of falling out of the postseason picture completely. On March 24, the Warriors started a crucial five-game road trip by losing to the Minnesota Timberwolves , which left them only a game over the Houston Rockets in the battle for 10th place.
However, the Warriors responded by winning their next four games -- all on the road -- before the home win against the Mavericks. During that streak, the Warriors held their opponents to 99.0 points per game, the franchise's fewest over a five-game span since January 2022.
"We've seen teams in this league for years that made the mistake of thinking they can just put a great offense out there and their defense sucks and think they're going to win," Green said after the Mavericks game. "As you run up against [a] team that is ... just competent on the offensive end but they play defense, you're going to lose."
Any postseason success will require Jackson-Davis, along with Green and veteran center Kevon Looney , to go up against a play-in field that includes All-Star big men such as the Kings' Domantas Sabonis or the Los Angeles Lakers ' Anthony Davis .
"Those assignments, I mean, it's all against some of the best players," Jackson-Davis told ESPN. "I know I'll have to lean on my veterans -- Draymond, Kevon. Those guys are really going to help me. And I know the preparation will be different, there will be a lot more of it. But I feel like I'm ready to tackle that challenge."
AFTER THE LOSS to the Timberwolves on March 24, Green had a harsh assessment of his team's defense.
"We have too many breakdowns," Green said. "You just can't win having breakdowns. When you have breakdowns, it changes momentum. And momentum in this league is not easy to get back."
The Warriors led the Wolves by as much as 12 points in the first half. They were up 70-65 with just over four minutes left in the third quarter when the Wolves then picked apart Golden State on a run that included six 3-pointers, including three by guard Nickeil Alexander-Walker . With Curry on the bench during that stretch and not returning until the 6:54 mark in the fourth, the Wolves went on a 32-19 run that then led to a 114-110 win.
"We're a very quiet team. So, you have issues on defense when you have bad communication," Green added. The Warriors' fourth-quarter defense -- which ranked 29th in defensive efficiency at the time -- had been a season-long issue. Defending the rim was one particular sore spot; Golden State was third worst in the league in field goal percentage allowed on layups and dunks in the fourth quarter.
Two days after the Wolves game, the Warriors bounced back with a 113-92 road win over the Miami Heat . With Jonathan Kuminga sitting out the following day's game in Orlando, Florida, because of knee tendonitis, Warriors coach Steve Kerr placed Jackson-Davis in the starting five. The rookie responded with eight points and 14 rebounds in the 101-93 win. Green was ejected four minutes into the game, but the duo quickly found a rhythm in the frontcourt over their next matchups.
In an April 4 win against the Rockets, Jackson-Davis spent time in the first half defending Houston guard Amen Thompson , while Green roamed as a paint defender. But in the second half, when Thompson began running more downhill and in transition, the Warriors' duo switched assignments.
"If I'm the 5, I'm the last line of defense," Green said after the game. "So the things that I do off of instinct or reading the game on the fly, it's harder to do. If I just go run over here to cover up something I see needs to be covered, that's leaving the rim unprotected ... but if I know the rim is protected by Trayce, I can just go do it."
Giving Green the ability to roam has helped Jackson-Davis hold opponents to 48% shooting in the paint as the contesting defender, which ranks ninth among players to contest more than 500 shots this season, per Second Spectrum tracking.
"Being able to tic-tac and interchange with him is great," Jackson-Davis said. "The things he teaches me, positioning and stuff of that nature, it's helped my game a lot."
Jackson-Davis and Green, who started 10 games together, finished the season with a 99.2 defensive efficiency as a duo, which ranks in the top 10 among all two-man lineups to play more than 225 minutes this season, according to ESPN Statistics & Information research.
"Trayce is wise beyond his years," Kerr told ESPN. "He is not a typical rookie these days, with four years of college experience and a lot of games played under his belt. He's gotten great experience the last couple of months."
AFTER SUNDAY'S WIN against the Utah Jazz to wrap up the regular season, all eyes turned to Tuesday's matchup: the play-in game between the 9-seed and 10-seed in Sacramento. The Kings provide an interesting defensive task for Golden State.
Led by their All-Star duo in Sabonis and guard De'Aaron Fox , rim protection and paint defense will be key for the Warriors.
"Everything starts with the head of the snake -- Sabonis and Fox -- and then everyone else gets their [offense] off of those two guys," Green said after the Jazz win. "We know [the Kings] well, they know us well, so there won't be any surprises."
The two teams met in the first round of the playoffs last season, with the Warriors eliminating the Kings in seven games. This season, three of their four meetings have been decided by one point, including Sacramento's 134-133 win in late January that saw the Kings hit 22 3-pointers on 48 attempts.
During the playoffs last season, Looney was primarily tasked with defending Sabonis. He was during this regular season, as well. According to Second Spectrum, Looney has defended Sabonis for 658 half-court matchups for their careers, including regular season and playoffs. That is the most by any player against Sabonis.
Even with Jackson-Davis taking over the majority of Looney's minutes in the rotation, the challenge remains the same in stopping Sacramento's star big man.
"At the center spot, you've got to be super talkative, real disruptive," Looney told ESPN. "Those matchups are super key, trying to stop two guys who are All-NBA. I think Trayce, Draymond, myself, we have the talent to do that."
Slowing down Sabonis and Fox will be no easy task, specifically on dribble handoffs. The pair ran 285 total handoffs together this season (fifth most in the NBA among all duos), but the Warriors are ranked sixth at defending them (0.97 points per direct play).
"Sabonis does all his damage pretty much within 10 to 15 feet ... and Trayce can affect some of those shots," Green said. "It starts with positioning, and Sabonis is great at creating angles. Trayce has got to use what he has and his strengths, which is his athleticism and his length."
And should the Warriors advance, it gives Kerr that flexibility to either rely on the rookie big man or a front-court veteran in what is a win-or-go-home setting.
"I have a lot of confidence in having Loon, knowing that for a rookie it's a lot to ask him to go into a playoff game and play against great players, which is going to be the case," Kerr said. "It's a good situation to be in."
- Cambridge Dictionary +Plus
Meaning of assignment – Learner’s Dictionary
Your browser doesn't support HTML5 audio
(Definition of assignment from the Cambridge Learner's Dictionary © Cambridge University Press)
Translations of assignment
Get a quick, free translation!

Word of the Day
have your hands full
to be so busy that you do not have time to do anything else

Binding, nailing, and gluing: talking about fastening things together

Learn more with +Plus
- Recent and Recommended {{#preferredDictionaries}} {{name}} {{/preferredDictionaries}}
- Definitions Clear explanations of natural written and spoken English English Learner’s Dictionary Essential British English Essential American English
- Grammar and thesaurus Usage explanations of natural written and spoken English Grammar Thesaurus
- Pronunciation British and American pronunciations with audio English Pronunciation
- English–Chinese (Simplified) Chinese (Simplified)–English
- English–Chinese (Traditional) Chinese (Traditional)–English
- English–Dutch Dutch–English
- English–French French–English
- English–German German–English
- English–Indonesian Indonesian–English
- English–Italian Italian–English
- English–Japanese Japanese–English
- English–Norwegian Norwegian–English
- English–Polish Polish–English
- English–Portuguese Portuguese–English
- English–Spanish Spanish–English
- English–Swedish Swedish–English
- Dictionary +Plus Word Lists
- Learner’s Dictionary Noun
- Translations
- All translations
Add assignment to one of your lists below, or create a new one.
{{message}}
Something went wrong.
There was a problem sending your report.

IMAGES
VIDEO
COMMENTS
assign: [verb] to transfer (property) to another especially in trust or for the benefit of creditors.
The meaning of ASSIGNMENT is the act of assigning something. How to use assignment in a sentence. Synonym Discussion of Assignment.
ASSIGNED definition: 1. past simple and past participle of assign 2. to give a particular job or piece of work to…. Learn more.
ASSIGNMENT definition: 1. a piece of work given to someone, typically as part of their studies or job: 2. a job that…. Learn more.
ASSIGNED meaning: 1. past simple and past participle of assign 2. to give a particular job or piece of work to…. Learn more.
Assignment definition: something assigned, as a particular task or duty. See examples of ASSIGNMENT used in a sentence.
Assign definition: to give or allocate; allot. See examples of ASSIGN used in a sentence.
1 to give someone something that they can use, or some work or responsibility assign something (to somebody) The two large classrooms have been assigned to us. The teacher assigned a different task to each of the children. assign somebody something We have been assigned the two large classrooms. The teacher assigned each of the children a different task.
assignment: 1 n an undertaking that you have been assigned to do (as by an instructor) Types: show 6 types... hide 6 types... school assignment , schoolwork a school task performed by a student to satisfy the teacher writing assignment , written assignment an assignment to write something classroom project a school task requiring considerable ...
assign: 1 v select something or someone for a specific purpose "The teacher assigned him to lead his classmates in the exercise" Synonyms: set apart , specify Types: dedicate set apart to sacred uses with solemn rites, of a church detail assign to a specific task Type of: choose , pick out , select , take pick out, select, or choose from a ...
7 meanings: 1. something that has been assigned, such as a mission or task 2. a position or post to which a person is assigned.... Click for more definitions.
assignment meaning, definition, what is assignment: a piece of work that is given to someone...: Learn more.
assignment - WordReference English dictionary, questions, discussion and forums. All Free. ... something assigned, as a particular task or duty: She completed the assignment and went on to other jobs. a position of responsibility, post of duty, or the like, to which one is appointed: ...
assigned: 1 adj appointed to a post or duty " assigned personnel" " assigned duties" Synonyms: allotted given as a task appointed selected for a job Antonyms: unassigned not assigned
Synonyms for ASSIGNMENT: task, job, duty, project, mission, chore, responsibility, function; Antonyms of ASSIGNMENT: dismissal, discharge, firing, expulsion ...
assign something to something Assign a different colour to each different type of information. assign something sth The painting cannot be assigned an exact date. assign something to somebody (law) to say that your property or rights now belong to somebody else. The agreement assigns copyright to the publisher. She has assigned the lease to her ...
1. : a job or duty that is given to someone : a task someone is required to do. [count] My assignment was to clean the equipment. = They gave me the assignment of cleaning the equipment. The students were given a homework assignment. The reporter's assignment is to interview the candidate. The reporter is here on an assignment.
What does the adjective assigned mean? There are three meanings listed in OED's entry for the adjective assigned. See 'Meaning & use' for definitions, usage, and quotation evidence. Entry status. OED is undergoing a continuous programme of revision to modernize and improve definitions. ... assignment, n. 1393- assignor, n ...
What this handout is about. The first step in any successful college writing venture is reading the assignment. While this sounds like a simple task, it can be a tough one. This handout will help you unravel your assignment and begin to craft an effective response. Much of the following advice will involve translating typical assignment terms ...
ASSIGNMENT meaning: 1. a piece of work given to someone, typically as part of their studies or job: 2. a job that…. Learn more.
ASSIGN meaning: 1. to give a particular job or piece of work to someone: 2. If you assign a time for a job or…. Learn more.
App distribution works best when the app is assigned before devices are configured or given to users. After the user receives the device and completes the Setup Assistant, Apple Business Manager can send the user an invitation by email or push notification. ... With device-based app assignment, there's no invitation process or requirement to ...
Recent innovations in single-cell RNA-sequencing (scRNA-seq) provide the technology to investigate biological questions at cellular resolution. Pooling cells from multiple individuals has become a common strategy, and droplets can subsequently be assigned to a specific individual by leveraging their inherent genetic differences. An implicit challenge with scRNA-seq is the occurrence of ...
Background: Accurate assignment of breed origin of alleles at a heterozygote locus may help to introduce a resilient or adaptive haplotype in crossbreeding. In this study, we developed and tested a method to assign breed of origin for individual alleles in crossbred dairy cattle. After generations of mating within and between local breeds as well as the importation of exotic bulls, five rounds ...
ASSIGN definition: 1. to give a particular job or piece of work to someone: 2. If you assign a time for a job or…. Learn more.
"Those assignments, I mean, it's all against some of the best players," Jackson-Davis told ESPN. "I know I'll have to lean on my veterans -- Draymond, Kevon. Those guys are really going to help me.
ASSIGNMENT definition: a piece of work or job that you are given to do: . Learn more.