Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals

Bioinformatics articles from across Nature Portfolio
Bioinformatics is a field of study that uses computation to extract knowledge from biological data. It includes the collection, storage, retrieval, manipulation and modelling of data for analysis, visualization or prediction through the development of algorithms and software.
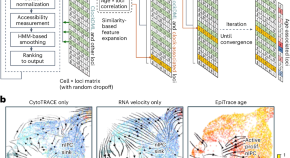
Decoding cell replicational age from single-cell ATAC-seq data
The replicational age of single cells provides a temporal reference for tracking cell fate transition trajectories. The computational framework EpiTrace measures cell age using single-cell ATAC-seq data, specifically by considering chromatin accessibility at clock-like genomic loci, enabling the reconstruction of the history of developmental and pathological processes.
Latest Research and Reviews
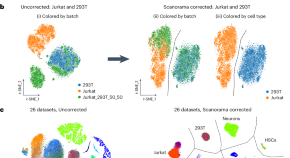
Scanorama: integrating large and diverse single-cell transcriptomic datasets
Scanorama is an effective tool for combining multiple single-cell RNA sequencing datasets, addressing technical variation introduced by differences in sample preparation, sequencing depth and experimental batches that can confound the analysis of diverse datasets.
- Brian L. Hie
- Bonnie Berger
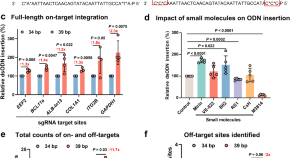
OliTag-seq enhances in cellulo detection of CRISPR-Cas9 off-targets
OliTag-seq, a specific and reproducible in-cellulo assay for CRISPR/Cas9 off-target analysis, can improve site cleavage efficiency and the identification of off-target sites.
- Zhi-Xue Yang
- Dong-Hao Deng
- Xiao-Bing Zhang
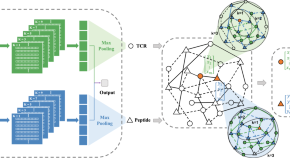
HeteroTCR: A heterogeneous graph neural network-based method for predicting peptide-TCR interaction
HeteroTCR extracts information on within-type (TCR-TCR or peptide-peptide) similarity and between-type (peptide-TCR) interaction from peptides and TCR input. It is robust across diverse datasets, affirming its potential in immunological applications.
- Mengnan Jiang
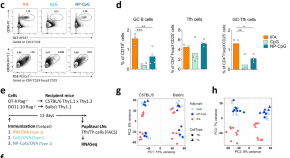
Specialized Tfh cell subsets driving type-1 and type-2 humoral responses in lymphoid tissue
- Saumya Kumar
- Afonso P. Basto
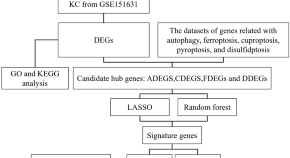
Bioinformatics analysis of signature genes related to cell death in keratoconus
- Jinghua Liu
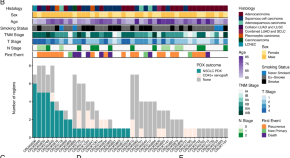
Representation of genomic intratumor heterogeneity in multi-region non-small cell lung cancer patient-derived xenograft models
Patient-derived xenografts are important tools for cancer drug development. Here, the authors develop models from 22 non-small cell lung cancer patients. They show genomic differences between models created from different spatial regions of tumours and a bottleneck on model establishment.
- Robert E. Hynds
- Ariana Huebner
- Charles Swanton
News and Comment

AI assistance for planning cancer treatment
Armed with the right data, advances in machine learning could help oncologists to home in quickly on the best treatment strategies for their patients.
- Michael Eisenstein
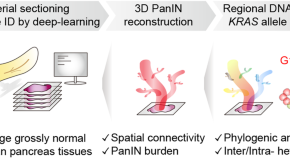
Illuminating the path to pancreatic cancer
- Hiroyuki Kato
- Nabeel Bardeesy
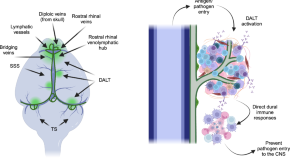
DALT: the brain’s border patrol
- Jang Hyun Park
- Jenolyn F. Alexander
- Jonathan Kipnis
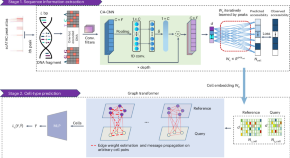
Annotating cell types in single-cell ATAC data via the guidance of the underlying DNA sequences
SANGO efficiently removed batch effects between the query and reference single-cell ATAC signals through the underlying genome sequences, to enable cell type assignment according to the reference data. The method achieved superior performance on diverse datasets and could detect unknown tumor cells, providing valuable functional biological signals.
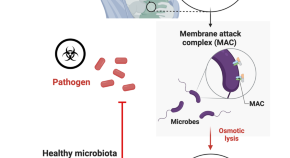
Complement(ing) the microbiome in infants through breastmilk
- Samuel P. Nobs
- Eran Elinav
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Bioinformatics: new tools and applications in life science and personalized medicine
Affiliations.
- 1 Centro de Investigação de Montanha (CIMO), Instituto Politécnico de Bragança, Campus de Santa Apolónia, 5300-253, Bragança, Portugal.
- 2 Centro de Investigação de Montanha (CIMO), Instituto Politécnico de Bragança, Campus de Santa Apolónia, 5300-253, Bragança, Portugal. [email protected].
- PMID: 33404829
- DOI: 10.1007/s00253-020-11056-2
While we have a basic understanding of the functioning of the gene when coding sequences of specific proteins, we feel the lack of information on the role that DNA has on specific diseases or functions of thousands of proteins that are produced. Bioinformatics combines the methods used in the collection, storage, identification, analysis, and correlation of this huge and complex information. All this work produces an "ocean" of information that can only be "sailed" with the help of computerized methods. The goal is to provide scientists with the right means to explain normal biological processes, dysfunctions of these processes which give rise to disease and approaches that allow the discovery of new medical cures. Recently, sequencing platforms, a large scale of genomes and transcriptomes, have created new challenges not only to the genomics but especially for bioinformatics. The intent of this article is to compile a list of tools and information resources used by scientists to treat information from the massive sequencing of recent platforms to new generations and the applications of this information in different areas of life sciences including medicine. KEY POINTS: • Biological data mining • Omic approaches • From genotype to phenotype.
Keywords: Applications; Bioinformatics; Life science; Personalized medicine; Sequencing; Tools.
Publication types
- Computational Biology*
- Data Mining
- Precision Medicine*
- Open access
- Published: 15 July 2022
Bioinformatics approaches and applications in plant biotechnology
- Yung Cheng Tan 1 ,
- Asqwin Uthaya Kumar ORCID: orcid.org/0000-0002-8785-6260 1 , 2 ,
- Ying Pei Wong 1 &
- Anna Pick Kiong Ling ORCID: orcid.org/0000-0003-0930-0619 1
Journal of Genetic Engineering and Biotechnology volume 20 , Article number: 106 ( 2022 ) Cite this article
11k Accesses
4 Citations
Metrics details
In recent years, major advance in molecular biology and genomic technologies have led to an exponential growth in biological information. As the deluge of genomic information, there is a parallel growth in the demands of tools in the storage and management of data, and the development of software for analysis, visualization, modelling, and prediction of large data set.
Particularly in plant biotechnology, the amount of information has multiplied exponentially with a large number of databases available from many individual plant species. Efficient bioinformatics tools and methodologies are also developed to allow rapid genome sequence and the study of plant genome in the ‘omics’ approach. This review focuses on the various bioinformatic applications in plant biotechnology, and their advantages in improving the outcome in agriculture. The challenges or limitations faced in plant biotechnology in the aspect of bioinformatics approach that explained the low progression in plant genomics than in animal genomics are also reviewed and assessed.
There is a critical need for effective bioinformatic tools, which are able to provide longer reads with unbiased coverage in order to overcome the complexity of the plant’s genome. The advancement in bioinformatics is not only beneficial to the field of plant biotechnology and agriculture sectors, but will also contribute enormously to the future of humanity.
Over the past decades, the term ‘bioinformatics’ has become a buzzword in all areas of research in biological science. With the continuous development and advancement in molecular biology, the explosive growth of biological information required a more organized, computerized system to collect, store, manage, and analyse the vast amount of biological data generated in the experiments from all fields [ 1 ]. Bioinformatics, as a new emerging interdisciplinary field for the past few decades, has many tools and techniques that are essential for efficient sorting and organizing of biological data into databases [ 1 , 2 ]. Bioinformatics can be referred as a computer-based scientific field which applies mathematics, biology, and computer science to form into a single discipline for the analyses and interpretation of genomics and proteomics data [ 2 , 3 ]. In short, the main components of bioinformatics are (a) the collection and analysis of database and (b) the development of software tools and algorithm as a tool for interpretation of biological data [ 2 ]. Bioinformatics played a crucial role in many areas of biology as its applications provide various types of data, including nucleotide and amino acid sequences, protein domains and structure as well as expression patterns from various organisms [ 3 ]. Similarly, the field of plant biotechnology has also taken advantages of bioinformatics, which provides full genomic information of various plant species to allow for efficient exploration into plants as biological resource to humans [ 1 , 3 , 4 ]. The intention of this article is to describe some of the key concepts, tools, and its applications in bioinformatics that are relevant to plant biotechnologies. The current challenges and limitations for improvement and continuous development of bioinformatics in plant science are also described.
Applications of bioinformatics in plant biotechnology
The introduction of bioinformatics and computational biology into the area of plant biology is drastically accelerating scientific invention in life science. With the aid of sequencing technology, scientists in plant biology have revealed the genetic architecture of various plant and microorganism species, such as proteome, transcriptome, metabolome, and even their metabolic pathway [ 1 ]. Sequence analysis is the most fundamental approach to obtain the whole genome sequence such as DNA, RNA, and protein sequence from an organism’s genome in modern science. The sequencing of whole genome permits the determination of organization of different species and provides a starting point to understand their functionality. A complete sequence data consists of coding and non-coding regions, which can act as a necessary precursor for any functional gene that determines the unique traits possessed by organisms. The resulting sequence includes all regions such as exons, introns, regulator, and promoter, which often leads to a vastly large amount of genome information [ 5 ]. With the emergence of next-generation sequencing (NGS) and some other omics technologies used to examine plants genomics, more and more sequenced plants genome will be revealed [ 1 , 6 , 7 , 8 ]. To deal with these vast amounts of data, the development and implementation of bioinformatics allow scientists to capture, store, and organize them in a systematic database [ 1 , 5 ].
Bioinformatics databases and tools for plant biotechnology
In the field of bioinformatics, there are a variety of options of databases and tools that are available to perform analysis related to plant biotechnology. Next-generation sequencing (NGS) and bioinformatics analysis on the plant genomes over the years have generated a large amount of data. All these data are submitted to various and multiple databases that are publicly available online. Each database is unique and has its focus. For instance, CottonGen, database is solely dedicated to obtaining genomics and breeding information of any cotton species of interest [ 9 ]. The establishment of such database eases the researchers who are working on cotton genomic studies by focussing on using just one database instead of searching through other available databases. However, some databases are established and designed to cater not only to one specific species or genus, but focus on all the plant species, such as the National Center for Biotechnology Information (NCBI) ( https://www.ncbi.nlm.nih.gov/ ) database, which as of 2021 possesses almost 21,000 plant genomes that are available for access [ 10 ]. Such a database is useful for studies that do not focus on one specific genus or species. This eases the researchers in accessing to all kinds of genomic data in one database. This section will briefly discuss some of the available plant genome databases, which are publicly accessible and not designated for one genus or species alone.
First would be the globally known and recognized database by all the researchers and biologists, which is the NCBI database. NCBI has been dedicated for gathering and analysing information about molecular biology, biochemistry, and genetics. In the NCBI database, one can download the genome information of the plant species of interest from either gene expression omnibus (GEO) ( https://www.ncbi.nlm.nih.gov/geo/ ) or sequence read archive (SRA) ( https://www.ncbi.nlm.nih.gov/sra ) by simply stating the scientific name of the plant in the search bar and the entire genomic information of the plant can then be obtained. The GEO and SRA comprise processed or raw gene expression data or RNA sequencing of plants that are reposited in the repository. For instance, to obtain the genomics of Rosa chinensis (Rose plant), by inputting the name in the search bar, it will direct to the search result page where the researcher can select the most recent or suitable datasets with specific accession number. Depending on the profiling platform used in each dataset, researchers could retrieve either gene symbols, Ensemble ID, open reading frame, chromosomal location, regulatory elements, etc. The information allows researcher to further analyse the subject of study using bioinformatics tools such as gene ontology ( http://geneontology.org/ ), Database for Annotation, Visualization and integration Discovery (DAVID) ( https://david.ncifcrf.gov/ ), Basic Local Alignment Search Tool (BLAST) ( https://blast.ncbi.nlm.nih.gov/Blast.cgi ), and others that is relevant for the study.
Another database that is available for accessing plant genome database is EnsemblPlants ( https://plants.ensembl.org/index.html ). Unlike the NCBI database, which is not only dedicated to plant genomes, EnsemblPlants is specifically dedicated to accessing plant genomes. EnsemblPlant is part of the Ensembl project that started in 1999, where the project aimed to automatically annotate the genome and integrate the outcome of the annotation with other publicly available biological data and establish an open access archive or database online for the use of the research community [ 11 ]. Ensembl project later launched the taxonomic specific websites designated for each taxon under their project that also includes the plants. The database is a user-friendly integrative platform, where it is continuously updated with the new addition of plant species every time a plant genome is completely sequenced. Compared to the NCBI database mentioned earlier, EnsemblPlant not only provides genome sequence, gene models, and functional annotation of the plant species of interest, but also includes the polymorphic loci, population structure, genotype, linkage, and phenotype information [ 11 , 12 ]. Unlike, NCBI, EnsemblPlant does also provide comparative genomics data of the plant species of interest. This indicates that the platform does not only offer genome sequence data but provide additional analytical data about the plant species of interest and help the researchers who are working on plant bioinformatics to save a lot of time by reducing the tedious work in running the analysis. Yet, the researchers could re-assess the data if necessary, depending on the stringency of their work.
Aside from the abovementioned databases that are widely used for retrieving plant genome sequence, there are still other plant databases such as PlantGDB, MaizeDIG, and Phytozome that can also be considered. Table 1 lists the available database and tools that are widely applied in plant biotechnology.
Biotechnology and bioinformatics for plant breeding
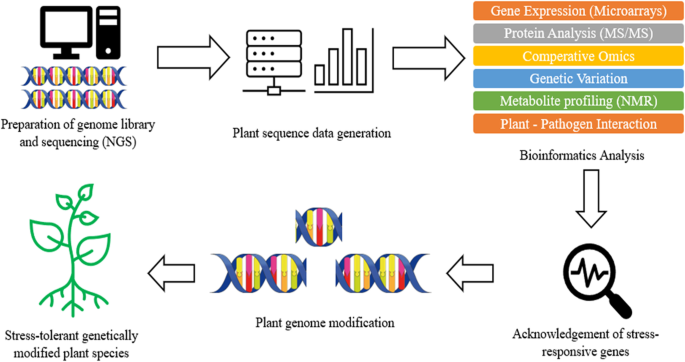
Plant breeding can be defined as the changing or improvement of desired traits in plants to produce improved new crop cultivars for the benefits of humankind [ 8 ]. Jhansi and Usha [ 13 ] mentioned a few benefits brought by genetically engineered plants such as improved quality, enhanced nutritional value, and maximized yield. The revolution of life science in molecular biology and genomics has enabled the leaps forward in plant breeding by applying the knowledge and biological data obtained in genomics research on crops [ 6 , 8 , 13 ]. In modern agriculture, transgenic technology on plants refers to genetic modification, which is done on plants or crops by altering or introducing foreign genes into the plant, to make them useful and productive and enhance their characteristic [ 13 , 14 ]. As mentioned above, the evolution of next-generation sequencing (NGS) and other sequencing technologies produces a large size of biological data which require databases to store the information. The accessibility of whole genome sequences in databases allows free association across genomes with respect to gene sequence, putative function, or genetic map position. With the aid of software, it is possible to formulate predictive hypothesis and incorporate the desired phenotypes from a complex combination into plants by looking at those genetic markers which score well and gives a higher reliability in breeding [ 2 , 15 ]. Other than genome sequence information, databases which store the information of metabolites also play a crucial role in the study of interaction with proteomics and genomics to reflect the changes in phenotype and specific function of an organism [ 1 ]. Some of the most widely used metabolomics databases for plants and crops such as Metlin ( http://metlin.scripps.edu ), provides multiple metabolite searching and about 240,000 metabolites, nearly 72,000 high-resolution MS/MS spectra, and PlantCyc ( https://plantcyc.org/ ), a database which stores information about biochemical pathway and their catalytic enzyme and genes from plants [ 1 , 16 ]. Moreover, single-nucleotide polymorphism markers also benefit from the revolution of NGS and other sequencing technologies. By using NGS, RNA sequencing (RNA-seq) allows direct measure of mRNA profile in order to identify known single-nucleotide polymorphism (SNP) [ 1 ]. SNP is the unique allelic variation within a genome of same species, which can be used as biological markers to locate the genes associated with desired traits in plants [ 17 , 18 ]. Besides, transcriptome resequencing using NGS allows rapid and inexpensive SNP discovery within a large, complex gene with highly repetitive regions of a genome such as wheat, maize, sugarcane, avocado, and black currant [ 17 ]. Figure 1 illustrates briefly the process involved in plant breeding using NGS and bioinformatics.
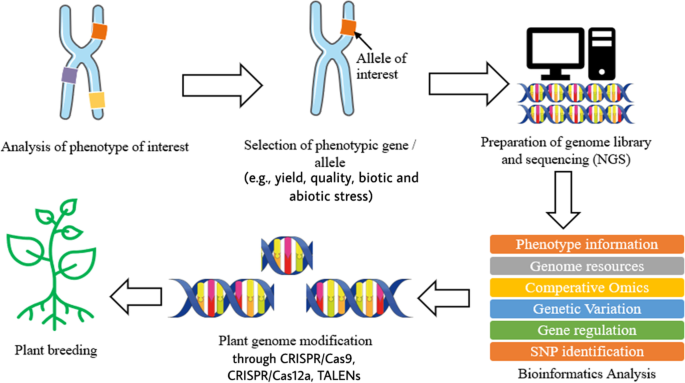
Brief process of plant breeding involving NGS and bioinformatics
Ever since the first transgenic rice production in 2000, there has been a significant revolution in crop genome sequencing projects, along with the advancement in technologies, rapidly increasing the pace in genetically modified organism (GMO) [ 2 , 13 , 19 ]. Among all the products in rice biotechnology, one of the most widely known GM rice is golden rice. Golden rice is a variety of rice engineered by introducing the biosynthetic pathway to produce β-carotene (pro-vitamin A) into staple food in order to resolve vitamin A deficiency. The World Health Organization has classified vitamin A deficiency as public health problem as it causes half a million of children to childhood blindness [ 13 ]. Vitamin A is an essential nutrient to humans as it helps with development of vision, growth, cellular differentiation, and proliferation of immune system; insufficient intake of vitamin A may lead to childhood blindness, anaemia, and reduced immune responsiveness against infection [ 20 ]. Being the first crop genome to be sequenced, rice has become the most suitable model to initiate the development and improvement of other species in genomic aspect [ 21 , 22 , 23 , 24 ]. The particular reason is due to its small genome size and diploidy, which enables rice to be an excellent model for other cereal crops with larger genomes, such as maize and wheat [ 21 , 23 ]. Song et al. [ 22 ] reported the complete genome sequence of two rice subspecies, japonica and indica , in 2005 that laid a strong foundation for molecular studies and plant breeding research [ 22 , 24 ]. With recent advancement in bioinformatics, it is now possible to run the sequence alignment between large and complex genome from other crop species with genomic data available from rice, by using different software or tools, in order to find out the shared conserved sequence through comparative genomics [ 2 , 7 ]. Vassilev et al. stated some of the most commonly used programmes such as BLAST and FASTA format allowed rapid sequence searching in databases and give the best possible alignment to each sequence [ 25 ]. The programming algorithm calculates the alignment score to measure the proportion of homology matching residue between sequence from related species [ 2 ].
Wheat, as the most widely grown consumed crops, together with rice and maize contributes more than 60% of the calories and protein for our daily life [ 26 , 27 ]. To meet the demands of human population growth, it is necessary to achieve more understanding in wheat research and breeding in order to accelerate the production of wheat yield by 2050 [ 26 , 27 , 28 ]. Despite its importance, the improvement of wheat has been challenging as the researchers have to overcome the complexity of the wheat genome such as highly repetitive and large polyploid in order to get a fully sequenced reference genome [ 26 , 29 ]. Advances in next-generation sequencing (NGS) platforms and other bioinformatics tools have revealed the extensive structural rearrangements and complex gene content in wheat, which revolutionized wheat genomics with the improvement of wheat yield and its adaptation to diversed environments [ 26 , 29 ]. The NGS platforms allow the swift detection of DNA markers from the huge genome data in a short period of time. These NGS-based approaches have undoubtedly revolutionized the allele discovery and genotype-by-sequencing (GBS). By providing a high-quality reference genome of wheat in databases, it allows more sequence comparison between wheat and other species to find out more homologous gene. Moreover, the development of sequencing technologies in both high-throughput genotyping and read length, combining with biological databases, allow the rapid development of novel algorithm to complex wheat genome [ 29 , 30 ]. For instance, genome-wide association studies (GWAS) are an approach used in genome research which allows rapid screening of raw data to select specific regions with agronomic traits [ 29 , 31 ]. It allows multiple genetic variants across genome to be tested to study the genotype-phenotype association; thus, this method can be used to facilitate improvement in crop breeding via genomic selection and genetic modification [ 16 , 29 ].
Maize, a globally important crop, not only has a wide variety of uses in terms of economic impact, but can also serve as genetic model species in genotype to phenotype relationship in plant genomic studies [ 32 , 33 ]. Besides, due to its extremely high level of gene diversity, maize has high potential in the improvement of yield to meet the demands of population growth [ 33 ]. Despite the combination of economic and genomic impact, the progress in generating a whole genome sequence in maize has been a computational challenge due to the presence of tremendous structural variation (SV) in its genome [ 34 ]. The introduction of NGS techniques in several crops including maize allowed the rapid de novo genome sequencing and production of huge amount genomics and phenomics information [ 1 , 35 ]. A better integration of data within multiple genome assemblies is much needed to study the connection between phenotype and genotype in order to achieve yield and quality improvement of maize [ 35 ]. Nowadays, some user-friendly online databases such as qTeller, MaizeDIG, and MaizeMine are designed to ease the comparison and visualization of relationships between genotypes and phenotypes [ 36 ]. MaizeGDB, a model organism database for maize, provides the access of data on genes, alleles, molecular markers, metabolic pathway information, phenotypic images with description, and more which are useful for maize research [ 35 , 36 ]. MaizeMine is a data mining resource under MaizeGDB, which was designed to accelerate the genomics analysis by allowing the researchers to better script their own research data in downstream analysis [ 36 ] whereas MaizeDIG is a genotype-phenotype database which allows the users to link the association of genotype with phenotype expressed by image [ 35 , 36 ]. Cho et al. [ 35 ] reported that with the accessibility via image search tool, the relationship between a gene and its phenotype features can be visualized within image. The integration and visualization of high-quality data with these tools enables quick prioritizing phenotype of interest in crops, which play a crucial role in the improvement of plant breeding.
Bioinformatics for studying stress resistance in plants
The understanding of the stress response on plants is vital for the improvement of breeding efforts in agriculture, and to predict the fate of natural plants under abiotic change especially in the current era of continuous climate change [ 37 ]. Stress response in plants can be divided into biotic and abiotic. Biotic stress mainly refers to negative influence caused by living organism such as virus, fungi, bacteria, insects, nematodes, and weeds [ 38 ] while abiotic stress refers to factors such as extreme temperature, drought, flood, salinity, and radiation which dramatically affect the crop yield [ 37 ]. NGS technologies and other potent computational tools, which allowed sequencing of whole genome and transcriptome, have led to the extensive studies of plants towards stress response on a molecular basis [ 1 , 2 , 37 ]. The tremendous amount of plant genome data obtained from genome sequencing allows the investigation of correlations between the molecular backbone of living organism and their adaptations towards the environment [ 16 ].
Biotic and abiotic stress management
How the plants and crops respond towards stress environment is the key to ensure their growth and development, and to avoid the great crop yield penalty caused by harsh condition [ 35 , 39 ]. Therefore, the utilization of bioinformatic tools is important to study and analyse the plant transcriptome in response to biotic and abiotic stress. Besides, the application of bioinformatics tools on plants and crops genome can benefit the agricultural community by searching the desired gene among genome from different species and elucidate their function on the crops [ 35 ]. The genome databases play a crucial role in storing and mining large and complex genome sequence from the plants. Besides data storage, some genome databases are also able to perform gene expression profiling to predict the pattern of gene expressed at the level of transcript in cell or tissues. By using in silico genomic technologies, the disease resistance gene-enzyme with their respective transcription factor, which plays a role in defence mechanism against stress, are able to be identified [ 40 , 41 ]. For instance, a large-scale transcriptome sequencing of chrysanthemum plants was carried out by Xu et al. [ 40 ] to study the dehydration stress in chrysanthemum plants. An online database called Chrysanthemum Transcriptome Database ( http://www.icugi.org/chrysanthemum ) was developed to allow the storage and distribution of transcriptome sequence and its analysis result among research community [ 40 ]. With the aid of different protein databases, the biochemical pathway and kinase activity of chrysanthemum in response to dehydration stress are able to be predicted [ 40 ]. Xu et al. [ 40 ] also reported a total of 306 transcription factor and 228 protein kinase that are important upstream regulator in plants when encountered with various biotic and abiotic stresses.
Bioinformatics approaches to study resistance to plant pathogen
One of the challenges in modern agriculture to supply the nutrition’s demand along with the world population growth is the crop loss due to disease. The study of plant pathogen plays an essential role in the study of plant diseases, including pathogen identification, disease aetiology, disease resistance, and economic impact, among others [ 41 ]. Plants protect themselves through a complex defence system against variety of pathogen, including insects, bacteria, fungi, and viruses. Plant-pathogen interaction is a multicomponent system mediated by the detection of pathogen-derived molecules in the form of protein, sugar, and polysaccharide, by pattern recognition receptor (PRRs) within the plants [ 42 , 43 , 44 , 45 ]. After the recognition of enemy molecules, signal transduction is carried out accordingly and plant immune systems will respond defensively through different pathways involving different genes [ 42 ]. According to Schneider et al. [ 46 ], the development of molecular plant pathology can be broadly divided into three eras, begins with the disease physiology starting from early 1900s until 1980s [ 46 ]. In the second era of molecular plant genetic studies, one or a few genes of bacterial pathogens were focused whereas the third era of plant genomic studies began in 2000 with the sequencing of genome, and the first complete genome of bacterial pathogen, Xylella fastidiosa , was obtained [ 46 ]. The recent advance in DNA sequence technologies allow researchers to study the immune system of plants on genomic and transcriptomics level [ 1 , 41 , 42 ]. Genomics has revealed the mystery and complexity and consequently the various information about phytopathogen. A clearer picture of plant-pathogen interactions in the context of transcriptomic and proteomics can be visualized through the application of different bioinformatics tools, which in turn made feasible the engineering resistance to microbial pathogen in plant [ 43 ].
PRGdb: bioinformatics web for plant pathogen resistance gene analysis
Plants have developed a wide range of defence mechanism against different pathogen and ultimately inhibit growth and spread of pathogen [ 47 , 48 ]. Plant defence system is mediated by resistance (R) gene [ 47 ]. R gene plays an important role in defence mechanism. They encode for protein that recognizes specific avirulent (Avr) pathogen proteins and initiated the defence mechanism through one or more signal transduction pathway in a hypersensitive response (HR) [ 41 , 47 , 48 ]. However, the essential components needed for protein to exert their resistance are still unidentified [ 48 ]. With the intention to study and identify more novel R gene, high-throughput genomic experiments and plant genomic sequence are essential to explore their function and new R gene discovery [ 47 ]. In 2009, Plant Disease Resistance Gene database (PRGdb), a comprehensive bioinformatics resource across hundreds of plant species, was launched in order to facilitate the plant genome research on discovery and predict plant disease resistance gene [ 47 , 48 ]. To date, PRGdb 3.0 has been released with 153 reference resistance genes and 177,072 annotated candidate pathogen receptor genes (PRGs) [ 49 ]. This database act as an important reference site and repository to all the research studies on exploration and use of plant resistance genes [ 48 , 49 ].
Apart from resistance gene storage, this easily accessible platform also allows different tools that are essential for exploration and discovery of novel R gene. For instance, the DRAGO 2.0 tool, which was built to explore known and novel disease resistance gene, can be launched on any transcriptome or proteome to annotate and predict PRG from DNA or amino acid with high accuracy [ 49 ]. Besides, BLAST search tools available in PRGdb provide comparison of different sequences which allowed the determination of gene homology and expression analysis. Apart from the database, plant pathology field also benefited from whole genome sequence technologies. The new DNA sequencing technologies such as NGS and Sanger sequencing allowed the study of genomics, proteomics, metabolomics, and transcriptomics on both the host plant and the pathogen [ 1 ]. The phytopathogen genomes which have been sequenced are expected to provide valuable information on the molecular basis for infection of plant host and explore the potential novel virulence factors [ 1 ]. Figure 2 illustrates a brief process involved in producing stress-resistant plant using bioinformatics approach.
Brief process involved in producing stress-resistant plant using bioinformatics approach
Metagenomics in plant biotechnology and Cas9 modification
The effects of environment microorganisms’ community, especially soil microorganism on plants, may contribute to plant’s growth and pathogenesis. Through metagenomics approaches, the soil microorganism community that contributed to plant growth may provide a great genomic insight into physiology and pathology [ 50 , 51 , 52 , 53 ]. In metagenomics approaches, the overall genetic materials obtained from soil are sequenced and advancing to microbial community analysis via data analytics [ 53 , 54 , 55 ]. The extracted genetic materials from the soil were subjected to high-throughput metagenomics analysis via various NGS approaches such as 16S rRNA sequencing, shotgun metagenomic sequencing, MiSeq sequencing [ 54 , 55 , 56 ] for microbial species identification, functional genomics study, and structural metagenomic analysis. A NGS produces huge genomics data for each study; thus, application of bioinformatics tools would add value in the metagenomics analysis as the target genes identified could advance into elucidation of plant growth, plant disease, soil contamination, and microbial taxonomy [ 52 ]. For example, the use of UNITE ( https://unite.ut.ee/ ) for fungi identification [ 57 ], SILVA ( https://www.arb-silva.de/ ) for 16S rRNA [ 58 ], and MGnify ( https://www.ebi.ac.uk/metagenomics/ ) possesses metagenomics data of microbiome [ 59 ]. These databases allow the researchers to retrieve and analyse the relevant metagenomic sequenced data for a specific study.
Since metagenomics analysis provides the greater output on plant-microbe interaction, the genes that are responsible for plant immunity may play a crucial role in protecting against disease-causing microorganism [ 60 , 61 ]. With the emergence of Clustered Regularly Interspaced Short Palindrome Repeats (CRISPR) gene editing technique, Cas9 modification could produce a better plant trait and disease-resistant plant [ 62 , 63 ]. The CRISPR/Cas9 system is employed in studying the functional genomics in plants in relation to plant-microbe interaction. CRISPR/Cas9 system facilitated the gene editing by creating a mutant through double-stranded break forming a targeted gene mutation and followed by genome repair [ 63 , 64 , 65 ]. The CRISPR/Cas9 modification on OsSWEET14 genes protects the Super Basmati Rice from bacterial blight causes by Xanthomonas oryzae pv. oryzae [ 66 ]. Gene editing to knockout OsMPK5 and OsERF922 genes in rice protects against Magnaporthe grisea and Magnaporthe oryzae , respectively [ 67 , 68 , 69 ]. Besides that, Cas9 modification on Cs WRKY22 and TcNPR3 increased host defence immunity through regulating salicylic acid in Citrus sinensis and Theobroma cacao , respectively [ 70 , 71 ]. Thus, CRISPR/Cas9 modification could be one of important science advancements to validate the metagenomics analysis on plant-microbe interaction.
Current challenges of bioinformatics applications in plant biotechnology
Despite the beneficial prospect of the bioinformatics applied in plant biotechnology, there are many challenges and limitations must be addressed in order to fully utilize their potentials [ 1 ]. Along with the rapid growth in plant genome data mining and database development, there are a few challenges faced by bioinformaticians and scientists which can be divided into number of areas as mentioned in the subsections below.
Bioinformatic data management and organization and synchronize update resources
Since the introduction of the next-generation sequencing (NGS), which is commercially available in 2004, enormous amount of data has been generated in plant genome research. Thousands of Gb of plants sequences are deposited in various public databases monthly [ 1 , 72 , 73 ]. Moreover, the constantly sequenced and re-sequenced of the plant genome has developed a vast amount of new genome sequence in all public databases. The increase in sequenced plant genome driven by technological improvement has led to a problem that arises along with the storage and update of a large amount of data [ 72 , 74 ]. The update process should occur in all the comparative databases, not just solely individual genome database [ 72 ]. With this, the synchronized update of genome data resources among different plant genomic platform is able to provide a strong, updated, reliable database community that all the plant researchers can rely on [ 72 ].
Complexity of plant genetic content
Other than the tremendous amount of genome sequence generated, the complexity of the plant genetic content is also a challenging issue faced by plant research community. Even though the arrival of next-generation sequencing technologies has allowed the rapid DNA sequencing for non-model or orphan plant species, the sequencing pace for plants is far from that of animal and microorganism [ 74 ]. The main factor which contributes to this situation is because sometimes the plant genome can be nearly hundred times larger than the currently sequenced animal and microorganism genome [ 73 ]. Needless to say, some of the plant genome even can have polyploidy, a duplication of an entire genome, which is estimated to occur in 80% of the plant species [ 73 , 75 ]. According to Schatz et al., the genome assembly in the case of large size plant genome with abundance of repetitive sequence can be metaphorically described as build-up of a large puzzle consisting of blue sky separated by nearly indistinguishable white clouds of small gene [ 73 ]. The particular reason for this is mainly because the sequence length in NGS is relatively shorter than in Sanger sequencing and required dedicated assembly algorithm [ 74 ]. Therefore, most plant genomes sequenced by NGS can only be used for establishing gene catalogues, interpreting the repeat content, glimpsing evolutionary mechanism, and performing on comparative genomics in early study [ 74 ].
Advance in sequencing technologies
There are two basic approaches to genome assembly, i.e. comparative genome assembly and de novo genome assembly [ 75 ]. It is important to distinguish between these two different approaches. Comparative is a reference-guided method which use a genome or transcriptome, or both, for guidance, whereas de novo assembly refers to reconstruction of a genome from organisms that have not been sequenced before [ 74 , 75 ]. Table 2 compares some of the available assembly and NGS technology available for genome sequencing. However, these two approaches are not completely exclusive due to a lack of bioinformatic tools designed to cope with the unique and challenging features of plant genomes [ 74 , 75 ]. One of the biggest challenges in the development of bioinformatic software is the algorithm development [ 76 ]. As is known, all the programmes or software in bioinformatic are very computationally intensive. As most of the assemblies available now solely rely on single assembly, a development in better algorithm in terms of resource requirement is essential for combining different assemblers by using a different underlying algorithm in order to give a more credible final assembly [ 74 , 76 ].
Database accessibility
To date, there are about 374,000 known plant species in the world [ 77 ]. The first full plant genome sequencing was completed on A rabidopsis thaliana through Sanger sequencing methods in 2000 [ 78 ]. Although introduction of molecular biology decades ago may have facilitated the species identification, obtaining the full plant genomic data remains challenging due to the genome complexity. The development of NGS platform may foster the plant genome sequencing, yet there are limited sequenced datasets reposited to the database. To date, there are only 29 plant genome databases accessible in PlantGDB genome browser allowing researchers to retrieve the information about gene structure, matched GSS contigs, similar protein, spliced alignments EST, etc. Besides, the PlaD database ( http://systbio.cau.edu.cn/plad/index.php ) that focuses on the microarray data of the plants developed by China Agricultural University comprises transcriptomic database for plant defence against pathogen. However, it is limited to Arabidopsis , rice, maize, and wheat [ 79 ]. The Plant Omics Data Center ( http://plantomics.mind.meiji.ac.jp/podc/ ) is another publicly available web-based plant database featuring omics data for co-expressed profile, regulatory network, and plant ontology information [ 80 ]. Although curated omics datasets could be retrieved from PODC, information are restricted for certain plants and crops such as Arabidopsis , tobacco, earthmoss, barrelclover, soybean, potato, rice, tomato, grape, maize, and sorghum. Furthermore, all these publicly available databases require constant updating with new released data or resequencing data so that the researcher could obtain the most updated version of genome datasets for their research.
The application of bioinformatics in plant biotechnology represents a fundamental shift in the way scientists study living organisms. Bioinformatics play a significant role in the development of agriculture sector as it helps to study the stress resistance and plant pathogen, which are critical in advancing crop breeding [ 75 ]. NGS and other sequencing technologies will make more plant genome data accessible in all public databases and enable the identification of genomic variants and prediction of protein structure and function [ 75 , 76 ]. Moreover, GWAS, which allows the identification of loci and allelic variation related to valuable traits, eased the crop modification and improvement [ 74 ]. In brief, the advance in bioinformatics application in plant biotechnology enables researchers to achieve fundamental and systematic understanding of economically important plant. However, despite all these exciting achievement by the application of bioinformatic on plant biotechnology, it is still a long way from automated full genome sequencing and assembly at a low cost [ 76 ]. There is a critical need for effective bioinformatic tools which are able to provide longer reads with unbiased coverage in order to overcome the complexity of the plant’s genome. To achieve this, an enhanced algorithm development is essential to enable data mining and analysis, comparison, and so on. Therefore, bioinformaticians and experts with mathematical and programming skills will play an important role in bringing fresh approaches and knowledge into bioinformatics, not only for the advancement in plant biotechnology and agriculture sector, but the future of humanity as well.
Availability of data and materials
Not applicable.
Abbreviations
Genome-wide association studies
Next-generation sequencing
Plant Disease Resistance Gene database
RNA sequencing
Single-nucleotide polymorphism
Gomez-Casati DF, Busi MV, Barchiesi J, Peralta DA, Hedin N, Bhadauria V (2018) Applications of bioinformatics to plant biotechnology. Curr Issues Mol Biol 27:89–104. https://doi.org/10.21775/cimb.027.089
Article Google Scholar
Zhang SY, Liu SL (2013) Bioinformatics. In: Maloy S, Hughes K (eds) Brenner’s Encyclopedia of Genetics, 2nd edn. Academic Press, London. https://doi.org/10.1016/B978-0-12-374984-0.00155-8
Chapter Google Scholar
Tiwari A, Singh P, Kumawat S (2020) Applications of bioinformatics in plant breeding system. Int J Curr Microbial App Sci. 11:2825–2831
Google Scholar
Rhee SY, Dickerson J, Xu D (2006) Bioinformatics and its applications in plant biology. Annu Rev Plant Biol 57:335–360. https://doi.org/10.1146/annurev.arplant.56.032604.144103
Normand EA, Van den Veyyer IB (2019) Next-generation sequencing for gene panels and clinical exomes. In: Leung PCK, Qiao J (eds) Human Reproductive and Prenatal Genetics, 1st edn. Academic Press, London. https://doi.org/10.1016/B978-0-12-813570-9.00025-5
Blätke MA, Szymanski JJ, Gladilin E, Scholz U, Beier S (2021) Editorial: advances in applied bioinformatics in crops. Front Plant Sci 12:640394. https://doi.org/10.3389/fpls.2021.640394
Kushwaha UKS, Deo I, Jaiswal JP, Prasad B (2017) Role of bioinformatics in crop improvement. Glob J Sci Front Res D Agric Vet 17(1):13–23
Caligari PDS, Brown J (2017) Plant Breeding, Practice. In: Thomas B, Murray BG, Murphy DJ (eds) Encyclopedia of Applied Plant Sciences, 2nd edn. Academic Press, London. https://doi.org/10.1016/B978-0-12-394807-6.00195-7
Yu J, Jung S, Cheng CH, Lee T, Zheng P, Buble K et al (2021) CottonGen: the community database for cotton genomics, genetics, and breeding research. Plants. 10(12):2805. https://doi.org/10.3390/plants10122805
Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC et al (2022) Database resources of the national center for biotechnology information. Nucleic Acids Res 50(D1):D20–D26. https://doi.org/10.1093/nar/gkab1112
Howe KL, Contreras-Moreira B, De Silva N, Maslen G, Akanni W, Allen J et al (2019) Ensembl Genomes 2020 – enabling non-vertebrate genomic research. Nucleic Acids Res 48(D1):D689–D695. https://doi.org/10.1093/nar/gkz890
Bolser D, Staines DM, Pritchard E, Kersey P (2016) Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data. In: Edwards D (ed) Plant Bioinformatics. Methods in Molecular Biology, vol 1374. Humana Press. https://doi.org/10.1007/978-1-4939-3167-5_6
Jhansi Rani S, Usha R (2013) Transgenic plants: Types, benefits, public concerns and future. J Pharm Res 6(8):879–883. https://doi.org/10.1016/j.jopr.2013.08.008
Barragán-Ocaña A, Reyes-Ruiz G, Olmos-Peña S, Gómez-Viquez H (2019) Transgenic crops: trends and dynamics in the world and in Latin America. Transgenic Res 28(3-4):391–399. https://doi.org/10.1007/s11248-019-00123-8
Platten JD, Cobb JN, Zantua RE (2019) Criteria for evaluating molecular markers: Comprehensive quality metrics to improve marker-assisted selection. PLoS One 14(1):e0210529. https://doi.org/10.1371/journal.pone.0210529
Filho HA, Machicao J, Bruno OM (2018) A hierarchical model of metabolic machinery based on the kcore decomposition of plant metabolic networks. PLoS One 13(5):e0195843. https://doi.org/10.1371/journal.pone.0195843
Mammadov J, Aggarwal R, Buyyarapu R, Kumpatla S (2012) SNP markers and their impact on plant breeding. Int J Plant Genomics 728398:1–11. https://doi.org/10.1155/2012/728398
Hoskins RA, Phan AC, Naeemuddin M, Mapa FA, Ruddy DA, Ryan JJ et al (2001) Single nucleotide polymorphism markers for genetics mapping in Drosophila melanogaster . Genome Res 11(6):1100–1113. https://doi.org/10.1101/gr.gr-1780r
Edwards D, Batley J (2010) Plant genome sequencing: applications for crop improvement. Plant Biotechnol J 8(1):2–9. https://doi.org/10.1111/j.1467-7652.2009.00459.x
Tang G, Qin J, Dolnikowski GG, Russell RM, Grusak MA (2009) Golden Rice is an effective source of vitamin A. Am J Clin Nutr 89(6):1776–1783. https://doi.org/10.3945/ajcn.2008.27119
Yu J, Hu S, Wang J, Wong GKS, Li S, Liu B et al (2002) A draft sequence of the rice genome ( Oryza sativa L. ssp. Indica ). Science. 296(5565):79–92. https://doi.org/10.1126/science.1068037
Song S, Tian D, Zhang Z, Hu S, Yu J (2018) Rice genomics: over the past two decades and into the future. Genomics Proteomics Bioinformatics 16(6):397–404. https://doi.org/10.1016/j.gpb.2019.01.001
Jackson SA (2016) Rice: The First Crop Genome. Rice. 9(14). https://doi.org/10.1186/s12284-016-0087-4
Jain R, Jenkins J, Shu S, Chern M, Martin JA, Copetti D et al (2019) Genome sequence of the model rice variety KitaakeX. BMC Genomics 20(905). https://doi.org/10.1186/s12864-019-6262-4
Vassilev D, Leunissen J, Atanassov A, Nenov A, Dimov G (2005) Application of bioinformatics in plant breeding. Biotechnol Biotechnol Equip 19(sup3):139–152. https://doi.org/10.1080/13102818.2005.10817293
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J et al (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature. 588(7837):277–283. https://doi.org/10.1038/s41586-020-2961-x
Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 361(6403). https://doi.org/10.1126/science.aar7191
Gill BS, Appels R, Borta-Oberholster AM, Buell CR, Bennetzen JL, Chalhoub B et al (2004) A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics. 168(2):1087–1096. https://doi.org/10.1534/genetics.104.034769
Babu P, Baranwal DK, Harikrishna PD, Bharti H, Joshi P et al (2020) Application of genomics tools in wheat breeding to attain durable rust resistance. Front Plant Sci 11:567147. https://doi.org/10.3389/fpls.2020.567147
Guan J, Garcia DF, Zhou Y, Appels R, Li A, Mao L (2020) The battle to sequence the bread wheat genome: a tale of the three kingdoms. Genomics Proteomics Bioinformatics 18(3):221–229. https://doi.org/10.1016/j.gpb.2019.09.005
Bolser D, Staines DM, Pritchard E, Kersey P (2016) Ensembl plants: integrating tools for visualizing, mining and analyzing plant genomics data. Methods Mol Biol 1374:115–140. https://doi.org/10.1007/978-1-4939-3167-5_6
Haberer G, Young S, Bharti AK, Gundlach H, Raymond C, Fuks G et al (2005) Structure and architecture of the maize genome. Plant Physiol 139(4):1612–1624. https://doi.org/10.1104/pp.105.068718
Li C, Song W, Luo Y, Gao S, Zhang R, Shi Z et al (2019) The HuangZaoSi maize genome provides insights into genomic variation and improvement history of maize. Mol Plant 12(3):402–409. https://doi.org/10.1016/j.molp.2019.02.009
Lu F, Romay MC, Glaubitz JC, Bradbury PJ, Elshire RJ, Wang T et al (2015) High-resolution genetic mapping of maize pan-genome sequence anchors. Nat Commun 6:6914. https://doi.org/10.1038/ncomms7914
Cho KT, Portwood JL, Gardiner JM, Harper LC, Lawrence-Dill CJ, Friedberg I et al (2019) MaizeDIG: maize database of images and genomes. Front Plant Sci 10:1050. https://doi.org/10.3389/fpls.2019.01050
Portwood JL, Woodhouse MR, Cannon EK, Gardiner JM, Harper LC, Schaeffer ML et al (2018) MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Res 47(D1):D1146–D1154. https://doi.org/10.1093/nar/gky1046
Ambrosino L, Colantuono C, Diretto G, Fiore A, Chiusano ML (2020) Bioinformatics resources for plant abiotic stress responses: state of the art and opportunities in the fast evolving -omics era. Plants. 9(5):591. https://doi.org/10.3390/plants9050591
Singla J, Krattinger SG (2016) Biotic stress resistance genes in wheat. Reference Module in Food Science. https://doi.org/10.1016/B978-0-08-100596-5.00229-8
Costa MCD, Farrant JM (2019) Plant resistance to abiotic stresses. Plants (Basel) 8(12):553. https://doi.org/10.3390/plants8120553
Xu Y, Gao S, Yang Y, Huang M, Cheng L, Wei Q et al (2013) Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genomics 14:662. https://doi.org/10.1186/1471-2164-14-662
Nishad R, Ahmed T, Rahman VJ, Kareem A (2020) Modulation of plant defense system in response to microbial interactions. Front Microbiol 11:1298. https://doi.org/10.3389/fmicb.2020.01298
Andersen EJ, Ali S, Byamukama E, Yen Y, Nepal MP (2018) Disease resistance mechanisms in plants. Genes (Basel) 9(7):339. https://doi.org/10.3390/genes9070339
Dong OX, Ronald PC (2019) Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol 180(1):26–38. https://doi.org/10.1104/pp.18.01224
Abdulkhair WM, Alghuthaymi MA (2016) Plant pathogens. In: Rigobelo EC (ed) Plant Growth, 1st edn. InTechOpen. https://doi.org/10.5772/65325 Available from: https://www.intechopen.com/chapters/52387
Gupta R, Lee SE, Agrawal GK, Rakwal R, Sangryeol P, Wang Y et al (2015) Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front Plant Sci 6:352. https://doi.org/10.3389/fpls.2015.00352
Schneider DJ, Collmer A (2010) Studying plant-pathogen interactions in the genomics era: beyond Molecular Koch’s postulates to systems biology. Annu Rev Phytopathol 48:457–479. https://doi.org/10.1146/annurev-phyto-073009-114411
Sanseverino W, Hermoso A, D’Alessandro R, Vlasova A, Andolfo G, Frusciante L et al (2013) PRGdb 2.0: towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res 41(Database Issue):D1167–D1171. https://doi.org/10.1093/nar/gks1183
Sanseverino W, Roma G, Simone MD, Faino L, Melito S, Stupka E et al (2010) PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res 38(Database Issue):D814–D821. https://doi.org/10.1093/nar/gkp978
Osuna-Cruz CM, Paytuvi-Gallart A, Donato AD, Sundesha V, Andolfo G, Cigliano RA et al (2018) PRGdb 3.0: a comprehensive platform for prediction and analysis of plant disease resistance genes. Nucleic Acids Res 46(D1):D1197–D1201. https://doi.org/10.1093/nar/gkx1119
Hily JM, Demanèche S, Poulicard N, Tannières M, Djennane S, Beuve M et al (2018) Metagenomic-based impact study of transgenic grapevine rootstock on its associated virome and soil bacteriome. Plant Biotechnol J 16(1):208–220. https://doi.org/10.1111/pbi.12761
Fadiji AE, Babalola OO (2020) Metagenomics methods for the study of plant-associated microbial communities: a review. J Microbiol Methods 70:105860. https://doi.org/10.1016/j.mimet.2020.105860
Piombo E, Abdelfattah A, Droby S, Wisniewski M, Spadaro D, Schena L (2021) Metagenomics approaches for the detection and surveillance of emerging and recurrent plant pathogens. Microorganisms. 9(1):188. https://doi.org/10.3390/microorganisms9010188
Chaudhary P, Khati P, Chaudhary A, Maithani D, Kumar G, Sharma A (2021) Cultivable and metagenomic approach to study the combined impact of nanogypsum and Pseudomonas taiwanensis on maize plant health and its rhizospheric microbiome. PLoS One 16(4):e0250574. https://doi.org/10.1371/journal.pone.0250574
Chukwuneme CF, Ayangbenro AS, Babalola OO (2021) Metagenomic analyses of plant growth-promoting and carbon-cycling genes in maize rhizosphere soils with distinct land-use and management histories. Genes (Basel) 12(9):1431. https://doi.org/10.3390/genes12091431
Zhao J, Ma J, Yang Y, Yu H, Zhang S, Chen F (2021) Response of soil microbial community to vegetation reconstruction modes in mining areas of the Loess Plateau, China. Front Microbiol 12:714967. https://doi.org/10.3389/fmicb.2021.714967
Babalola OO, Fadiji AE, Ayangbenro AS (2020) Shotgun metagenomic data of root endophytic microbiome of maize ( Zea mays L.). Data Brief 31(105893). https://doi.org/10.1016/j.dib.2020.105893
Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D et al (2019) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47(D1):D259–D264. https://doi.org/10.1093/nar/gky1022
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(Database issue):D590–D596. https://doi.org/10.1093/nar/gks1219
Mitchell AL, Almeida A, Beracochea M, Boland M, Burgin J, Cochrane G et al (2020) MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48(D1):D570–D578. https://doi.org/10.1093/nar/gkz1035
Musidlak O, Buchwald W, Nawrot R (2014) Plant defense responses against viral and bacterial pathogen infections. Focus on RNA-binding proteins (RBPs). Herba Polonica 60:60–73. https://doi.org/10.1515/hepo-2015-0005
Silva MS, Arraes FBM, Campos MDA, Grossi-de-Sa M, Fernandez D, Cândido EDS et al (2018) Review: potential biotechnological assets related to plant immunity modulation applicable in engineering disease-resistant crops. Plant Sci 270:72–84. https://doi.org/10.1016/j.plantsci.2018.02.013
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P et al (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23(10):1229–1232. https://doi.org/10.1038/cr.2013.114
Wada N, Ueta R, Osakabe Y, Osakabe K (2020) Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol 20:234. https://doi.org/10.1186/s12870-020-02385-5
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31(8):691–693. https://doi.org/10.1038/nbt.2655
Langner T, Kamoun S, Belhaj K (2018) CRISPR crops: plant genome editing toward disease resistance. Annu Rev Phytopathol 56:479–512. https://doi.org/10.1146/annurev-phyto-080417-050158
Zafar K, Khan MZ, Amin I, Mukhtar Z, Yasmin S, Arif M et al (2020) Precise CRISPR-Cas9 mediated genome editing in super basmati rice for resistance against bacterial blight by targeting the major susceptibility gene. Front Plant Sci 11:575. https://doi.org/10.3389/fpls.2020.00575
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6(6):1975–1983. https://doi.org/10.1093/mp/sst119
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y et al (2016) Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11(4):e0154027. https://doi.org/10.1371/journal.pone.0154027
Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T et al (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37(11):1344–1350. https://doi.org/10.1038/s41587-019-0267-z
Wang L, Chen S, Peng A, Xie Z, He Y, Zou X (2019) CRISPR/CAS9 -mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange ( Citrus sinensis (L.) Osbeck). Plant Biotechnol Rep 13(5):501–510. https://doi.org/10.1007/s11816-019-00556-x
Fister AS, Landherr L, Maximova SN, Guiltinan MJ (2018) Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 Enhances defense response in theobroma cacao. Front Plant Sci 9:268. https://doi.org/10.3389/fpls.2018.00268
Ong Q, Nguyen P, Thao NP, Le L (2016) Bioinformatics approach in plant genomic research. Curr Genomics 17(4):368–378. https://doi.org/10.2174/1389202917666160331202956
Schatz MC, Witkowski J, McCombie WR (2012) Current challenges in de novo plant genome sequencing and assembly. Genome Biol 13(4):243. https://doi.org/10.1186/gb-2012-13-4-243
Claros MG, Bautista R, Guerrero-Fernández D, Benzerki H, Seoane P, Fernández-Pozo N (2012) Why assembling plant genome sequences is so challenging. Biology (Basel) 1(2):439–459. https://doi.org/10.3390/biology1020439
Kyriakidou M, Tai HH, Anglin NL, Ellis D, Strömvik MV (2018) Current strategies of polyploid plant genome sequence assembly. Front Plant Sci 9:1660. https://doi.org/10.3389/fpls.2018.01660
Mathur M (2018) Bioinformatics challenges: a review. Int J Adv Sci Res 3(6):29–33
Fazan L, Song YG, Kozlowski G (2020) The woody planet: from past triumph to manmade decline. Plants (Basel) 9(11):1593. https://doi.org/10.3390/plants9111593
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 408(6814):796–815. https://doi.org/10.1038/35048692
Qi H, Jiang Z, Zhang K, Yang S, He F, Zhang Z (2018) PlaD: a transcriptomics database for plant defense responses to pathogens, providing new insights into plant immune system. Genomics Proteomics Bioinformatics 16(4):283–293. https://doi.org/10.1016/j.gpb.2018.08.002
Ohyanagi H, Takano T, Terashima S, Kobayashi M, Kanno M, Morimoto K et al (2015) Plant Omics Data Center: an integrated web repository for interspecies gene expression networks with NLP-based curation. Plant Cell Physiol 56(1):e9. https://doi.org/10.1093/pcp/pcu188
Download references
Acknowledgements
The authors wish to thank Prof. Hoe I. Ling of Columbia University (New York, USA) for his editorial input and proofread the manuscript.
Author information
Authors and affiliations.
Division of Applied Biomedical Sciences and Biotechnology, School of Health Sciences, International Medical University, 126 Jalan Jalil Perkasa 19, Bukit Jalil, 57000, Kuala Lumpur, Malaysia
Yung Cheng Tan, Asqwin Uthaya Kumar, Ying Pei Wong & Anna Pick Kiong Ling
School of Biosciences and Biotechnology, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600, Bangi, Malaysia
Asqwin Uthaya Kumar
You can also search for this author in PubMed Google Scholar
Contributions
YCT designed the content and was a major contributor in writing the manuscript. AUK and YPW edited the manuscript. APKL designed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Anna Pick Kiong Ling .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Tan, Y.C., Kumar, A.U., Wong, Y.P. et al. Bioinformatics approaches and applications in plant biotechnology. J Genet Eng Biotechnol 20 , 106 (2022). https://doi.org/10.1186/s43141-022-00394-5
Download citation
Received : 30 November 2021
Accepted : 05 July 2022
Published : 15 July 2022
DOI : https://doi.org/10.1186/s43141-022-00394-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Bioinformatics
- Biotic and abiotic
- Plant breeding
- Plant sequencing
- Plant pathogen
- PRGdb sequence analysis
- Search Menu
- Sign in through your institution
- Author Guidelines
- Submission Site
- Open Access
- About Briefings in Bioinformatics
- Journals Career Network
- Editorial Board
- Advertising and Corporate Services
- Self-Archiving Policy
- Journals on Oxford Academic
- Books on Oxford Academic
Browse issues

Cover image

Volume 25, Issue 4, July 2024
Sharing sensitive data in life sciences: an overview of centralized and federated approaches.
- View article
NIPT-PG: empowering non-invasive prenatal testing to learn from population genomics through an incremental pan-genomic approach
- Supplementary data
VISH-Pred: an ensemble of fine-tuned ESM models for protein toxicity prediction
Comparison of software packages for detecting unannotated translated small open reading frames by ribo-seq, attention mechanism models for precision medicine, predicting rna polymerase ii transcriptional elongation pausing and associated histone code, predicting functional utr variants by integrating region-specific features, mutational signatures of colorectal cancers according to distinct computational workflows, spin: sex-specific and pathway-based interpretable neural network for sexual dimorphism analysis, benchmarking mapping algorithms for cell-type annotating in mouse brain by integrating single-nucleus rna-seq and stereo-seq data, rtcpredictor: identification of read-through chimeric rnas from rna sequencing data, a comprehensive benchmarking of machine learning algorithms and dimensionality reduction methods for drug sensitivity prediction, accurate prediction of antibody function and structure using bio-inspired antibody language model, irgsea: the integration of single-cell rank-based gene set enrichment analysis, a multi-view graph contrastive learning framework for deciphering spatially resolved transcriptomics data, complementary multi-modality molecular self-supervised learning via non-overlapping masking for property prediction, idmir: identification of dysregulated mirnas associated with disease based on a mirna–mirna interaction network constructed through gene expression data, spancmg: improving spatial domains identification of spatial transcriptomics using neighborhood-complementary mixed-view graph convolutional network, multi-modal domain adaptation for revealing spatial functional landscape from spatially resolved transcriptomics, pipet: predicting relevant subpopulations in single-cell data using phenotypic information from bulk data, a hybrid demultiplexing strategy that improves performance and robustness of cell hashing, themis: advancing precision oncology through comprehensive molecular subtyping and optimization, efficient sars-cov-2 variant detection and monitoring with spike screen next-generation sequencing, prediction of disease-free survival for precision medicine using cooperative learning on multi-omic data, correction to: diagnostic prediction of portal vein thrombosis in chronic cirrhosis patients using data-driven precision medicine model, email alerts.
- Recommend to your Library
Affiliations
- Online ISSN 1477-4054
- Copyright © 2024 Oxford University Press
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
- Open access
- Published: 01 May 2012
Cancer bioinformatics: A new approach to systems clinical medicine
- Duojiao Wu 1 ,
- Catherine M Rice 3 &
- Xiangdong Wang 1 , 2
BMC Bioinformatics volume 13 , Article number: 71 ( 2012 ) Cite this article
33k Accesses
48 Citations
13 Altmetric
Metrics details
Cancer is one of the commonest causes of patient death in the clinic and a complex disease occurring in multiple organs per system, multiple systems per organ, or both, in the body. The poor diagnoses, therapies and prognoses of the disease could be mainly due to the variation of severities, durations, locations, sensitivity and resistance against drugs, cell differentiation and origin, and understanding of pathogenesis. With increasing evidence that the interaction and network between genes and proteins play an important role in investigation of cancer molecular mechanisms, it is necessary and important to introduce a new concept of Systems Clinical Medicine into cancer research, to integrate systems biology, clinical science, omics-based technology, bioinformatics and computational science to improve diagnosis, therapies and prognosis of diseases. Cancer bioinformatics is a critical and important part of the systems clinical medicine in cancer and the core tool and approach to carry out the investigations of cancer in systems clinical medicine. “Thematic Series on Cancer Bioinformatics” gather the strength of BMC Bioinformatics , BMC Cancer , Genome Medicine and Journal of Clinical Bioinformatics to headline the application of cancer bioinformatics for the development of bioinformatics methods, network biomarkers and precision medicine. The Series focuses on new developments in cancer bioinformatics and computational systems biology to explore the potential of clinical applications and improve the outcomes of patients with cancer.
Expectations of methodologies
Cancer bioinformatics is one of multiple ways to concentrate bioinformatics methods in cancer, according to the specificity of disease metabolisms, signaling, communication, and proliferations. Clinical bioinformatics, an emerging science combining clinical informatics, bioinformatics, medical informatics, information technology, mathematics, and omics science together [ 1 ], can be considered to be one of critical elements addressing clinical relevant challenges in early diagnosis, efficient therapies, and predictive prognosis of patients with cancer. There is a need to develop cancer bioinformatics-specific methodologies or introduce new and advanced bioinformatics tools to answer the specific question of cancer. For example, the Semantic Web technology was used to understand high throughput clinical data and develop quantitative semantic models retrieved from Corvus, a data warehouse which provides a uniform interface to various forms of Omics data, based on systematic biological knowledge and by application of SPARQL endpoint [ 2 ]. Semantic models, containing genomic, transcriptomic and epigenomic data from melanoma samples with Gene Ontology data and regulatory networks constructed from transcription factor binding information, were applied for the interplay between a cell molecular state and its response to anti-cancer therapy. Multivariate assays, a process to characterize error introduced in the assay results from the intrinsic error in sample preparation and measurement of the contributing factors, were used to help and guide clinicians understanding the application to PAM50 centroid-based genomic predictors for breast cancer treatment plans and providing the uncertainty information in a usable way [ 3 ].
The applicability, specificity, and integration of methodologies, software, computational tools, and databases which can be used to explore the molecular mechanisms of cancer and identify and validate novel biomarkers, network biomarkers, and individualized medicine in cancer should be seriously considered. miRTrail is an integrative tool for analyzing comprehensive interactions of genes and miRNAs based on expression profiles to generate more robust and reliable results on deregulated pathogenic processes. It was suggested that miRTrail may open avenues for investigating the regulatory interactions between genes and miRNAs for human diseases, including cancer, by integrating information on 20.000 genes, almost 1.000 miRNAs, and roughly 280.000 putative interactions [ 4 ]. It would be helpful to explore the potential computational mode correlating such regulatory interactions between genes and miRNAs with clinical phenotypes, e.g. the variation of gene interactions among tumor locations, phages, differentiations, patient symptoms, or responses to therapies. Medical imaging should be one of important factors to be considered in the application of cancer bioinformatics, since the imaging in clinical pathology, ultrasonic, computerized tomography, nuclear magnetic resonance imaging, and positron emission tomography is one of the most necessary and important approaches in the “early and accurate” detection and diagnosis of cancer. Bioinformatic analyses of morphological features of masses and other abnormalities in medical images were initiated by selective extraction of target features by mathematical morphology and enhancement of the extracted features by two contrast modification techniques [ 5 ]. The algorithm described by Haustein and Schumacher in the Thematic Series on Cancer Bioinformatics in Journal of Clinical Bioinformatics [ 6 ] can simulate tumor growth and detect the formation of some metastases in advance of clinical detection in cells, on basis of clinical breast cancer data.
It may be a non-relative question or a future expectation how experts in cancer bioinformatics can help clinicians to establish the potential picture of gene or protein interactions and mechanisms correlated with tumor-associated shapes, densities, or locations. A Commentary by von der Heyde and Beissbarth in the Thematic Series on Cancer Bioinformatics in BMC Medicine [ 7 ] discusses the recent insights into mechanisms of cetuximab resistance in head and neck cancers resulting from novel analysis of the EGFR pathway.
New strategies of biomarkers
Cancer bioinformatics is expected to play a more important role in the identification and validation of biomarkers, specific to clinical phenotypes related to early diagnoses, measurements to monitor the progress of the disease and the response to therapy, and predictors for the improvement of patient’s life quality. Of gene-, protein-, peptide-, chemical- or physic-based variables in cancer, biomarkers were investigated from a single one to multiple markers, from the expression to functional indication, and from the network to dynamic network. Network biomarkers as a new type of biomarkers with protein-protein interactions were investigated with the integration of knowledge on protein annotations, interaction, and signaling pathway. Alterations of network biomarkers can be monitored and evaluated at different stages and time points during the development of diseases, named dynamic network biomarkers, as one of the new strategies. Dynamic network biomarkers were expected to be correlated with clinical informatics, including patient complaints, history, therapies, clinical symptoms and signs, physician’s examinations, biochemical analyses, imaging profiles, pathologies and other measurements [ 8 ].
Systems clinical medicine is recommended as one of new strategies for the development of cancer biomarkers. Systems clinical medicine is coined as the integration of systems biology, clinical phenotypes, high-throughout technologies, bioinformatics and computational science to improve diagnosis, therapies and prognosis of diseases. Cancer biomarkers should possess the characters of networks, dynamics, interactions, and specificities to disease diagnosis, therapy and prognosis. Understanding the interaction between clinical informatics and bioinformatics is the first and critical step to discover and develop the new diagnostics and therapies for diseases. Such strategy has been described in other diseases like acute rejection after renal transplantation or lung diseases [ 9 , 10 ]. In brief, human samples from clinical studies under clear and strict criteria of participating recruitments are collected and harvested with an entire profile of clinical informatics translated from clinical descriptions. Gene and/or protein profiles of defined samples are analyzed and dynamic networks and interactions between genes and/or proteins can be figured out by bioinformatics and systems biology.
Selected disease-specific networks and dynamic networks of genes and/or proteins in patients are correlated with each of clinical phenotypes by the computational mode, to validate and optimize disease-special biomarkers. However, a number of challenges in the application of systems clinical medicine are encountered and need to be overcome; e.g. the optimal system to translate the information of clinical descriptions to clinical informatics, bioinformatics analysis oriented with disease severity, duration, location, sensitivity to therapies, and progress, or computational mode to integrate all elements from clinical and high-throughout data for precision conclusions. It is also a challenge to find out the variation and significance between molecular networks, between networks of molecules and clinical phenotypes, and between gene and/or protein interactions, in addition to the expression of genes and proteins. Cun and Fröhlich in the Thematic Series on Cancer Bioinformatics in BMC Bioinformatics report that incorporating protein network and interaction data improve the ability to interpret gene signatures in a study to stratify breast cancer patients, evidenced by findings that R weighted Recursive Feature Elimination and average pathway expression were most effective at generating interpretable signatures in those methods tested [ 11 ].
Monitoring and prediction of precision medicine
Systems cancer medicine has been proposed as a new strategy towards realization of predictive, preventive, personalized and participatory (P4) medicine [ 12 – 15 ]. Tian et al. [ 15 ] recently proposed that a virtual cloud of billions of data generated from high-throughout technologies in patients would be figured out, including one or more disease-perturbed networks in cells of the relevant organ in the disease. Disease-perturbed molecular networks may indicate the abnormality of early signals and the functioning, to finally carry out P4 medicine in cancer. However, cancer clinical bioinformatics is an important way to reach systems clinical medicine by combining clinical measurements and signs with human cancer tissue-generated bioinformatics, understanding clinical symptoms and signs, disease development and progress, and therapeutic strategy, and mapping relationships that integrate discrete elements that collectively direct global function within a particular -omic category, with clinical examinations, pathology, biochemical analysis, imaging and therapies [ 1 , 8 ]. Ren and colleagues in the Thematic Series on Cancer Bioinformatics in BMC Bioinformatics have developed an algorithm named Optimization Tool for Clustering and Classification for multiple types of measurements, including proteomic and next generation sequencing data types [ 16 ]. Such method could successfully and effectively discover class of unknown cancer samples as class prediction in both breast cancer and leukemia data sets.
Cancer bioinformatics plays an important role in monitoring and predicting the efficiency and effectiveness of the precision medicine, which provides the safest and most effective therapeutic strategy based on the gene and protein variations of each subject. The semantic heterogeneity of the data generated from microarrays, proteomics, epigenetics and next generation sequencing, provided an ontology-based solution for querying distributed databases over service-oriented, model-driven infrastructures by integrating molecular, pathology, radiology and clinical data in an efficient manner [ 17 ]. A recent study performed a forward-genetic screen guided by genomic analysis of human hepatic cellular carcinoma, and found that a common genetic alteration in liver cancer (11q13.3 amplification) resulted in activation of FGF19 which caused the selective sensitivity to FGF19 inhibition through subsequent analysis with mouse models and RNAi [ 18 ]. It is expected to develop accurate tools for delivering the right treatment to the right patient in the right time, based on molecular network characters of each patient’s tumor. Cancer bioinformatics and systems biology are expected to improve prevention, diagnosis and treatment through therapy design. The classical techniques of statistics and bioinformatics for analysis of the genome, biological sequences, large-scale ‘omic’ data sets and protein three-dimensional structure could form an indispensable backbone for computational cancer research [ 19 ].
In conclusion, cancer bioinformatics as an emerging strategy is one of the most critical and useful approaches to systems clinical medicine for clinical research and applications and improve the outcomes of patients with cancer. The Thematic Series on Cancer Bioinformatics provides a unique and outstanding platform and opportunity for scientists to integrate omics science, bioinformatics tools and data, clinical research, disease-specific biomarkers, dynamic networks, with precision medicine, together fighting cancer and improving the life quality of patients with cancer.
Wang XD, Liotta L: Clinical bioinformatics: a new emerging science. J Clin Bioinforma 2011, 1(1):1. 10.1186/2043-9113-1-1
Article PubMed Central PubMed Google Scholar
Holford ME, McCusker JP, Cheung KH, Krauthammer M: A semantic web framework to integrate cancer omics data with biological knowledge. BMC Bioinformatics 2012, 13(Suppl 1):S10. 10.1186/1471-2105-13-S10-S10
Ebbert MTW, Bastien RRL, Boucher KM, Martín M, Carrasco E, Caballero R, Stijleman IJ, Bernard PS, Facelli JC: Characterization of uncertainty in the classification of multivariate assays: application to PAM50 centroid-based genomic predictors for breast cancer treatment plans. J Clin Bioinforma 2011, 1: 37. 10.1186/2043-9113-1-37
Laczny C, Leidinger P, Haas J, Ludwig N, Backes C, Gerasch A, Kaufmann M, Vogel B, Katus HA, Meder B, et al .: miRTrail - a comprehensive webserver for analyzing gene and miRNA patterns to enhance the understanding of regulatory mechanisms in diseases. BMC Bioinformatics 2012, 13(1):36. 10.1186/1471-2105-13-36
Article PubMed Central CAS PubMed Google Scholar
Kimori Y: Mathematical morphology-based approach to the enhancement of morphological features in medical images. J Clin Bioinforma 2011, 1: 33. 10.1186/2043-9113-1-33
Haustein V, Schumacher U: A dynamic model for tumour growth and metastasis formation. J Clin Bioinforma 2012. (MS: 1377215016594165) in pre-accept
Google Scholar
von der Heyde S, Beissbarth T: A new analysis approach of epidermal growth factor receptor pathway activation patterns provides insights into cetuximab resistance mechanisms in head and neck cancer. BMC Medicine 2012. (MS: 2092284597711620) in pre-accept
Wang XD: Role of clinical bioinformatics in the development of network-based Biomarkers. J Clin Bioinforma 2011, 1: 28. 10.1186/2043-9113-1-28
Wu DJ, Zhu D, Xu M, Rong RM, Tang QY, Wang XD, Zhu TY: Analysis of Transcriptional Factors and Regulation Networks in Patients with Acute Renal Allograft Rejection. J Proteome Res 2011, 10(1):175–181. 10.1021/pr100473w
Article CAS PubMed Google Scholar
Chen H, Song ZJ, Qian MJ, Bai CX, Wang XD: Selection of disease-specific biomarkers by integrating inflammatory mediators with clinical informatics in AECOPD patients: a preliminary study. J Cell Mol Med 2011. Aug 25. doi: 10.1111/j.1582–4934.2011.01416.x
Cun Y, Fröhlich H: Prognostic Gene Signatures for Patient Stratification in Breast Cancer - Accuracy, Stability and Interpretability of Gene Selection Approaches Using Prior Knowledge on Protein-Protein Interactions. BMC Bioinformatics 2012. (MS: 1321151249583179 in pre-accept)
Chen H, Wang Y, Bai C, Wang XD: Alterations of plasma inflammatory biomarkers in the healthy and chronic obstructive pulmonary disease patients with or without acute exacerbation. J Proteomics 2012., 10:
Hood L, Friend SH: Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol 2011, 8: 184–187. 10.1038/nrclinonc.2010.227
Article PubMed Google Scholar
Hood L, Heath JR, Phelps ME, Lin B: Systems biology and new technologies enable predictive and preventative medicine. Science 2004, 306(5696):640–643. 10.1126/science.1104635
Tian Q, Price ND, Hood L: Systems cancer medicine: towards realization of predictive, preventive, personalized and participatory (P4) medicine. J Intern Med 2012, 271(2):111–121. 10.1111/j.1365-2796.2011.02498.x
Ren X, Wang Y, Wang J, Zhang XS: A unified computational model for revealing and predicting subtle subtypes of cancers. BMC Bioinformatics 2012. (MS: 1910002661647107 in pre-accept).
González-Beltrán A, Tagger B, Finkelstein A: Federated ontology-based queries over cancer data. BMC Bioinformatics 2012, 13(Suppl 1):S9. 10.1186/1471-2105-13-S11-S9
Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, Zhao A, Busuttil RW, Yee H, Stein L, et al .: Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell 2011, 19(3):347–358. 10.1016/j.ccr.2011.01.040
Sylvia Nagl (Ed): Cancer bioinformatics; from therapy design to treatment . Publisher: John Wiley & Sons, ; 2006:Volume 30, Issue 2, 0 pp 287.
Download references
Author information
Authors and affiliations.
Qingpu Branch, Fudan University Zhongshan Hospital, Shanghai, China
Duojiao Wu & Xiangdong Wang
Department of Pulmonary Medicine, Fudan University Zhongshan Hospital, Shanghai, China
Xiangdong Wang
BMC Bioinformatics Executive Editor, BioMed Central, London, UK
Catherine M Rice
You can also search for this author in PubMed Google Scholar

Corresponding author
Correspondence to Xiangdong Wang .
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Reprints and permissions
About this article
Cite this article.
Wu, D., Rice, C.M. & Wang, X. Cancer bioinformatics: A new approach to systems clinical medicine. BMC Bioinformatics 13 , 71 (2012). https://doi.org/10.1186/1471-2105-13-71
Download citation
Received : 26 April 2012
Accepted : 01 May 2012
Published : 01 May 2012
DOI : https://doi.org/10.1186/1471-2105-13-71
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]
Suggestions or feedback?
MIT News | Massachusetts Institute of Technology
- Machine learning
- Social justice
- Black holes
- Classes and programs
Departments
- Aeronautics and Astronautics
- Brain and Cognitive Sciences
- Architecture
- Political Science
- Mechanical Engineering
Centers, Labs, & Programs
- Abdul Latif Jameel Poverty Action Lab (J-PAL)
- Picower Institute for Learning and Memory
- Lincoln Laboratory
- School of Architecture + Planning
- School of Engineering
- School of Humanities, Arts, and Social Sciences
- Sloan School of Management
- School of Science
- MIT Schwarzman College of Computing
Ultrasound offers a new way to perform deep brain stimulation
Press contact :.
Previous image Next image
Deep brain stimulation, by implanted electrodes that deliver electrical pulses to the brain, is often used to treat Parkinson’s disease and other neurological disorders. However, the electrodes used for this treatment can eventually corrode and accumulate scar tissue, requiring them to be removed.
MIT researchers have now developed an alternative approach that uses ultrasound instead of electricity to perform deep brain stimulation, delivered by a fiber about the thickness of a human hair. In a study of mice, they showed that this stimulation can trigger neurons to release dopamine, in a part of the brain that is often targeted in patients with Parkinson’s disease.
“By using ultrasonography, we can create a new way of stimulating neurons to fire in the deep brain,” says Canan Dagdeviren, an associate professor in the MIT Media Lab and the senior author of the new study. “This device is thinner than a hair fiber, so there will be negligible tissue damage, and it is easy for us to navigate this device in the deep brain.”
In addition to offering a potentially safer way to deliver deep brain stimulation, this approach could also become a valuable tool for researchers seeking to learn more about how the brain works.
MIT graduate student Jason Hou and MIT postdoc Md Osman Goni Nayeem are the lead authors of the paper, along with collaborators from MIT’s McGovern Institute for Brain Research, Boston University, and Caltech. The study appears today in Nature Communications .
Deep in the brain
Dagdeviren’s lab has previously developed wearable ultrasound devices that can be used to deliver drugs through the skin or perform diagnostic imaging on various organs . However, ultrasound cannot penetrate deeply into the brain from a device attached to the head or skull.
“If we want to go into the deep brain, then it cannot be just wearable or attachable anymore. It has to be implantable,” Dagdeviren says. “We carefully customize the device so that it will be minimally invasive and avoid major blood vessels in the deep brain.”
Deep brain stimulation with electrical impulses is FDA-approved to treat symptoms of Parkinson’s disease. This approach uses millimeter-thick electrodes to activate dopamine-producing cells in a brain region called the substantia nigra. However, once implanted in the brain, the devices eventually begin to corrode, and scar tissue that builds up surrounding the implant can interfere with the electrical impulses.
The MIT team set out to see if they could overcome some of those drawbacks by replacing electrical stimulation with ultrasound. Most neurons have ion channels that are responsive to mechanical stimulation, such as the vibrations from sound waves, so ultrasound can be used to elicit activity in those cells. However, existing technologies for delivering ultrasound to the brain through the skull can’t reach deep into the brain with high precision because the skull itself can interfere with the ultrasound waves and cause off-target stimulation.
“To precisely modulate neurons, we must go deeper, leading us to design a new kind of ultrasound-based implant that produces localized ultrasound fields,” Nayeem says. To safely reach those deep brain regions, the researchers designed a hair-thin fiber made from a flexible polymer. The tip of the fiber contains a drum-like ultrasound transducer with a vibrating membrane. When this membrane, which encapsulates a thin piezoelectric film, is driven by a small electrical voltage, it generates ultrasonic waves that can be detected by nearby cells.
“It’s tissue-safe, there’s no exposed electrode surface, and it’s very low-power, which bodes well for translation to patient use,” Hou says.
In tests in mice, the researchers showed that this ultrasound device, which they call ImPULS (Implantable Piezoelectric Ultrasound Stimulator), can provoke activity in neurons of the hippocampus. Then, they implanted the fibers into the dopamine-producing substantia nigra and showed that they could stimulate neurons in the dorsal striatum to produce dopamine.
“Brain stimulation has been one of the most effective, yet least understood, methods used to restore health to the brain. ImPULS gives us the ability to stimulate brain cells with exquisite spatial-temporal resolution and in a manner that doesn’t produce the kind of damage or inflammation as other methods. Seeing its effectiveness in areas like the hippocampus opened an entirely new way for us to deliver precise stimulation to targeted circuits in the brain,” says Steve Ramirez, an assistant professor of psychological and brain sciences at Boston University, and a faculty member at B.U.’s Center for Systems Neuroscience, who is also an author of the study.
A customizable device
All of the components of the device are biocompatible, including the piezoelectric layer, which is made of a novel ceramic called potassium sodium niobate, or KNN. The current version of the implant is powered by an external power source, but the researchers envision that future versions could be powered a small implantable battery and electronics unit.
The researchers developed a microfabrication process that enables them to easily alter the length and thickness of the fiber, as well as the frequency of the sound waves produced by the piezoelectric transducer. This could allow the devices to be customized for different brain regions.
“We cannot say that the device will give the same effect on every region in the brain, but we can easily and very confidently say that the technology is scalable, and not only for mice. We can also make it bigger for eventual use in humans,” Dagdeviren says.
The researchers now plan to investigate how ultrasound stimulation might affect different regions of the brain, and if the devices can remain functional when implanted for year-long timescales. They are also interested in the possibility of incorporating a microfluidic channel, which could allow the device to deliver drugs as well as ultrasound.
In addition to holding promise as a potential therapeutic for Parkinson’s or other diseases, this type of ultrasound device could also be a valuable tool to help researchers learn more about the brain, the researchers say.
“Our goal to provide this as a research tool for the neuroscience community, because we believe that we don’t have enough effective tools to understand the brain,” Dagdeviren says. “As device engineers, we are trying to provide new tools so that we can learn more about different regions of the brain.”
The research was funded by the MIT Media Lab Consortium and the Brain and Behavior Foundation Research (BBRF) NARSAD Young Investigator Award.
Share this news article on:
Related links.
- Canan Dagdeviren
- Conformable Decoders Group
- School of Architecture and Planning
Related Topics
- Neuroscience
- Brain and cognitive sciences
- Medical devices
- Parkinson's
Related Articles

A new ultrasound patch can measure how full your bladder is

Soft optical fibers block pain while moving and stretching with the body

A wearable ultrasound scanner could detect breast cancer earlier
Previous item Next item
More MIT News

Improving working environments amid environmental distress
Read full story →

A data-driven approach to making better choices

Paying it forward

John Fucillo: Laying foundations for MIT’s Department of Biology

Researchers demonstrate the first chip-based 3D printer

The unexpected origins of a modern finance tool
- More news on MIT News homepage →
Massachusetts Institute of Technology 77 Massachusetts Avenue, Cambridge, MA, USA
- Map (opens in new window)
- Events (opens in new window)
- People (opens in new window)
- Careers (opens in new window)
- Accessibility
- Social Media Hub
- MIT on Facebook
- MIT on YouTube
- MIT on Instagram
The state of AI in early 2024: Gen AI adoption spikes and starts to generate value
If 2023 was the year the world discovered generative AI (gen AI) , 2024 is the year organizations truly began using—and deriving business value from—this new technology. In the latest McKinsey Global Survey on AI, 65 percent of respondents report that their organizations are regularly using gen AI, nearly double the percentage from our previous survey just ten months ago. Respondents’ expectations for gen AI’s impact remain as high as they were last year , with three-quarters predicting that gen AI will lead to significant or disruptive change in their industries in the years ahead.
About the authors
This article is a collaborative effort by Alex Singla , Alexander Sukharevsky , Lareina Yee , and Michael Chui , with Bryce Hall , representing views from QuantumBlack, AI by McKinsey, and McKinsey Digital.
Organizations are already seeing material benefits from gen AI use, reporting both cost decreases and revenue jumps in the business units deploying the technology. The survey also provides insights into the kinds of risks presented by gen AI—most notably, inaccuracy—as well as the emerging practices of top performers to mitigate those challenges and capture value.
AI adoption surges
Interest in generative AI has also brightened the spotlight on a broader set of AI capabilities. For the past six years, AI adoption by respondents’ organizations has hovered at about 50 percent. This year, the survey finds that adoption has jumped to 72 percent (Exhibit 1). And the interest is truly global in scope. Our 2023 survey found that AI adoption did not reach 66 percent in any region; however, this year more than two-thirds of respondents in nearly every region say their organizations are using AI. 1 Organizations based in Central and South America are the exception, with 58 percent of respondents working for organizations based in Central and South America reporting AI adoption. Looking by industry, the biggest increase in adoption can be found in professional services. 2 Includes respondents working for organizations focused on human resources, legal services, management consulting, market research, R&D, tax preparation, and training.
Also, responses suggest that companies are now using AI in more parts of the business. Half of respondents say their organizations have adopted AI in two or more business functions, up from less than a third of respondents in 2023 (Exhibit 2).
Gen AI adoption is most common in the functions where it can create the most value
Most respondents now report that their organizations—and they as individuals—are using gen AI. Sixty-five percent of respondents say their organizations are regularly using gen AI in at least one business function, up from one-third last year. The average organization using gen AI is doing so in two functions, most often in marketing and sales and in product and service development—two functions in which previous research determined that gen AI adoption could generate the most value 3 “ The economic potential of generative AI: The next productivity frontier ,” McKinsey, June 14, 2023. —as well as in IT (Exhibit 3). The biggest increase from 2023 is found in marketing and sales, where reported adoption has more than doubled. Yet across functions, only two use cases, both within marketing and sales, are reported by 15 percent or more of respondents.
Gen AI also is weaving its way into respondents’ personal lives. Compared with 2023, respondents are much more likely to be using gen AI at work and even more likely to be using gen AI both at work and in their personal lives (Exhibit 4). The survey finds upticks in gen AI use across all regions, with the largest increases in Asia–Pacific and Greater China. Respondents at the highest seniority levels, meanwhile, show larger jumps in the use of gen Al tools for work and outside of work compared with their midlevel-management peers. Looking at specific industries, respondents working in energy and materials and in professional services report the largest increase in gen AI use.
Investments in gen AI and analytical AI are beginning to create value
The latest survey also shows how different industries are budgeting for gen AI. Responses suggest that, in many industries, organizations are about equally as likely to be investing more than 5 percent of their digital budgets in gen AI as they are in nongenerative, analytical-AI solutions (Exhibit 5). Yet in most industries, larger shares of respondents report that their organizations spend more than 20 percent on analytical AI than on gen AI. Looking ahead, most respondents—67 percent—expect their organizations to invest more in AI over the next three years.
Where are those investments paying off? For the first time, our latest survey explored the value created by gen AI use by business function. The function in which the largest share of respondents report seeing cost decreases is human resources. Respondents most commonly report meaningful revenue increases (of more than 5 percent) in supply chain and inventory management (Exhibit 6). For analytical AI, respondents most often report seeing cost benefits in service operations—in line with what we found last year —as well as meaningful revenue increases from AI use in marketing and sales.
Inaccuracy: The most recognized and experienced risk of gen AI use
As businesses begin to see the benefits of gen AI, they’re also recognizing the diverse risks associated with the technology. These can range from data management risks such as data privacy, bias, or intellectual property (IP) infringement to model management risks, which tend to focus on inaccurate output or lack of explainability. A third big risk category is security and incorrect use.
Respondents to the latest survey are more likely than they were last year to say their organizations consider inaccuracy and IP infringement to be relevant to their use of gen AI, and about half continue to view cybersecurity as a risk (Exhibit 7).
Conversely, respondents are less likely than they were last year to say their organizations consider workforce and labor displacement to be relevant risks and are not increasing efforts to mitigate them.
In fact, inaccuracy— which can affect use cases across the gen AI value chain , ranging from customer journeys and summarization to coding and creative content—is the only risk that respondents are significantly more likely than last year to say their organizations are actively working to mitigate.
Some organizations have already experienced negative consequences from the use of gen AI, with 44 percent of respondents saying their organizations have experienced at least one consequence (Exhibit 8). Respondents most often report inaccuracy as a risk that has affected their organizations, followed by cybersecurity and explainability.
Our previous research has found that there are several elements of governance that can help in scaling gen AI use responsibly, yet few respondents report having these risk-related practices in place. 4 “ Implementing generative AI with speed and safety ,” McKinsey Quarterly , March 13, 2024. For example, just 18 percent say their organizations have an enterprise-wide council or board with the authority to make decisions involving responsible AI governance, and only one-third say gen AI risk awareness and risk mitigation controls are required skill sets for technical talent.
Bringing gen AI capabilities to bear
The latest survey also sought to understand how, and how quickly, organizations are deploying these new gen AI tools. We have found three archetypes for implementing gen AI solutions : takers use off-the-shelf, publicly available solutions; shapers customize those tools with proprietary data and systems; and makers develop their own foundation models from scratch. 5 “ Technology’s generational moment with generative AI: A CIO and CTO guide ,” McKinsey, July 11, 2023. Across most industries, the survey results suggest that organizations are finding off-the-shelf offerings applicable to their business needs—though many are pursuing opportunities to customize models or even develop their own (Exhibit 9). About half of reported gen AI uses within respondents’ business functions are utilizing off-the-shelf, publicly available models or tools, with little or no customization. Respondents in energy and materials, technology, and media and telecommunications are more likely to report significant customization or tuning of publicly available models or developing their own proprietary models to address specific business needs.
Respondents most often report that their organizations required one to four months from the start of a project to put gen AI into production, though the time it takes varies by business function (Exhibit 10). It also depends upon the approach for acquiring those capabilities. Not surprisingly, reported uses of highly customized or proprietary models are 1.5 times more likely than off-the-shelf, publicly available models to take five months or more to implement.
Gen AI high performers are excelling despite facing challenges
Gen AI is a new technology, and organizations are still early in the journey of pursuing its opportunities and scaling it across functions. So it’s little surprise that only a small subset of respondents (46 out of 876) report that a meaningful share of their organizations’ EBIT can be attributed to their deployment of gen AI. Still, these gen AI leaders are worth examining closely. These, after all, are the early movers, who already attribute more than 10 percent of their organizations’ EBIT to their use of gen AI. Forty-two percent of these high performers say more than 20 percent of their EBIT is attributable to their use of nongenerative, analytical AI, and they span industries and regions—though most are at organizations with less than $1 billion in annual revenue. The AI-related practices at these organizations can offer guidance to those looking to create value from gen AI adoption at their own organizations.
To start, gen AI high performers are using gen AI in more business functions—an average of three functions, while others average two. They, like other organizations, are most likely to use gen AI in marketing and sales and product or service development, but they’re much more likely than others to use gen AI solutions in risk, legal, and compliance; in strategy and corporate finance; and in supply chain and inventory management. They’re more than three times as likely as others to be using gen AI in activities ranging from processing of accounting documents and risk assessment to R&D testing and pricing and promotions. While, overall, about half of reported gen AI applications within business functions are utilizing publicly available models or tools, gen AI high performers are less likely to use those off-the-shelf options than to either implement significantly customized versions of those tools or to develop their own proprietary foundation models.
What else are these high performers doing differently? For one thing, they are paying more attention to gen-AI-related risks. Perhaps because they are further along on their journeys, they are more likely than others to say their organizations have experienced every negative consequence from gen AI we asked about, from cybersecurity and personal privacy to explainability and IP infringement. Given that, they are more likely than others to report that their organizations consider those risks, as well as regulatory compliance, environmental impacts, and political stability, to be relevant to their gen AI use, and they say they take steps to mitigate more risks than others do.
Gen AI high performers are also much more likely to say their organizations follow a set of risk-related best practices (Exhibit 11). For example, they are nearly twice as likely as others to involve the legal function and embed risk reviews early on in the development of gen AI solutions—that is, to “ shift left .” They’re also much more likely than others to employ a wide range of other best practices, from strategy-related practices to those related to scaling.
In addition to experiencing the risks of gen AI adoption, high performers have encountered other challenges that can serve as warnings to others (Exhibit 12). Seventy percent say they have experienced difficulties with data, including defining processes for data governance, developing the ability to quickly integrate data into AI models, and an insufficient amount of training data, highlighting the essential role that data play in capturing value. High performers are also more likely than others to report experiencing challenges with their operating models, such as implementing agile ways of working and effective sprint performance management.
About the research
The online survey was in the field from February 22 to March 5, 2024, and garnered responses from 1,363 participants representing the full range of regions, industries, company sizes, functional specialties, and tenures. Of those respondents, 981 said their organizations had adopted AI in at least one business function, and 878 said their organizations were regularly using gen AI in at least one function. To adjust for differences in response rates, the data are weighted by the contribution of each respondent’s nation to global GDP.
Alex Singla and Alexander Sukharevsky are global coleaders of QuantumBlack, AI by McKinsey, and senior partners in McKinsey’s Chicago and London offices, respectively; Lareina Yee is a senior partner in the Bay Area office, where Michael Chui , a McKinsey Global Institute partner, is a partner; and Bryce Hall is an associate partner in the Washington, DC, office.
They wish to thank Kaitlin Noe, Larry Kanter, Mallika Jhamb, and Shinjini Srivastava for their contributions to this work.
This article was edited by Heather Hanselman, a senior editor in McKinsey’s Atlanta office.
Explore a career with us
Related articles.

Moving past gen AI’s honeymoon phase: Seven hard truths for CIOs to get from pilot to scale

A generative AI reset: Rewiring to turn potential into value in 2024

Implementing generative AI with speed and safety
To revisit this article, visit My Profile, then View saved stories .
- Backchannel
- Newsletters
- WIRED Insider
- WIRED Consulting
Will Knight
OpenAI Offers a Peek Inside the Guts of ChatGPT

ChatGPT developer OpenAI’s approach to building artificial intelligence came under fire this week from former employees who accuse the company of taking unnecessary risks with technology that could become harmful.
Today, OpenAI released a new research paper apparently aimed at showing it is serious about tackling AI risk by making its models more explainable. In the paper , researchers from the company lay out a way to peer inside the AI model that powers ChatGPT. They devise a method of identifying how the model stores certain concepts—including those that might cause an AI system to misbehave.
Although the research makes OpenAI’s work on keeping AI in check more visible, it also highlights recent turmoil at the company. The new research was performed by the recently disbanded “superalignment” team at OpenAI that was dedicated to studying the technology’s long-term risks.
The former group’s coleads, Ilya Sutskever and Jan Leike—both of whom have left OpenAI —are named as coauthors. Sutskever, a cofounder of OpenAI and formerly chief scientist, was among the board members who voted to fire CEO Sam Altman last November, triggering a chaotic few days that culminated in Altman’s return as leader.
ChatGPT is powered by a family of so-called large language models called GPT, based on an approach to machine learning known as artificial neural networks. These mathematical networks have shown great power to learn useful tasks by analyzing example data, but their workings cannot be easily scrutinized as conventional computer programs can. The complex interplay between the layers of “neurons” within an artificial neural network makes reverse engineering why a system like ChatGPT came up with a particular response hugely challenging.
“Unlike with most human creations, we don’t really understand the inner workings of neural networks,” the researchers behind the work wrote in an accompanying blog post . Some prominent AI researchers believe that the most powerful AI models, including ChatGPT, could perhaps be used to design chemical or biological weapons and coordinate cyberattacks. A longer-term concern is that AI models may choose to hide information or act in harmful ways in order to achieve their goals.
OpenAI’s new paper outlines a technique that lessens the mystery a little, by identifying patterns that represent specific concepts inside a machine learning system with help from an additional machine learning model. The key innovation is in refining the network used to peer inside the system of interest by identifying concepts, to make it more efficient.
OpenAI proved out the approach by identifying patterns that represent concepts inside GPT-4, one of its largest AI models. The company released code related to the interpretability work, as well as a visualization tool that can be used to see how words in different sentences activate concepts, including profanity and erotic content, in GPT-4 and another model. Knowing how a model represents certain concepts could be a step toward being able to dial down those associated with unwanted behavior, to keep an AI system on the rails. It could also make it possible to tune an AI system to favor certain topics or ideas.

By Andy Greenberg

By Scharon Harding, Ars Technica

By Paresh Dave
By Emily Mullin
Even though LLMs defy easy interrogation, a growing body of research suggests they can be poked and prodded in ways that reveal useful information. Anthropic, an OpenAI competitor backed by Amazon and Google, published similar work on AI interpretability last month. To demonstrate how the behavior of AI systems might be tuned, the company's researchers created a chatbot obsessed with San Francisco's Golden Gate Bridge . And simply asking an LLM to explain its reasoning can sometimes yield insights .
“It’s exciting progress,” says David Bau , a professor at Northeastern University who works on AI explainability, of the new OpenAI research. “As a field, we need to be learning how to understand and scrutinize these large models much better.”
Bau says the OpenAI team’s main innovation is in showing a more efficient way to configure a small neural network that can be used to understand the components of a larger one. But he also notes that the technique needs to be refined to make it more reliable. “There’s still a lot of work ahead in using these methods to create fully understandable explanations,” Bau says.
Bau is part of a US government-funded effort called the National Deep Inference Fabric , which will make cloud computing resources available to academic researchers so that they too can probe especially powerful AI models. “We need to figure out how we can enable scientists to do this work even if they are not working at these large companies,” he says.
OpenAI’s researchers acknowledge in their paper that further work needs to be done to improve their method, but also say they hope it will lead to practical ways to control AI models. “We hope that one day, interpretability can provide us with new ways to reason about model safety and robustness, and significantly increase our trust in powerful AI models by giving strong assurances about their behavior,” they write.
You Might Also Like …
Navigate election season with our WIRED Politics Lab newsletter and podcast
Don’t think breakdancing is an Olympic sport ? The world champ agrees (kinda)
How researchers cracked an 11-year-old password to a $3M crypto wallet
The uncanny rise of the world’s first AI beauty pageant
Give your back a break: Here are the best office chairs we’ve tested

Lauren Goode

Kate Knibbs

Reece Rogers

An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Genet Mol Biol
- v.45(2); 2022
The past, present and future of genomics and bioinformatics: A survey of Brazilian scientists
Mariana rocha.
1 Technological University Dublin, School of Computer Science, Dublin, Ireland.
Luisa Massarani
2 Fundação Oswaldo Cruz, Instituto Nacional de Comunicação Pública da Ciência e Tecnologia, Rio de Janeiro, RJ, Brazil.
Sandro José de Souza
3 Universidade Federal do Rio Grande do Norte, Instituto do Cérebro, Natal, RN, Brazil.
4 Universidade Federal do Rio Grande do Norte, Centro Multiusuário de Bioinformática, Natal, RN, Brazil.
5 Sichuan University, Institutes of Systems Genetics, West China Hospital, Chengdu, China.
Ana Tereza R. de Vasconcelos
6 Laboratório Nacional de Computação Científica, Laboratório de Bioinformática, Petrópolis, RJ, Brazil.
Authors contributions: ATRV conceived and the study, ATRV and LM conducted the data collection process, MR analyzed the data, MR, ATRV, LM, and SJS wrote the manuscript. All authors read and approved the final version.
Associated Data
Brazil has one of the highest rates of scientific production, occupying the ninth position among countries with genome-sequencing projects. Considering the rapid development of this research area and the diversity of professionals involved, the present study aims to understand the expectations, past experiences and the current scenario of Brazilian research in bioinformatics and genomics. The present research was carried out by analyzing the perceptions of 576 researchers in genomics and bioinformatics in Brazil through content and sentiment analysis techniques. This group of participants is equivalent to 48% of the members of the research community. The results suggest that most researchers have a positive perception of the potential of this research area. However, there is concern about the lack of funding for investing in equipment and professional training. As part of a wish list for the future, researchers highlighted the need for higher funding, formal education, and collaboration among research networks. When asked about genomics and bioinformatics in other countries, the participants recognize that sequencing technologies and infrastructure are more accessible, allowing better data volume expansion.
Introduction
The year 2021 marked the 20 th anniversary of the publication of two drafts of the human genome sequence: one by the Human Genome Project ( Lander et al., 2001 ) and the other by a company, Celera Genomics ( Venter et al., 2001 ). From discovering the DNA to sequencing all 3 billion letters of a human genome, research in genomics and bioinformatics has provided insights into various biological features, such as cellular metabolism, molecular biology, species evolution, and pathology ( Weissenbach, 2016 ). Furthermore, sequencing the genome is relevant not only for biologists but also for different actors in science. For example, the advent of next-generation sequencing (NGS), a direct consequence of the first two decades of genomics, led to an exponential influx of data and increased the computational challenge associated with data processing and analysis ( Gauthier et al., 2019 ). In addition, developing countries increased the scientific productivity in the area, providing more ethnic diversity in genome studies, a relevant need mainly when genomics technologies are used to identify rare diseases or predict human responses to new drugs ( Bustamante et al., 2011 ).
In spite of the fact that a bias still exists, genomics studies focusing on specific populations have been advancing in the last few years. In the first years of the genomics era, most geneticists directed their research to analyze European populations and their needs. Nowadays, the growth of genomics studies over racial and ethnics minorities provided the possibility of spreading the benefits of the area, including initiatives such as the Special Programme for Research and Training in Tropical Diseases (TDR), a program for scientific collaboration that support efforts to combat tropical diseases ( Morel, 2000 ). Take, for example, the research environment for bioinformatics and genomics research in Latin America. This region, composed of 20 countries and 14 dependent territories, had a relevant growth in scientific productivity, showing a continuous increase from 2000 ( De Las Rivas et al., 2019 ). This trend is not particular to Latin American countries, as studies around life science technologies started to grow world-wide during the early 2000s ( De Las Rivas et al ., 2019 ). However, Latin American countries’ scientific production in genomics was particularly benefited by the establishment of a number of research networks. In 2009, the Iberoamerican Society for Bioinformatics (SoIBIO) was launched ( De Las Rivas et al. , 2019 ) and, since 2010, the International Society for Computational Biology (ISCB) began organizing the ISCB Latin America Conference on Bioinformatics. Both initiatives are intended to contribute to the field’s growth within this region and support scientific innovation across Latin America. However, the fast development of research in the area caught Latin American researchers unprepared a decade ago. The need for sophisticated technology and large-scale database management made it challenging to deliver results at the same level as European and north-American countries countries, resulting in difficulties in communicating those on good or regular impact journals ( Ramírez et al., 2002 ). Recent studies suggest the challenges have not changed. For example, recent research conducted in Mexico revealed that experts in Bioinformatics believe the lack of technological infrastructure and human resources are deficiencies in the research area ( Armenta-Medina et al., 2020 ). Another study carried out by consulting experts in the field revealed that researchers from different regions of Brazil deal with unequal access to sequencing platforms, especially for the country’s northern area ( Bicudo, 2016 ). Due to these difficulties, Latin American countries would need to double their research effort to approach the average world scientific production in the field ( De Las Rivas et al. , 2019 ).
In Latin America, Brazil has a high rate of scientific productivity in genomics and bioinformatics. In an analysis of scientific papers on bioinformatics published by Latin American authors between 1991 and 2016, De Las Rivas et al. (2019 ) identified 2119 publications - more than half of those (1068) were published by Brazilian authors. The turning point in the history of genomics in Brazil was the complete sequencing of the citrus pathogen Xylella fastidiosa in 2000 ( Simpson et al., 2000 ). For the first time, a phytopathogen was sequenced, revealing pathogenicity mechanisms. This discovery supported developing solutions to control the damage caused on oranges, grapevines, citrus and coffee commercial production. Nevertheless, this result stimulated the development of new sequencing projects in the country ( Simpson, 2001 ). A relevant aspect that is usually related to the success of this research field in Brazil was the setup of research networks ( Xavier et al., 2008 ). Some examples are the Organization for Nucleotide Sequencing and Analysis (ONSA), launched by the São Paulo Research Foundation (FAPESP) in 1997 ( Simpson and Perez, 1998 ), and the Brazilian National Genome Network, launched by the Brazilian National Council for Scientific and Technological Development (CNPq) in 2000 ( Simpson et al. , 2004 ), first to be undertaken on a national scale. Many other regional initiatives have been launched in the past two decades, which allowed the country to have an installed capacity not only for sequencing but also for the analysis and interpretation of data. Recently, Brazilian researchers have been quick to characterize the first sample of the SARS-CoV-2 virus, which caused the COVID-19 pandemic ( Araujo et al., 2020 ; Jesus et al. , 2020 ), and to study the spread of the virus in the country since March 2020 ( Candido et al., 2020 ; Silva Francisco Jr et al., 2020 ; Buss et al., 2021 ; Faria et al., 2021 ; Voloch et al., 2021 ). According to ( Chasapi et al., 2020 ), Brazil is part of the ranking listing of 40 leading countries in bioinformatics, and occupies position 23 of the top 1% of highly cited papers. Although research in bioinformatics and genomics in Brazil had positive and relevant features during the last two decades, the country still faces challenges common to developing countries, such as the lack of funding, depending on researchers’ creative approaches to achieve their research goals ( Patrinos et al., 2020 ). This study aims to understand the expectations, past experiences, and the current scenario of Brazilian research in bioinformatics and genomics by gathering researchers’ perceptions about the field.
Research design
The present study investigates the experts’ perceptions of the past, present, and future of genomics and bioinformatics in Brazil. The approach adopted to achieve this goal was to gather and evaluate the perceptions of Brazilian professionals working in the field. Perception can be defined as how “we see things”, and it influences people’s opinion and understanding about a situation ( Given and Saumure, 2008 ). Our hypotheses were based on the fact that understanding expert perceptions about their field, especially when considering a diverse group of professionals, can provide valuable information about the challenges, achievements, and relevance of that research area.
The current study was developed to address the following research question (RQ): What do Brazilian professionals working in the field think about the past, the present, and the future of genomics and bioinformatics? To better structure our work and considering this research’s multidimensional character, the main RQ was divided into three sub-questions to better structure our work and consider this research’s multidimensional character. Table 1 describes the sub-questions, objectives, and methods to be adopted to answer each of those. The methodology is described in detail in the data analysis section.
Data collection
The data was collected through an online questionnaire ( Appendix ) containing 25 questions, 19 closed-ended and six open-ended. The first closed-ended question asks how long the respondent is working in the area of genomics and bioinformatics. If the participant selected the option that states s/he is not working in the area, the questionnaire was then ended. This procedure allowed the exclusion of researchers that are not from the aforementioned area. Participants were recruited by convenience sampling, being contacted by email or phone. The questionnaire was shared with postgraduate courses in the area, short courses and specializations, national networks of professionals, research groups, and contacts in the industry. The questionnaire was available from February to August 2020. The data could be provided anonymously, but researchers had the option of providing their names and email for future contact.
Participants’ profiles
To answer the SQ1, the first phase of data analysis focused on gathering an overview of participants’ profiles through a descriptive analysis of the data. This analysis considered only the data generated by the answers provided on the close-ended questions. Aspects such as participants’ gender, the state of Brazil where they live, number of years working in the area of genomics and/or bioinformatics, and other details were included.
Participants’ perceptions
The data generated by the answers to the open-ended questions were evaluated by two main methods to answer SQ2. First, semantic and lexical analysis of the answers provided by the participants was conducted using the QDA Miner software, developed by Provalis Research of Montreal, Canada. QDA Miner is a full-featured software package for coding, searching, and analyzing mixed-model data ( Lewis and Maas, 2007 ). Using this software, we attempted to identify the words most commonly used by the researchers, considering their frequency on the participant’s answers. This was done after “stemming the words”, a linguistics procedure to reduce all words with the same stem to a common form ( Lovins, 1968 ). Also, common words, also known as stop words, were excluded from the pool. This process allowed us to focus on the meaningful words, removing very common ones such as “a”, “with”, “at” and “on”. We used a built-in list provided by WordStat, but also added extra words, such as “bioinformatics” and “genomics”, which appeared many times as this was the focus of the survey.
Besides retrieving the frequency of the words, we also used QDA Miner to evaluate the groups of words that tend to show up together in the same sentence, forming clusters. Data clustering is performed based on Jaccard’s coefficient, a statistics measurement to identify similarities between texts ( Niwattanakul et al., 2013 ). We also used the WordStat text mining package from Provalis Research to perform a content analysis of the answers to two questions of the questionnaire. Content analysis is a systematic, objective, quantitative analysis of text characteristics ( Neuendorf and Kumar, 2015 ). The first question aimed to collect what the respondents believed to be milestones of the development of genomics and bioinformatics in Brazil. In the second question, each participant was asked to create a wish list containing their expectations for the future in the field. The responses were carefully read so patterns could be identified and codes created. After that, the responses were classified according to the codebook created. The codes were used to classify each sentence of a response. The same answer could contain a number of codes, and the same code could appear twice in the same response (case) if identified in different sentences.
Sentiment analysis was the second main method applied to evaluate the open-ended questions. Sentiment analysis is a technique that classifies sentiments/opinions identified in texts. These sentiments are usually classified as positive, negative, or neutral. This process helps determine how a specific population perceives a context, a product, public policies, and other social aspects ( Prabowo and Thelwall, 2009 ). We adopted the lexical-based sentiment analysis, where the data collected was classified according to a predefined list of words, where each word is associated with a specific sentiment ( Gonçalves et al., 2013 ). The analysis was made based on the OpLexicon ( Souza et al., 2011 ), a Portuguese language lexicon constituted around 15,000 words classified by their morphological category and with polarities positive, negative, and neutral. Unlike other dictionaries, OpLexicon is composed not only of adjectives but also of different types of words, providing better accuracy ( Souza and Vieira, 2012 ). Before classifying the data, a number of procedures were performed. The pre-processing phase included the use of Python libraries to access, clean, and manipulate the dataset. The dataset was stored on Google Sheet and gspread (https://docs.gspread.org/en/v4.0.1/) library was used to access it. The data analysis library pandas was used to transform the dataset into a data frame, avoiding missing values. The dataset was then converted into lowercase text using the method lower() , and the Python module string (https://pandas.pydata.org) was used to clean the dataset by identifying and removing punctuations. The module re (https://docs.python.org/3/library/re.html) performed a tokenization process, separating each response of the dataset into individual words. To simplify the text, the package Natural Language Toolkit (NLTK, https://www.nltk.org) was used to remove stop words, such as “a”, “with”, “at”, and “on”. As the dataset was ready for analysis, urllib (https://docs.python.org/3/library/urllib.html) package was used for accessing the file containing the OpLexicon dictionary and io (https://docs.python.org/3/library/io.html) module was used to prepare this file for the analysis. Finally, matplotlib (https://matplotlib.org) was adopted for the visualization of the final results. This pre-phase process is relevant considering how the classification using OpLexicon works. Each word (token) of the text is classified according to the OpLexicon dictionary - positive words receive a score of 1, negative words receive a score of -1, and neutral words receive a score of 0. After that, all the scores of the words in the response are summed up and, if the resulting value is higher than 0, the response is positive; if lower than 0, the response is negative; if equals to 0, the response is neutral. Both the pre-process and sentiment analysis phases were carried out and are available on Google Colab (https://colab.research.google.com/drive/19UF9PhYCgd6JvryVjiC_vZWe5K8DlD1a), a free, cloud-hosted Jupyter notebook that allows developers to write, execute, and share Python code.
A total of 576 participants answered the questionnaire. To estimate the approximate number of Bioinformaticians in Brazil, we cross-referenced the ones who answered this survey, those who are or were part of the Brazilian Association of Bioinformatics and Computational Biology (AB3C) in the last 10 years, and the number of masters and doctoral students that finished or were/are enrolled in graduate courses (800 students). It is worth noting that we removed the duplicity to have a number closer to reality. Considering these criteria, we estimated that the area includes around 1200 researchers and students in Brazil, and our pool of respondents represents approximately 48% of the population active in the field. After collecting the data, the first step consisted of a data cleaning process to remove empty and duplicates. This resulted in a collection of 541 responses.
From the collected answers, 50.1% were provided by female researchers, 49.5% by male researchers, and 0.4% of participants preferred not to declare their gender. Most of the participants were between 35-39 years old (90 participants, 16.6%), followed by participants between 25-29 years old (86 participants, 16.1%). These data are shown in Figure 1A .

Most of the participants currently live in Brazil (430 researchers, 97.5%), with most of them concentrating in the São Paulo state (38.0%). The results also show that 11 participants live in other countries, such as the United States, Argentina, and China. Figure 1 B illustrates the distribution of the participants around Brazil. When answering how long they are working in the field, 30.0% of participants said to be working in the area for less than 5 years, while 27% of participants do research in Genomics and/or Bioinformatics for over 15 years ( Figure 1 C). Most of the respondents (94.0%) believe their research is extremely relevant to the area.
Participants were asked about their previous/current education, considering aspects such as if they attended private or public institutions and how long they have been graduated. When referring to their high school education, 47.0% of the respondents said to have attended a public institution, 41.0% attended a private institution, and 11.0% had part of their high school studies in public and part in a private institution. When talking about their university studies, 89.0% attended public institutions, 7.0% had part of their studies in public and part in a private institution, and 4.0% attended private university institutions. Considering the time since they have finished their studies, most of the participants finished their courses over 10 years ago ( Table 2 ).
The most popular areas of formal education among researchers in bioinformatics and genomics is biological science (35.0%), followed by biotechnology (13.0%) and biodiversity (7.0%). The majority of participants had their first contact with the area during their bachelor’s years (22.8%), but without attending a discipline or engaging in research work. Around 37.6% of participants had their first contact with the field while doing research in their doctoral years, while 22% had it during their master’s years. Around 11.0% studied genomics and/or bioinformatics for the first time during their master’s years, while approximately 8.8% did it during their doctoral studies. One-fourth of the respondents (25.0%) had a scholarship during their bachelor’s, master’s, doctoral degree, and post-doctoral position.
The participants were also asked about their work experience. The majority of them work in the university or another public institution in an area related to genomics and/or bioinformatics (59.0%), and around 10.0% of participants are not working at the moment. Around 8% of the respondents work with genomics and/or bioinformatics in the private sector. Most of them collaborate with other national (51.0%) or international (35.0%) research groups, while a minority has no collaboration (10.0%).
Around 91% of the participants said to access or generate sequencing data (DNA, RNA, or proteins) in their research, and 28.9% of the participants make use of data that is available in public databases, while only 6.2% hire private services. Furthermore, 87.0% of those believe that the generation of sequencing data changed the area in the last ten years. Most of the participants access bioinformatics tools as a user (36%), while 27.0% generate databases and 11.0% generate bioinformatics programs.
As mentioned in section 3.2.2, the open-ended questions of the questionnaire were evaluated by applying content analysis and sentiment analysis techniques.
Perceptions about the genomics and/or bioinformatics current situation in Brazil
The first question to be evaluated was: How do you see the field in the area of genomics and/or bioinformatics in Brazil? This question aimed to gather participants’ perceptions about the current situation of the field in Brazil, and it resulted in 525 valid responses after data cleaning. Few procedures were used to analyze the data. After performing the stemming processing over the data, the most common stems were identified. The most frequent stem is “grow” (18.3% of the cases), which is found in words such as “growing”, “growth”, and the verb “to grow”. When looking at this word in context, we find positive perceptions about the field in Brazil, such as the following:
Case 145: A field that is GROWING more and more and is recognized as very relevant to medicine, agriculture, pharmaceuticals, among others.
In some other cases, the stem “grow” shows up as part of a positive but concerned perception. The following example highlights the potential of the research area’s growth in Brazil, but the participant demonstrates to be worried about the lack of financial investment:
Case 57: Brazil has great potential to GROW in the area and could be a great producer and exporter of knowledge and research in the areas of genomics and bioinformatics, but there is a lack of incentives and funding.
The next most popular stem is “develop” (13.0% of cases), occurring in responses related to the development of the area in Brazil. Some comments highlight aspects relevant to the attraction of professionals to the research area, such as the creation of new courses:
Case 34: Exponential DEVELOPMENT. The impact is currently occurring at the undergraduate and internships level. More courses are being offered in the earlier training periods. It has attracted professionals from outside Biology, especially Physicists and professionals with computer training.
Another popular stem is “promis” (10.8% of cases), which occurs as part of words like “promising”. The high frequency of this stem seems to be related to the stem “grow”, as most of the cases state the quick development of the field in Brazil, as illustrated by the following example:
Case 120: It is one of the most PROMISING fields in science since the number of data increases with each new platform developed, and, in contrast, the number of well-qualified bioinformatics/statistics professionals is still very limited.
The following word cloud illustrates the most frequent whole words, which occur in the analysis at least 15 times ( Figure 2 ).

The analysis also allowed us to identify clusters of words that co-occurred in the same sentence. A sentence is included in the cluster when containing at least two of the words that are part of that cluster. The following image illustrates the clusters identified ( Figure 3 ).

Table 3 illustrates an example of each cluster.
The answers to the question were also evaluated using sentiment analysis based on the OpLexicon dictionary. The results show that around 47.0% of the comments about the current scenario of the field in Brazil are positive, 38.0% are neutral, and 15.0% are negative. Table 4 illustrates examples of each sentiment classification.
Perceptions about the genomics and/or bioinformatics future situation in Brazil
The second question to be evaluated was: What is your vision for the future for genomics and/or bioinformatics in Brazil . This question aimed to gather participants’ perceptions about the future situation of the field in Brazil, and it resulted in 507 valid responses after data cleaning. The analysis shows that the most common stem is “research” (19.3% of cases). Some researchers highlighted the potential of research in the field, but the need for more investments and funding:
Case 2: I believe it will improve, but as long as the remuneration and investment in RESEARCH are low, the tendency is to continue to lose recently graduated researchers abroad.
Others expanded the needs of research funding to other areas as well:
Case 197: Any vision of the future in the RESEARCH area in Brazil is very obscure and not very encouraging with the current government. I do not see a promising future for science in the country as a whole, including bioinformatics. However, I think that bioinformatics may be less affected than other areas that need funds for data generation and sequencing.
Furthermore, the second most common stem is “invest” (11.1% of cases), which occurs in words such as “investment” and the verb “to invest”. These stems occur in responses where the need for investment in the field is highlighted:
Case 2: I believe it will improve, but as long as the remuneration and INVESTMENT in research are low, the tendency is to continue to lose recently graduated researchers abroad.
Others suggest a positive view of the future of the area in Brazil considering investments already made:
Case 39: Extremely promising, with the emergence of new private-sector research and INVESTMENTS groups in the creation of new companies specializing in the area.
The next most frequent stem is “develop” (10.8% of cases), which occurs in words like “development”. Some responses highlighted the need of developing national technologies:
Case 7: Stop relying so much on technology and professionals from abroad, and be more consolidated in Brazil, both in the existence of professionals able to execute and DEVELOP new technologies in the area
Others believe this development is possible if there is enough investment:
Case 229: If there is support for research, very promising, with international partnerships for the DEVELOPMENT of products and shared databases
Figure 4 illustrates the most frequent whole words in the responses about the future of genomics and/or bioinformatics in Brazil.

The analysis was also extended to identify clusters with words co-occurring in the same sentence ( Figure 5 ). Table 5 illustrates an example of each cluster.

The sentiment analysis of the responses about the future of genomics and/or bioinformatics in Brazil resulted in around 48.0% of positive comments, 15.0% of negative comments, 37.0% of neutral comments. Table 6 illustrates examples of each sentiment classification.
Perceptions about genomics and/or bioinformatics situation in the world
The third question to be evaluated was: How do you see the field in the area of genomics and/or bioinformatics in the world? This question aimed to gather participants’ perceptions about the field of genomics and/or bioinformatics outside Brazil, and resulted in 507 valid responses after data cleaning. Once again, stemmed words were used to gather the most common words used by the respondents. The most frequent stem is “grow” (22.3% of the cases). A number of responses focused on talking about the area in the world by comparing the productivity of Brazilian science with other countries’:
Case 111: As there is a greater investment by private companies (and government, in some places in the world), I believe that the area of genomics/bioinformatics will have a more advanced GROWTH when compared to Brazil. There will be the development of new sequencing platforms and greater “popularisation” of their use. Along with this, I believe that there will also be a greater demand (and also development) for more advanced computational architectures than the current one.
Others highlighted the progress and growth of the area in emerging countries:
Case 143: Exponential GROWTH with the great development of tools, new technologies, and applications in the most diverse areas of knowledge. Expansion in Latin America and emerging countries.
The next most popular stem was “data” (9.5% of cases), mostly focusing on the amount of data being generated by the current analysis. Once again, some participants compared the Brazilian capacity of generating data with the other countries’ capacity:
Case 339: More advanced than in Brazil, mainly due to the ease in generating data in countries with cutting-edge science.
Others shared their experiences in working in other countries. These comments tend to emphasize how advanced the research in genomics and bioinformatics can be in some countries, highlighting the possibilities of development when researchers have easy access to the technologies they need:
Case 355: Very developed. I had the opportunity to do an internship in a Microbiome laboratory in the Czech Republic that only worked with cutting-edge genomics and bioinformatics. Researchers in the laboratory made it possible long-term work, and therefore, very relevant. In other areas, such as oncology, genomics and bioinformatics are considered the tools for decoding cancer. Large sequencing projects are common in the United States and Europe, with the possibility of free access to at least part of the DATA.
However, there seems to be a concern in relation to the large amount of data being generated by countries with access to cutting-edge technologies. Some researchers seem to worry about how this data is being analyzed and if these analyses can provide insightful results:
Case 19: I think it is well developed, but I have read many articles, mainly with sequencing data for bacterial genomes, with which I work, with doubtful or little clarified DATA. You see many articles now with studies on everything, many data being published but few with more in-depth and relevant content.
Others stated that the lack of proper analysis might be due to the lack of trained researchers, leading to poor data analysis:
Case 152: Promisingly and expanding. I note that the generation of genomic DATA is no longer a problem and can be done quickly and cheaply abroad, but there is a lack of people to do the data analysis (bioinformatics) with quality.
The following word cloud illustrates the most frequent words in the responses about the situation of genomics and/or bioinformatics in the world ( Figure 6 ).

The following shows the cluster of words that co-occurred in the same sentence in the responses about the situation of the field outside Brazil ( Figure 7 ), followed by an example of each cluster in Table 7 .

The sentiment analysis of the responses about the situation of genomics and/or bioinformatics outside Brazil resulted in around 48.0% of positive comments, 7.0% of negative comments, 45.0% of neutral comments. Table 8 illustrates examples of each sentiment classification.
Perceptions about the milestones of genomics and/or bioinformatics in Brazil
One of the questions present in the questionnaire was related to the main achievements of the field in Brazil: What are the scientific and technological milestones in the area of genomics and/or bioinformatics that you consider important in Brazil in the last 20 years?
This question resulted in 474 valid responses after data cleaning, and the responses were evaluated by content analysis. Based on a careful review of the responses, codes were retrieved and used to classify the texts. A number of 32 participants stated that they do not know or remember what the milestones are, and 4 participants said there are no milestones in the area in Brazil. These responses were excluded from the analysis. The most frequent code is related to programs and projects developed in the area (87.0%). Among those, the most cited project (22.3% of cases) was the ONSA network. The next most frequent category is the sequencing of specific organisms carried out by Brazilian researchers, such as human cancer, sugar cane, and other agriculture-related organisms.
Table 9 summarises the results of this content analysis, showing the main milestones of the field in Brazil.
The milestones highlighted by the participants of this study are in line with the historical achievements of the genomics and bioinformatics research in Brazil, as illustrated by Figure 8 .

After stating what the past milestones of the field of genomics and/or bioinformatics in Brazil are, the participants stated what they wish for the field in the future by answering the question: If you had a “wish list” of what could be done to improve research in genomics and/or bioinformatics in Brazil, what would you change? For example, sequencing platform, bioinformatics, training, collaborations, financing? Once again, we performed content analysis classifying the text based on codes retrieved from the responses ( Table 10 ). In line with some of the results identified previously, most of the participants responded they hope to get more financial investments in the field (56.8%), besides a reduction of research input prices and taxes, facilitating their purchase. Furthermore, a significant number of respondents reinforced the need for investments in formal education by creating more courses and specific training in the area of genomics and/or bioinformatics (48.2%).
The present study aimed to deliver an overview of the field of genomics and bioinformatics based on the perception of Brazilian experts. The field is relatively new and combines a variety of other areas, such as biology, computer science, statistics and mathematics, as an effort to answer biological queries. When looking at the demographics outcomes of our study, the results suggest there is a balance in terms of participants’ gender. This is a positive result, especially considering that, in the world, only 28.0% of researchers in Science, Technology, Engineering and Mathematics (STEM) are female ( UNESCO, 2018 ). However, few of the respondents are over 50 years old, which might suggest that the field is still young in Brazil. There is a concentration of researchers in São Paulo and very few outside the South and Southeast regions of Brazil. That is in line with other areas of research, especially considering the high concentration of research funding in these areas. According to the GEOCAPES - Georeferenced Information System , the number of research scholarships in the South and Southeast areas of Brazil can be up to 10 times higher than in the North and Northeast areas. A recent report about science production in Brazil shows that 70% of the expenses with research and development are concentrated in São Paulo ( Centro de Gestão e Estudos Estratégicos, CGEE, 2021 . This can also be attributed to the relevance of FAPESP for the state, considering the foundation provides funding support not only for research and innovation, but also for granting scholarships for graduate students and tools for the insertion of researchers in companies. While setting up any new program, whether academic or service orientated, is a challenging task, together with some established resources that will aid in the development of a bioinformatics program in different Brazilian regions. In an era driven by data science, the need for bioinformatics research and service activities within academic institutes is essential to ensure equal opportunities for competitive research funding. Brazil has attempted to decrease the difference between regions. For example, it is mandatory that at least 30% of all science and technology funding go to the North, Northeast and Midwest regions of the country (Decree-Law 719/69). Among the thirteen bioinformatics networks funded by Capes from 2014 to 2019, one, one and two were from the North, Midwest and Northeast regions, respectively. A direct consequence of this action was the establishment of the Ms/PhD program at the Federal University of Rio Grande do Norte in 2016. Since the importance of genomics and bioinformatics continues to grow, initiatives like the one above are important and should be continued”.
Most participants have a background in Biological Sciences and tend to have their first contact with the area during their PhD. Since computer scientists and mathematicians are important for the development of the area, the above information is a sign that specific policies for higher involvement of these professionals should be developed. “Using and producing genetic material” was selected by 91.0% of the participants. This illustrates the importance of data access and generation to this field. However, as highlighted by the respondents in the open-ended questions, this large amount of data needs to be adequately evaluated to generate proper insights. The reduction of the costs of biological systems profiling leads the field to a path of “big data”, and the development of efficient methodologies, such as machine learning techniques and computational power, are necessary to generate valuable results ( Greene et al., 2014 ; Yin et al., 2017 ; Nazipova et al., 2018 ; Aron et al., 2021 ).
The views of the field in Brazil are positive, both for the current situation and for the future. However, compared to the opinions about the field abroad, the respondents had almost double negative opinions. The positive views can be identified by the perceptions of a growing, promising research field. It is clear there is a need for funding investments for the future, as stated in the participants’ wish list. Nevertheless, this scenario seems to be unlikely - the year 2021 was hit by a reduction in science funding ( Pires, 2020 ; Quintans-Júnior et al., 2021 ). This can be a disadvantage not only for the research field but also for the economy, as the global market for genomics is expected to reach USD 54.4 billion by 2025. Even with financial challenges, Brazilian researchers managed to work and develop relevant programs and projects in the area, which were highlighted by the participants when talking about the milestones in the field. National projects generate important results considering the specificities of the country’s industry and population, allowing researchers to search for personalized solutions ( Salzano, 2018 ; Giugliani et al., 2019 ).
Although research output in genomics and bioinformatics has significantly increased in the last decade in Latin America, impact and quality is still a matter of concern. A comparative analysis of the data presented here with similar initiatives in other Latin American countries ( Blas et al., 2011 ; Bicudo, 2016 ; De Las Rivas et al., 2019 ; Zambrano-Mila et al., 2019 ; Armenta-Medina et al., 2020 ) has revealed some interesting patterns. One of them is the recognition for a better characterization of the genetic structure of the region’s population, a theme that can be deeply explored by genomics and bioinformatics. Another common theme among different countries is the lack of computational infrastructure, which could be minimized by establishing new transnational networks (and an improvement in funding of the existing networks) with common infrastructure. The establishment of undergraduate bioinformatics electives can be implemented within the long-term context of building significant capacity to create a graduate bioinformatics program. Investments in infrastructure support can make important contributions to the advancement of biomedical research, agriculture, among others, through the association of different applications of bioinformatics techniques. Ongoing support through Networking also presents opportunities for collaborative research. This type of initiative proved to be extremely important for the emergence and maintenance of the area in Brazil and should be permanently encouraged. Finally, genomics/bioinformatics communities in several countries recognized the huge potential of both areas and the need for continuous educational efforts.
The study does not aim to be exhaustive and has limitations. First, we only covered around half of the population expected to be working in the field, so results reflect the perceptions of a sample of the professionals active in the area. Furthermore, there are still limitations related to text mining studies in the Portuguese language, especially due to the limitation of the lexicon available. Nevertheless, this study contributes to the body of knowledge, offering details about the current situation and future expectations of the professionals from the field considering a diverse analysis that includes multiple methods. This type of analysis allows the development of public policy and industry initiatives that can support the development of the field based on the perceptions of their stakeholders. This manuscript and the accompanying data will be forwarded to the major funding agencies in Brazil.
Acknowledgments
We would like to thank those who took the time to answer the survey. This research was supported by FINEP (grant no. 01.16.0078.00). This work was developed in the frameworks of the Brazilian National Institute of Public Communication of Science and Technology, with support by the National Council for Scientific and Technological Development (CNPq, 465658/2014-8) and the Research Support Carlos Chagas Filho Foundation of Rio de Janeiro (FAPERJ, E-26/200.899/2018). This survey was carried out to celebrate 20 years of genomics and bioinformatics in Brazil, which took place in December 2020 (https://bioinfo.imd.ufrn.br/genobio20/). A.T.R.V. is supported by CNPq (303170/2017-4) and FAPERJ (26/202.903/20). S.J.S. is supported by CNPq (309475/2020-1). L.M. is supported by CNPq (304156/2020-5) and FAPERJ (E-26/202.816/2017).
Supplementary Material
The following online material is available for this article:
- Araujo DB, Machado RRG, Amgarten DE, Malta F de M, de Araujo GG, Monteiro CO, Candido ED, Soares CP, de Menezes FG, Pires ACC, et al. SARS-CoV-2 isolation from the first reported patients in brazil and establishment of a coordinated task network. Mem Inst Oswaldo Cruz. 2020; 115 :e200342. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Armenta-Medina D, de Leon-Castañeda CD, Valderrama-Blanco B. Bioinformatics in Mexico: A diagnostic from the academic perspective and recommendations for a public policy. PLoS One. 2020; 15 :e0243531. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Aron S, Jongeneel CV, Chauke PA, Chaouch M, Kumuthini J, Zass L, Radouani F, Kassim SK, Fadlelmola FM, Mulder N. Ten simple rules for developing bioinformatics capacity at an academic institution. PLoS Comput Biol. 2021; 17 :e1009592. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Bicudo E. Genomics politics through space and time: The case of bioinformatics in Brazil. Public Health Genomics. 2016; 19 :81–92. [ PubMed ] [ Google Scholar ]
- Blas MM, Curioso WH, Garcia PJ, Zimic M, Carcamo CP, Castagnetto JM, Lescano AG, Lopez DM. Training the biomedical informatics workforce in Latin America: Results of a needs assessment. BMJ Open. 2011; 1 :e000233 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Buss LF, Prete CA, Jr, Abrahim CMM, Mendrone A, Jr, Salomon T, De Almeida-Neto C, França RFO, Belotti MC, Carvalho MPSS, Costa AG, et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021; 371 :288–292. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Bustamante CD, Burchard EG, De La Vega FM. Genomics for the world. Nature. 2011; 475 :163–165. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Candido DS, Claro IM, de Jesus JG, Souza WM, Moreira FRR, Dellicour S, Mellan TA, du Plessis L, Pereira RHM, Sales FCS, et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020; 369 :1255–1260. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Centro de Gestão e Estudos Estratégicos (CGEE) Boletim Anual OCTI. Vol. 1. Brasília: 2021. Panorama da ciência brasileira: 2015-2020.200 [ Google Scholar ]
- Chasapi A, Promponas VJ, Ouzounis CA. The bioinformatics wealth of nations. Bioinformatics. 2020; 36 :2963–2965. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- De Las Rivas J, Bonavides-Martínez C, Campos-Laborie FJ. Bioinformatics in Latin America and SoIBio impact, a tale of spin-off and expansion around genomes and protein structures. Brief Bioinform. 2019; 20 :390–397. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DS, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021; 372 :815–821. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Gauthier J, Vincent AT, Charette SJ, Derome N. A brief history of bioinformatics. Brief Bioinform. 2019; 20 :1981–1996. [ PubMed ] [ Google Scholar ]
- Giugliani L, Vanzella C, Zambrano MB, Donis KC, Wallau TKW, da Costa FM, Giugliani R. Clinical research challenges in rare genetic diseases in Brazil. Genet Mol Biol. 2019; 42 :305–311. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Given LM, Saumure K, editors. The SAGE encyclopedia of qualitative research methods. SAGE Publications; Los Angeles: 2008. 1014 [ Google Scholar ]
- Gonçalves P, Araújo M, Benevenuto F, Cha M. Comparing and combining sentiment analysis methods. COSN. 2013; 13 :27–37. [ Google Scholar ]
- Greene CS, Tan J, Ung M, Moore JH, Cheng C. Big data bioinformatics. J Cell Physiol. 2014; 229 :1896–1900. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409 :860–921. [ PubMed ] [ Google Scholar ]
- Lewis RB, Maas SM. QDA Miner 2.0: Mixed-model qualitative data analysis software. Field Methods. 2007; 19 :87–108. [ Google Scholar ]
- Lovins JB. Development of a stemming algorithm. Mech Transl Comput Linguist. 1968; 11 :22–31. [ Google Scholar ]
- Morel CM. Reaching maturity - 25 years of the TDR. Parasitol Today. 2000; 16 :522–528. [ PubMed ] [ Google Scholar ]
- Nazipova NN, Isaev EA, Kornilov V V, Pervukhin DV, Morozova AA, Gorbunov AA, Stinin MN. Big data in bioinformatics. Math Biol Bioinf. 2018; 12 :102–119. [ Google Scholar ]
- Neuendorf KA, Kumar A. Content analysis. Encycl Polit Commun. 2015; 1 :1–10. [ Google Scholar ]
- Niwattanakul S, Singthongchai J, Naenudorn E, Wanapu S. Using of Jaccard coefficient for keywords similarity. Lect Notes Eng Comput Sci. 2013; 2202 :380–384. [ Google Scholar ]
- Patrinos GP, Pasparakis E, Koiliari E, Pereira AC, Hünemeier T, Pereira LV, Mitropoulou C. Roadmap for establishing large-scale genomic medicine initiatives in low- and middle-income countries. Am J Hum Genet. 2020; 107 :589–595. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Prabowo R, Thelwall M. Sentiment analysis: A combined approach. J Informetr. 2009; 3 :143–157. [ Google Scholar ]
- Quintans-Júnior LJ, Albuquerque GR, Oliveira SC, Silva RR. Brazil’s research budget: Endless setbacks. EXCLI J. 2021; 19 :1322–1324. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ramírez JL, González A, Cantú JM, Chavez-Crooker P, Leiva JC, Blamey JM, Cortes H, Holmes D. Latin American Genome Initiative, the creation of a network and web based resource to aid and nurture genome biology in developing countries. Electron J Biotechnol. 2002; 5 :203–204. [ Google Scholar ]
- Salzano FM. The evolution of science in a Latin-American country: Genetics and genomics in Brazil. Genetics. 2018; 208 :823–832. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Silva Francisco R, Jr, Benites LF, Lamarca AP, de Almeida LGP, Hansen AW, Gularte JS, Demoliner M, Gerber AL, Guimarães AP de C, Antunes AKE, et al. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2020; 296 :198345 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Simpson AJG. Genome sequencing networks. Nat Rev Genet. 2001; 2 :979–983. [ PubMed ] [ Google Scholar ]
- Simpson AJG, Perez JF. ONSA, the São Paulo Virtual Genomics Institute. Organization for nucleotide sequencing and analysis. Nat Biotechnol. 1998; 16 :795–796. [ PubMed ] [ Google Scholar ]
- Simpson AJG, Camargo AA, Ferro JA, Parra J, Vasconcelos AT. Coordinated, network-based research as a strategic component of science in Brazil. Genet Mol Res. 2004; 3 :18–25. [ PubMed ] [ Google Scholar ]
- Simpson AJG, Reinach FC, Arruda P, Abreu FA, Acencio M, Alvarenga R, Alves LMC, Araya JE, Baia GS, Baptist CS, et al. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa Cosortium of the Organization for Nucleotide Sequencing and Analysis. Nature. 2000; 406 :151–159. [ PubMed ] [ Google Scholar ]
- Souza M, Vieira R. Sentiment analysis on twitter data for portuguese language. Lect Notes Comput Sci. 2012; 7243 :241–247. [ Google Scholar ]
- Souza M, Vieira R, Chishman R, Alves IM. Proceedings of the 8th Brazilian Symposium in Information and Human Language Technology. 2011. Construction of a Portuguese Opinion Lexicon from multiple resources; pp. 59–66. [ Google Scholar ]
- UNESCO . Decifrar o código: Educação de meninas e mulheres em ciências, tecnologia, engenharia e matemática (STEM) UNESCO; Brasília: 2018. 84 [ Google Scholar ]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001; 291 :1304–1351. [ PubMed ] [ Google Scholar ]
- Voloch CM, da Silva Francisco R, Jr, de Almeida LGP, Cardoso CC, Brustolini OJ, Gerber AL, Guimarães APC, Mariani D, da Costa RM, Ferreira OC, Jr, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol. 2021; 95 :e00119-21 [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Weissenbach J. The rise of genomics. C R Biol. 2016; 339 :231–239. [ PubMed ] [ Google Scholar ]
- Xavier ERC, Capanema BPX, Ruiz JC, Oliveira G, Meyer R, D’Afonseca V, Miyoshi A, Azevedo V. Brazilian genome sequencing projects: State of the art. Recent Pat DNA Gene Seq. 2008; 2 :111–132. [ PubMed ] [ Google Scholar ]
- Yin Z, Lan H, Tan G, Lu M, Vasilakos AV, Liu W. Computing platforms for big biological data analytics: Perspectives and challenges. Comput Struct Biotechnol J. 2017; 15 :403–411. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Zambrano-Mila MS, Agathos SN, Reichardt JKV. Human genetics and genomics research in Ecuador: Historical survey, current state, and future directions. 64 Hum Genomics. 2019; 13 [ PMC free article ] [ PubMed ] [ Google Scholar ]
Internet Resources
- Concessão de Bolsas de pós-graduação da Capes no Brasil. 2019. [29 August 2021]. GEOCAPES - Sistema de Informações Georreferenciadas (2019) Concessão de Bolsas de pós-graduação da Capes no Brasil, https://geocapes.capes.gov.br/geocapes/ [ Google Scholar ]
- Jesus JG de, Sacchi C, Claro I, Salles F, Manulli E, Silva D da, Paiva TM de, Pinho M, Afonso AMS, Mathias A, et al. First cases of coronavirus disease (COVID-19) in Brazil, South America (2 genomes, 3rd March 2020) [29 August 2021]; Genome Reports. 2020 Genome Reports, https://virological.org/t/first-cases-of-coronavirus-disease-covid-19-in-brazil-south-america-2-genomes-3rd-march-2020/409 . [ Google Scholar ]
- Pires B. Ciência brasileira sofre com cortes de verbas e encara cenário dramático para pesquisas em 2021. [29 August 2021]; El País. 2020 El País, https://brasil.elpais.com/brasil/2020-12-31/ciencia-brasileira-sofre-com-cortes-de-verbas-e-encara-cenario-dramatico-para-pesquisas-em-2021.html . [ Google Scholar ]
Life-cycle Forces make Monetary Policy Transmission Wealth-centric
This paper adds life-cycle features to a New Keynesian model and shows how this places financial wealth at the center of consumption/saving decisions, thereby enriching the determinants of aggregate demand and affecting the transmission of monetary policy. As retirement preoccupations strengthen, the potency of conventional monetary policy declines and depends more on the response of asset prices (supporting central banks closely monitoring the impact of monetary policy on asset prices). Especially “low/high for long” policies are shown to often have only muted effects on economic activity due to offsetting income and substitution effects of interest rates, in a way that can be compounded by Quantitative Easing. We also show why the presence of life-cycle forces can favor a monetary policy strategy which stabilizes asset prices in response to financial shocks. Being explicit about the role of retirement savings in aggregate demand therefore offers new perspectives on several aspects of monetary interventions.
We thank Ricardo Caballero, Ben Moll, Thijs Knaap, Jean-Paul L'Huillier, Tatiana Kirsanova, Alessandro Rebucci, audiences at the 2023 SED conference in Cartagena, the 5th WMMF Conference in Warsaw, the 2023 PSE Macro Days, the Reserve Bank of Australia, and the Bank of England for useful comments and discussions. The views expressed in this paper are those of the authors, and not necessarily those of the BIS, the Bank of England or its committees. The views expressed herein are those of the authors and do not necessarily reflect the views of the National Bureau of Economic Research.
MARC RIS BibTeΧ
Download Citation Data
More from NBER
In addition to working papers , the NBER disseminates affiliates’ latest findings through a range of free periodicals — the NBER Reporter , the NBER Digest , the Bulletin on Retirement and Disability , the Bulletin on Health , and the Bulletin on Entrepreneurship — as well as online conference reports , video lectures , and interviews .


IMAGES
VIDEO
COMMENTS
Bioinformatics is a field of study that uses computation to extract knowledge from biological data. It includes the collection, storage, retrieval, manipulation and modelling of data for analysis ...
Bisulfite sequencing (BS-Seq) is a fundamental technique for characterizing DNA methylation profiles. Genotype calling from bisulfite-converted BS-Seq data allows allele-specific methylation analysis and the c... Yance Feng and Fei Gao. BMC Bioinformatics 2024 25 :206. Research Published on: 5 June 2024. Full Text.
These articles reflect current trend and development in bioinformatics research. The supplement to BMC Bioinformatics was proposed to launch during the BIOCOMP'19—The 2019 International Conference on Bioinformatics and Computational Biology held from July 29 to August 01, 2019 in Las Vegas, Nevada. In this congress, a variety of research ...
Bioinformatics is an official journal of the International Society for Computational Biology, the leading professional society for computational biology and bioinformatics. Members of the society receive a 15% discount on article processing charges when publishing Open Access in the journal. Read papers from the ISCB. Find out more.
Bioinformatics and Biology Insights is an open access, peer-reviewed journal that considers articles on bioinformatics methods and their applications, which must pertain to biological insights. All papers should be easily amenable to biologists and, as such, help bridge the gap between theories and applications. View full journal description
This is an editorial report of the supplements to BMC Bioinformatics that includes 6 papers selected from the BIOCOMP'19—The 2019 International Conference on Bioinformatics and Computational Biology. These articles reflect current trend and development in bioinformatics research. Keywords: Bioinformatics, Biomarkers, Human disease, Microbiome.
Publishes scientific papers and review articles on new developments in bioinformatics and computational biology. Shorter papers report biologically interesting discoveries using computational methods and explore their applications.
Recent Advances of Deep Learning in Bioinformatics and Computational Biology. Binhua Tang 1,2 * † Zixiang Pan 1 † Kang Yin 1 Asif Khateeb 1. 1 Epigenetics & Function Group, Hohai University, Nanjing, China. 2 School of Public Health, Shanghai Jiao Tong University, Shanghai, China. Extracting inherent valuable knowledge from omics big data ...
We focused our search on the most relevant journals for bioinformatics and translational informatics with electronic publication dates on or after January 1, 2021. The journals surveyed for best papers are as follows: Journal of the American Medical Informatics Association (JAMIA), Journal of Biomedical Informatics (JBI), PLoS Computational ...
Note the related bioinformatics tools papers [20,21] published in the Frontiers in Genetics special issue "Bioinformatics of Genome Regulation and Systems Biology" , and BMC Genomics issue . The research topic on gene expression regulation in Frontiers in Genetics is continued in 2020.
Bioinformatics combines the methods used in the collection, storage, identification, analysis, and correlation of this huge and complex information. All this work produces an "ocean" of information that can only be "sailed" with the help of computerized methods. The goal is to provide scientists with the right means to explain normal biological ...
Song et al. reported the complete genome sequence of two rice subspecies, japonica and indica, in 2005 that laid a strong foundation for molecular studies and plant breeding research [22, 24]. With recent advancement in bioinformatics, it is now possible to run the sequence alignment between large and complex genome from other crop species with ...
Briefings in Bioinformatics | 25 | 4 | May 2024. Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
An innovative journal that provides a forum for new discoveries in bioinformatics. It focuses on how new tools and applications can bring insights to specific biological problems. ... Research Topics. Submission open Integrating Bioinformatics and AI to Natural Product-based Drug Discovery and Development. Leandro de Mattos Pereira; Cristiano ...
Systems clinical medicine is recommended as one of new strategies for the development of cancer biomarkers. Systems clinical medicine is coined as the integration of systems biology, clinical phenotypes, high-throughout technologies, bioinformatics and computational science to improve diagnosis, therapies and prognosis of diseases.
Long‐Term Administration of Omeprazole‐Induced Hypergastrinemia and Changed Glucose Homeostasis and Expression of Metabolism‐Related Genes. Alina Kabaliei,Vitalina Palchyk,Olga Izmailova,Viktoriya Shynkevych,Oksana Shlykova,Igor Kaidashev, First Published:  30 May 2024. Abstract.
The research team acknowledges, however, that this connection is speculative and awaits specific experimental validation. "The understanding that the subcellular details of the NMDA receptor can lead to increased gamma oscillations was the basis for a new theory about how ketamine may work for treating depression," Kopell says.
Integration of bioinformatics data on gene expression, mutagenesis, and pathway interaction analysis form a trend in recent medical genomics studies [12,13]. Immunohistochemistry research in cancer was presented in papers by Anastasiya Snezhkina and colleagues [ 22 , 23 ] in the journal issues of the "Medical Genetics, Genomics and ...
Seeing its effectiveness in areas like the hippocampus opened an entirely new way for us to deliver precise stimulation to targeted circuits in the brain," says Steve Ramirez, an assistant professor of psychological and brain sciences at Boston University, and a faculty member at B.U.'s Center for Systems Neuroscience, who is also an author ...
If 2023 was the year the world discovered generative AI (gen AI), 2024 is the year organizations truly began using—and deriving business value from—this new technology.In the latest McKinsey Global Survey on AI, 65 percent of respondents report that their organizations are regularly using gen AI, nearly double the percentage from our previous survey just ten months ago.
Broadband Internet Access, Economic Growth, and Wellbeing. Kathryn R. Johnson & Claudia Persico. Working Paper 32517. DOI 10.3386/w32517. Issue Date May 2024. Between 2000 and 2008, access to high-speed, broadband internet grew significantly in the United States, but there is debate on whether access to high-speed internet improves or harms ...
Today, OpenAI released a new research paper apparently aimed at showing it is serious about tackling AI risk by making its models more explainable. In the paper, researchers from the company lay ...
Issue Date May 2024. In a collaborative project with ten central banks, we have investigated the causes of the post-pandemic global inflation, building on our earlier work for the United States. Globally, as in the United States, pandemic-era inflation was due primarily to supply disruptions and sharp increases in the prices of food and energy ...
According to (Chasapi et al., 2020), Brazil is part of the ranking listing of 40 leading countries in bioinformatics, and occupies position 23 of the top 1% of highly cited papers. Although research in bioinformatics and genomics in Brazil had positive and relevant features during the last two decades, the country still faces challenges common ...
Life-cycle Forces make Monetary Policy Transmission Wealth-centric. Paul Beaudry, Paolo Cavallino & Tim Willems. Working Paper 32511. DOI 10.3386/w32511. Issue Date May 2024. This paper adds life-cycle features to a New Keynesian model and shows how this places financial wealth at the center of consumption/saving decisions, thereby enriching ...