- Publications
- Conferences & Events
- Professional Learning
- Science Standards
- Awards & Competitions
- Instructional Materials
- Free Resources
- American Rescue Plan
- For Preservice Teachers

NCCSTS Case Collection
- Science and STEM Education Jobs
- Interactive eBooks+
- Digital Catalog
- Regional Product Representatives
- e-Newsletters
- Bestselling Books
- Latest Books
- Popular Book Series
- Prospective Authors
- Web Seminars
- Exhibits & Sponsorship
- Conference Reviewers
- National Conference • Denver 24
- Leaders Institute 2024
- National Conference • New Orleans 24
- Submit a Proposal
- Latest Resources
- Professional Learning Units & Courses
- For Districts
- Online Course Providers
- Schools & Districts
- College Professors & Students
- The Standards
- Teachers and Admin
- eCYBERMISSION
- Toshiba/NSTA ExploraVision
- Junior Science & Humanities Symposium
- Teaching Awards
- Climate Change
- Earth & Space Science
- New Science Teachers
- Early Childhood
- Middle School
- High School
- Postsecondary
- Informal Education
- Journal Articles
- Lesson Plans
- e-newsletters
- Science & Children
- Science Scope
- The Science Teacher
- Journal of College Sci. Teaching
- Connected Science Learning
- NSTA Reports
- Next-Gen Navigator
- Science Update
- Teacher Tip Tuesday
- Trans. Sci. Learning
MyNSTA Community
- My Collections
Case Study Listserv
Permissions & Guidelines
Submit a Case Study
Resources & Publications
Enrich your students’ educational experience with case-based teaching
The NCCSTS Case Collection, created and curated by the National Center for Case Study Teaching in Science, on behalf of the University at Buffalo, contains over a thousand peer-reviewed case studies on a variety of topics in all areas of science.
Cases (only) are freely accessible; subscription is required for access to teaching notes and answer keys.
Subscribe Today
Browse Case Studies
Latest Case Studies

Development of the NCCSTS Case Collection was originally funded by major grants to the University at Buffalo from the National Science Foundation , The Pew Charitable Trusts , and the U.S. Department of Education .
Using Case Studies with Large Classes
Why Use Case Studies?
Case studies are powerful tools for teaching. They explore the story behind scientific research to understand the phenomenon being studied, the question the scientist asked, the thinking they used to investigate it, and the data they collected to help students better understand the process and content of science.
A strength of this approach is that it gives students the chance to consider how they would investigate a topic. Their answers are often similar to what the researchers being studied did. But students also come up with novel perspectives and unique approaches to the problems.
Many BioInteractive resources lend themselves to a case study approach. In most instances, what I ultimately decide is to convert the resource into a case study. For example, the video Animated Life: Mary Leakey is an excellent tool to get students thinking about the logic scientists use to study fossils and extinct species. Data Point resources are also a rich source of figures and questions that can be copied and pasted into a presentation to provide a brief case study that introduces a topic.
The Challenge for Large Classes
Many BioInteractive activities are structured in a way that they are particularly useful for smaller groups and classes. And by smaller, I am thinking of fewer than 50 students. To some colleagues, that may seem to be a large class size. Indeed, in many instances, it probably is more than is optimal.
However, when I refer to large classes, what I am thinking of are the large introductory classes encountered in many colleges and universities in which enrollment can range from 100 to 500 or more depending on the institution. Classes of this size present instructors with the dual challenges of not just numbers but also anonymity. It’s logistically unmanageable to share and distribute printed copies of handouts or worksheets.
How to Scale Up
So how can an instructor promote the interaction that is essential to the success of these types of case study activities in such a large group? These are issues I grappled with when I went from teaching at a small liberal arts college where my classes were smaller than 30 to teaching at a large university with classes of several hundreds. I have found what I think are four parts to an effective solution.
1. Define a learning objective.
First and foremost, whether I have 30 or 300 students, I try to think about why I want to use a particular BioInteractive resource. I consider what it is that I want the students to do or think about while using the resource. How do I want them to be different after completing the assignment? In essence, I define the learning objective so I can determine the most effective platform and approach to deliver the lesson utilized in the resource.
2. Create presentations with strategic pause points.
PowerPoint is a common tool for delivering material in large classrooms. It is quite easy to take images and questions from BioInteractive resource PDFs and insert them into slides. After reading the teaching notes and text in the student handouts, it’s relatively simple to develop the story that weaves the slides together in an interrupted case study. This is a style of case study that progressively leads students through the information with carefully planned “reveals” of information and strategically placed questions as stopping points to ponder the material along the way.
Videos are also fabulous resources to use during interrupted case studies in class. For example, I regularly use the video Niche Partitioning and Species Coexistence , which describes Dr. Rob Pringle’s work on niche partitioning in the savanna, as the core of a video case study in class. After the class watches the video for a few minutes, I stop and ask students about the phenomenon being studied and approaches that could be used to answer different questions.
I often use the following questions/prompts:
- Why would anyone care about factors shaping species presence or absence?
- Think about what factors could be important influences on shaping species richness in a community.
How can we use modern techniques to study what an animal is eating when we can’t watch the animal eat? The video does an excellent job of addressing these topics and showing how researchers developed a creative approach to applying molecular techniques to answer ecological questions. How awesome is it that one video can help students tie together the central dogma, ecological theory, and community concepts! Depending on how much an instructor wants to structure the video case study in advance, it is even possible to embed small video clips and questions directly into a PowerPoint presentation.
3. Have students use clickers.
How should we tell the scientific story to large numbers of students and engage them in it? Clickers are a particularly helpful tool for asking questions about experiments, concepts, or results, because they present students with a specific moment when they need to choose among different options for a survey of their opinion or decide among right and wrong answers in a multiple-choice question.
For example, I typically start a case study with survey questions asking students to identify what they think is the most important item on a list of potential phenomena or to give their feedback about an issue in a Likert-scale response. Later, as the case study develops, I ask more specific questions about the experiment that require students to predict experimental outcomes or interpret a figure. For example, when I use the video The Effects of Fungicides on Bumble Bee Colonies , I show students several bar graphs with possible outcomes for the experiment and have them pick which they think the researchers will observe. After revealing the actual results, I ask them questions about interpreting the results and whether the results support the experimental hypothesis. I always allow students to talk and help one another during clicker questions to enhance their interaction and give them a choice to go along with a group opinion or answer based on their individual thinking.
4. Flip the classroom.
Another effective way to use BioInteractive resources in large classes is to use videos to flip a class session. BioInteractive animations and short films are rich with information that can pique interest, start discussions, or provide fundamental information. For example, I recently had my students watch the Genes as Medicine short film outside of class time. I asked them to then imagine they were an alien that found this video clip and to consider what information it would give them about life on Earth. This sparked a lively discussion about what life is to start the next class meeting that was more interesting than me going through a checklist of terms and definitions. Students had to uncover the characteristics of life from the video for themselves.
Benefits and Takeaways
What I hope these hints and suggestions from my own experiences show is how relatively simple it can be to scale up these resources to engage a class of any size. When they first encounter case studies, students can be a little unsure about this approach that requires them to talk to one another in a setting where they are expecting to be a face in the crowd. However, after they experience one or two case studies, I can see groups of students talking and exchanging ideas about the case. They are no longer passive listeners sitting in a room but instead have become active problem solvers seeking answers together. I can leave the stage and mingle through the room to listen to their discussions and encourage them as they develop their answers. This also gives me an opportunity to interact with students besides those sitting in the front row and to further develop a sense of community and connection, solving one of the challenges with big classes: anonymity.
It has been my experience that students quickly adapt to and begin to enjoy this approach. Rather than sitting in class watching yet another series of PowerPoint slides flash by, they are thinking and talking about science with one another. After my students talk things through with their neighbors and “shoulder buddies” during a case study, I find that they are more likely to speak up in class during the case study and at other points during the course.
At the beginning of the semester, I can barely get anyone to answer a question. After a few case studies, students begin asking and answering questions (even when we aren’t doing case studies), and the level of participation by different students in the room is noticeably higher. So in addition to case studies being a more interesting way for me as the teacher to present material to students and explore different biological topics, this approach also has the added benefits of helping build confidence within individual students and community among students, which makes a more rewarding and exciting learning environment for everyone.
Educational case studies based on examples of simulated or real research data can engage students in the process of thinking like a scientist, even when it is not possible to get into the field or laboratory to actually run an experiment. They can help overcome the challenges of data analysis and interpretation that are at the core of science education experiences. The collections of different resources available through HHMI BioInteractive provide a menu of modules for instructors to choose from that do just that. They get students to explore important biological topics from a variety of different approaches and look at the world through the lenses of different scientists. Regardless of what the actual format of a resource is when I encounter it, I know that it is possible to scale it up in some way to meet the needs of my classes.
Come join a conversation about this blog post at our Facebook group!
Phil Gibson is a professor at the University of Oklahoma, where he enjoys teaching his students that learning a little botany never hurt anyone and is probably good for them in the long run. When he’s not thinking about new resources to use in class, he enjoys hiking with his family, listening to music, and cooking outrageously large breakfasts on the weekends.
Related Articles
Cindy Gay presents BioInteractive's short video clip demonstrating the fruit fly courtship dance and its role in selection. She describes how she uses the clip in her AP® Biology curriculum, and how it connects to topics ranging from animal behavior to sympatric speciation.
Kim Parfitt describes two activities (now merged into the activity “Scientific Inquiry and Data Analysis Using WildCam Gorongosa”) associated with the WildCam Gorongosa project. She also discusses a short film on lion populations in Gorongosa that she uses to introduce the topic.
In this video Educator Voices post, hear from St. John Fisher College professor Kaitlin Bonner about how she uses a publicly available data set, along with BioInteractive’s elephant resources, to have her students investigate data.
Let your curiosity lead the way:
Request Info
- Arts & Sciences
- Graduate Studies in A&S
Case Studies in Biology: Climate and Health Exploration Course
This is an online course offered in the summer and open to current high school students.
As scientists, we take a lot of STEM classes, including biology, chemistry, physics, and math. But we often don’t have time to connect all of this information together. That’s where case studies are so incredibly helpful especially to organizations such as the CDC and World Health Organization. This course will use real world examples to help teach students about the scientific process and how theories and hypotheses are developed. Sometimes the answers aren’t clear, and even experts can’t agree. Using case studies focused on climate and it's connection to health, we will analyze data and apply biology concepts to learn about how to form a solid argument, supported by evidence from published research. This is your chance to learn how to conduct systematic literature reviews and meta-analyses to analyze scientific controversies and develop your own theories. Students with an interest in both biology and environmental science are welcome.
Prerequisite : none
Experience WashU From Home
Our online Exploration Courses are ideal for students looking for the flexibility of an online experience to build college readiness skills. Courses provide students an introduction to many of the majors, fields of study, and interdisciplinary programs offered by the College of Arts & Sciences.

- Introduction to Environmental Science Exploration Course
- Environmental Studies Institute

Case Studies in Systems Biology
- © 2021
- Pavel Kraikivski 0
Academy of Integrated Science, Division of Systems Biology, Virginia Polytechnic Institute and State University, Blacksburg, USA
You can also search for this editor in PubMed Google Scholar
- One of the first book on systems biology for the classroom
- Offers flexible way to choose the topics
- Each chapter can be used as an independent project
- Features contributions from key scientists in the field, such as Drs. John Tyson, Alex Mogilner nad Jae Kyoung Kim
14k Accesses
4 Citations
14 Altmetric
This is a preview of subscription content, log in via an institution to check access.
Access this book
- Available as EPUB and PDF
- Read on any device
- Instant download
- Own it forever
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Other ways to access
Licence this eBook for your library
Institutional subscriptions
Table of contents (18 chapters)
Front matter, mitotic cycle regulation. i. oscillations and bistability.
- John J. Tyson
Mitotic Cycle Regulation. II. Traveling Waves
Cell cycle regulation. bifurcation theory, glycolytic oscillations.
Pavel Kraikivski
NF-κB Spiky Oscillations
Tick, tock, circadian clocks.
- Jae Kyoung Kim
Spruce Budworm and the Forest
- Lauren M. Childs
Modeling cAMP Oscillations in Budding Yeast
- Amogh Jalihal
Synchronization of Oscillatory Gene Networks
The lac operon, fate decisions of cd4+ t cells.
- Andrew Willems, Tian Hong
Stochastic Gene Expression
Collective molecular motor transport.
- Christopher Miles, Alex Mogilner
Principle of Cooperativity in Olfactory Receptor Selection
- Jianhua Xing, Hang Zhang
Applying Quantitative Systems Pharmacology (QSP) Modeling to Understand the Treatment of Pneumocystis
- Tongli Zhang
Virus Dynamics
- Stanca M. Ciupe, Jonathan E. Forde
Identifying Virus-Like Regions in Microbial Genomes Using Hidden Markov Models
- Frank O. Aylward
Computational Software
Back matter.
- Dynamic modeling
- Biological Networks
- Bio-systems
- Genetic Networks
- Synthetic biology
- Signaling Pathways
About this book
This book provides case studies that can be used in Systems Biology related classes. Each case study has the same structure which answers the following questions: What is the biological problem and why is it interesting? What are the relevant details with regard to cell physiology and molecular mechanisms? How are the details put together into a mathematical model? How is the model analyzed and simulated? What are the results of the model? How do they compare to the known facts of the cell physiology? Does the model make predictions? What can be done to extend the model? The book presents a summary of results and references to more relevant sources.
The volume contains the classic collection of topics and studies that are well established yet novel in the systems biology field.
Editors and Affiliations
About the editor, bibliographic information.
Book Title : Case Studies in Systems Biology
Editors : Pavel Kraikivski
DOI : https://doi.org/10.1007/978-3-030-67742-8
Publisher : Springer Cham
eBook Packages : Computer Science , Computer Science (R0)
Copyright Information : Springer Nature Switzerland AG 2021
Hardcover ISBN : 978-3-030-67741-1 Published: 07 October 2021
Softcover ISBN : 978-3-030-67744-2 Published: 07 October 2022
eBook ISBN : 978-3-030-67742-8 Published: 06 October 2021
Edition Number : 1
Number of Pages : VIII, 310
Number of Illustrations : 24 b/w illustrations, 80 illustrations in colour
Topics : Systems Biology , Gene Function , Stem Cells
- Publish with us
Policies and ethics
- Find a journal
- Track your research
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- CBE Life Sci Educ
- v.15(4); Winter 2016
A Case Study Documenting the Process by Which Biology Instructors Transition from Teacher-Centered to Learner-Centered Teaching
Gili marbach-ad.
† College of Computer, Mathematical and Natural Sciences, University of Maryland, College Park, MD 20742
Carly Hunt Rietschel
‡ College of Education, University of Maryland, College Park, MD 20742
Associated Data
A case study approach was used to obtain an in-depth understanding of the change process of two university instructors who were involved with redesigning a biology course to implement learner-centered teaching. Implications for instructors wishing to transform their teaching and for administrators who wish to support them are provided.
In this study, we used a case study approach to obtain an in-depth understanding of the change process of two university instructors who were involved with redesigning a biology course. Given the hesitancy of many biology instructors to adopt evidence-based, learner-centered teaching methods, there is a critical need to understand how biology instructors transition from teacher-centered (i.e., lecture-based) instruction to teaching that focuses on the students. Using the innovation-decision model for change, we explored the motivation, decision-making, and reflective processes of the two instructors through two consecutive, large-enrollment biology course offerings. Our data reveal that the change process is somewhat unpredictable, requiring patience and persistence during inevitable challenges that arise for instructors and students. For example, the change process requires instructors to adopt a teacher-facilitator role as opposed to an expert role, to cover fewer course topics in greater depth, and to give students a degree of control over their own learning. Students must adjust to taking responsibility for their own learning, working collaboratively, and relinquishing the anonymity afforded by lecture-based teaching. We suggest implications for instructors wishing to change their teaching and administrators wishing to encourage adoption of learner-centered teaching at their institutions.
This is the analogy I thought of, the first semester was where you drop a ball on a hard floor, and at first it bounces really high, then the next bounce is a little lower, hopefully it’s going to be a dampened thing, where we make fewer and fewer changes. Alex
It seems to take a village to send a course in a new direction!! Julie
INTRODUCTION
This study documents the process by which instructors transition from teacher-centered instruction to emphasizing learner-centered teaching in an introductory biology course. Weimer (2013 ) defines teacher-centered instruction as lecture-based teaching wherein students are “passive recipients of knowledge” (p. 64). She characterizes learner-centered teaching as “teaching focused on learning—what the students are doing is the central concern of the teacher” (p. 15). Weimer delineates five principles of learner-centered teaching, which are 1) to engage students in their learning, 2) to motivate and empower students by providing them some control over their own learning, 3) to encourage collaboration and foster a learning community, 4) to guide students to reflect on what and how they learn, and 5) to explicitly teach students skills on how to learn. Of note, various terms are used in the literature to refer to strategies that are related to learner-centered teaching (e.g., active learning, student-centered teaching).
The literature suggests that teacher-centered instruction as opposed to learner-centered teaching promotes memorization ( Hammer, 1994 ) rather than desired competencies like knowledge application, conceptual understanding, and critical thinking emphasized in national reports (American Association for the Advancement of Science [AAAS], 2011). Further, lecture-based teaching fails to promote understanding of the collaborative, interdisciplinary nature of scientific inquiry ( Handelsman et al ., 2007 ). Notably, female and minority students have expressed feelings of alienation and disenfranchisement in classrooms using teacher-centered instruction ( Okebukola, 1986 ; Seymour and Hewitt, 1997 ).
A recommended practice that can support implementation of learner-centered teaching is the use of the backward design ( Wiggins and McTighe, 2005 ). The backward design model involves articulation of learning goals, designing an assessment that measures achievement of the learning goals, and developing activities that are aligned with the assessment and learning goals.
Despite robust evidence documenting the superiority of learner-centered teaching over teacher-centered instruction (as reviewed by Freeman et al ., 2014 ), instructors continue to adhere to teacher-centered instruction. A recent study showed that the majority of faculty members participating in professional development programs designed to help them adopt learner-centered teaching practices continue to rely on lecture-based pedagogy as indicated by classroom observational data ( Ebert-May et al ., 2011 ). Possible reasons for such loyalty to lecturing include the following: 1) instructors’ own personal experiences with lecture as undergraduates ( Baldwin, 2009 ); 2) personal beliefs that transmission of knowledge to students through lecture is the best way to teach ( Wieman et al ., 2010 ); 3) the perception that lecture preparation is more time-effective than preparing learner-centered activities ( Dancy and Henderson, 2010 ); 4) student resistance to active learning ( Henderson and Dancy, 2007 ; Seidel and Tanner, 2013 ; Bourrie et al ., 2014 ); 5) initial difficulties are often encountered when transitioning to learner-centered teaching, requiring several iterations to perfect a new teaching style; 6) learner-centered teaching encourages instructors to cover fewer topics in greater depth to promote meaningful learning ( Weimer, 2013 ), and many instructors are uncomfortable with such loss of content coverage ( Fink, 2013 ); and 7) the learner-centered instructor must change his/her role from an expert who delivers knowledge to a “teacher-facilitator,” giving a degree of control over the learning process to students, and many instructors are uncomfortable with the unpredictability and vulnerability that comes with relinquishing control in the classroom ( Weimer, 2013 ). Further, universities oftentimes fail to incentivize and encourage faculty members to prioritize teaching to a similar degree as research ( Fairweather et al ., 1996 ). It has been argued that the professional culture of science assigns higher status to research over teaching, encouraging scientists to adopt a professional identity based on research that typically ignores teaching ( Brownell and Tanner, 2012 ).
Given that many instructors face challenges and intimidation while implementing learner-centered teaching in their classrooms, there is a need to explore their experiences and learn what support instructors need as they engage in the process of transforming their courses. Science education researchers have recently emphasized the critical need “to better understand the process by which undergraduate biology instructors decide to incorporate active learning teaching strategies, sustain use of these strategies, and implement them in a way that improves student outcomes” ( Andrews and Lemons, 2015 , p. 1).
Case studies have been shown as a useful tool to understand change processes ( Yin, 2003 ). A case study approach represents a qualitative method of inquiry that allows for in-depth description and understanding of the experience of one or more individuals ( Creswell, 2003 ; Merriam, 2009 ). Yin (2003 , p. 42) provides a rationale for using single, longitudinal case studies that document participants’ perspectives at two or more occasions to show how conditions and processes change over time. In this study, we used a case study approach to obtain an in-depth understanding of the change process of two university instructors (Julie and Alex) who were involved with redesigning a biology course. The instructors sought to transform the course from a teacher-centered, lecture-style class to one that incorporated learner-centered teaching. We interviewed the two instructors on multiple occasions; we also interviewed a graduate teaching assistant (GTA) and an undergraduate learning assistant (ULA) to gain their perspectives on teaching the course. We explored the motivation, challenges, and thought processes of the instructors during the interviews. We used several data sources in addition to the interviews to build the case study, including class observations by external observers and student feedback data.
Given that faculty members have difficulty changing their teaching, there are recommendations to use theoretical models of change to examine processes of change ( Connolly and Seymour, 2015 ). We looked for theoretical models of change ( Ellsworth, 2000 ; Rogers, 2003 ; Kezar et al ., 2015 ) and found that the innovation-decision model ( Rogers, 2003 ) has recently been used by science education researchers ( Henderson, 2005 ; Bourrie et al ., 2014 ; Andrews and Lemons, 2015 ). Therefore, we decided to use this model to theoretically approach our data. Specifically, we decided to use the adapted model developed by Andrews and Lemons (2015) , which they modified to represent the change process that biology instructors experience when redesigning a course. This model includes the following stages: 1) knowledge, in which the instructor learns about the innovation and how it functions; 2) persuasion/decision, in which the instructor develops an attitude, positive or negative, toward the innovation and decides whether or not to adopt the innovation; 3) implementation, when the instructor behaviorally implements the innovation; and 4) reflection, in which instructor considers the benefits and challenges of using the innovation. On the basis of reflection, an instructor decides to stay with the present version of the implementation or to start the process once again in an iterative manner by seeking new knowledge (see Figure 1 ). According to Rogers (2003) , a condition to begin the change process is that an instructor must be dissatisfied with his or her current teaching approach. Such dissatisfaction is one contributing factor leading an instructor to begin seeking new knowledge about new teaching strategies. Other external and internal factors usually influence an instructor’s decision to change his or her teaching, including release time, institutional commitment, and instructor attitude ( Andrews and Lemons, 2015 ).

Innovation-decision model adapted from Rogers (2003) , Henderson (2005) , and Andrews and Lemons (2015) .
Context of the Study
This study was conducted at a research-intensive university on the East Coast of the United States. The instructors cotaught Principles of Biology III: Organismal Biology (BSCI207). BSCI207 follows two prerequisite courses, BSCI105 and BSCI106. BSCI105 covers molecular and cellular biology, while BSCI106 covers ecology, evolution, and diversity. BSCI207 requires students to synthesize concepts and principles taught in prerequisite courses, apply them across contexts in biology, and generally engage in higher-order learning (e.g., interdisciplinarity, conceptual understanding, quantitative reasoning). The course enrolls between 100 and 200 students per semester.
In Fall 2013, the provost’s office distributed a call for grant proposals encouraging instructors to redesign their courses to incorporate evidence-based teaching approaches. The call specifically required applicants to design experimental studies to evaluate their course redesign approaches in comparison with their usual teaching approaches. Julie and Alex applied for the grant and were funded. Their proposed evidence-based teaching approach was to incorporate a series of small-group active-engagement (GAE) exercises throughout the semester. The traditional section would retain the usual three 50-minute lectures per week schedule. The experimental section would replace one 50-minute lecture with a shortened 20-minute lecture followed by a 30-minute GAE exercise with content matched to the traditional class occurring that day.
The instructors designed the GAEs to accomplish a series of learning goals that were consistent with Weimar’s five principles of learner-centered teaching. For example, one of the GAE goals was to foster collaboration among students in order to mimic the scientific process of inquiry. This goal was in accord with Weimer’s (2013 ) learner-centered teaching principle of collaboration, creating a learning community with a shared learning agenda, and modeling how experts learn. To accomplish this goal, the instructors implemented the GAEs in a small-group setting and required students to exchange ideas and achieve consensus on a single worksheet.
A second goal, which accords with Weimer’s (2013 ) framework, was to engage students in their learning and motivate them to take responsibility and control over their learning process. For example, one of the GAEs asked students to complete a humorous, fictional case study involving a spaceship captain and deadly neurotoxins. In this activity, students needed to use mathematical equations to calculate membrane potentials and to create simulations of conditions that impact membrane potential. Another activity was to collaboratively create a plot of ion transport rate versus concentration. Students were given a computer simulation that they used to generate data; they then entered the data into a Google documents Excel spreadsheet. This created a classroom database that was used to build the plot, which the instructor displayed using the lecture hall projector at the end of class. This activity involved multiple components of learner-centered teaching, including collaboration, student engagement, and student responsibility for learning. Detailed descriptions of a selection of GAEs are published elsewhere ( Carleton et al ., in press, 2017 ; Haag and Marbach-Ad, in press, 2017 ).
The provost grant offered funding that could be used for various purposes. The instructors decided to use the funding for summer salary to develop GAEs and to pay for support from a science education expert. Grant awardees were required to participate in Faculty Learning Communities (FLCs) and teaching workshops arranged by the campus teaching and learning center.
In Fall 2014, the instructors started to implement their experiment. Jeffrey, a third instructor, joined Alex and Julie to teach both sections; each of them was responsible for teaching several topics associated with their specific research expertise. In the GAE class, students were divided into small groups to complete a learning activity pertaining to the course topic. In total, 12 GAE sessions were held during the semester. Both GAE and traditional classes were taught in large auditoriums. For each GAE session, students self-selected into groups of three to five students. Four GTAs circulated among the groups to facilitate group work. Students were asked to leave empty rows around their respective groups to allow GTAs to move throughout the groups. This same topic was covered only by lecture format in the traditional class.
In Fall 2015, the instructors no longer conducted a comparative experiment while teaching. Julie and Alex continued to coteach the course with the GAE format with many modifications to the activities and other aspects of the course (see Results ). Jeffrey continued to teach a different section of the course independently. Henceforth, we will describe the experience of Julie and Alex in their process of transforming the course.
Teaching Staff
Julie and Alex are associate professors. Lisa is a doctoral-level teaching assistant (TA) in the biology department. Lisa was a GTA in the Fall 2014 and Fall 2015 semesters. Jason was a freshman student in the GAE section of the Fall 2014 semester. In Fall 2015, Jason served as a guided study session (GSS) peer leader in BSCI207. GSS leaders are students who have taken a course on implementing evidence-based teaching approaches, and who have also completed the course they are tutoring with a high grade. GSS students are expected to facilitate small-group discussions outside class. Jason also volunteered to attend all GAE sessions to help facilitate.
Data Collection Instruments
Yin (2003) notes that multiple data sources are important in building case studies. As such, we use interview data, class observations, student feedback on the course, and information written in the grant proposal.
Interview Protocol.
Julie and Alex were interviewed independently immediately following Fall 2014 for 20 minutes each. Julie was also interviewed independently in the beginning of Fall 2015 for 1 hour. Julie and Alex were interviewed together immediately following Fall 2015 for approximately 1 hour. Lisa and Jason were also interviewed following Fall 2015 for 20–30 minutes each. We used semistructured interview protocols (see the Supplemental Material) with additional questions to probe for clarification. The questions probed participants’ motivation for change, attitudes toward change, barriers and challenges, administrative supports, details about the implementation, and teaching philosophies.
Class Observations.
Two independent raters conducted class observations. Each year, raters attended six classes. In Fall 2014, they observed GAE class sessions and the parallel, content-matched class sessions that took place in the traditional class (overall 12 sessions). This procedure allowed the raters to compare the class sessions covering the same material but with differing teaching approaches (i.e., learner-centered vs. teacher-centered instruction). The two raters attended each class session together. Once in the class, the raters used a rubric to evaluate the class. In Fall 2014, raters used a rubric based on a previously constructed rubric that was created by the biology department for peer observations ( http://extras.springer.com/2015/978-3-319-01651-1 , in SM-Evaluation of teaching performance.pdf). In Fall 2015, to better document group work, the raters used the rubric developed by Shekhar and colleagues (2015) .
Student Feedback.
Students were invited to reflect on GAEs by providing anonymous written feedback on note cards following the activity. We use some of these data in the present study.
Data Analysis
Interviews were conducted by a science education researcher, audiotaped, and transcribed. A science education researcher and a doctoral student in counseling psychology separately analyzed the interviews and the note cards to define emergent themes. Then, they negotiated the findings until they could agree upon the themes ( Maykut and Morehouse, 1994 ). The instructors were shown the interpretation of data to verify accuracy of interpretations. We present the results in accordance with the adapted Rogers (2003) model presented in Andrews and Lemons (2015) . We slightly adapted the Andrews and Lemons (2015) model to the iterative process through which our instructors progressed to modify the course (see Figure 1 ).
Motivation for Change
Before 2014, the traditional BSCI207 class as taught was a three-credit course with three 50-minute lectures per week. Alex described the traditional course:
Before the GAEs came into being, we taught in the very standard, traditional lecture. We used mostly PowerPoint to show text and images, occasionally we would bring a prop in, like sometimes I would bring a piece of a tree to gesture towards as I was lecturing about water transport or something like that. But it was basically standard lecture.
The instructors were dissatisfied with the traditional lecture format for the following reasons:
- Evidence for inferiority of teacher-centered instruction compared with learner-centered teaching . The instructors expressed awareness of the empirical data documenting the superiority of learner-centered teaching over teacher-centered instruction, “There’s a lot of research that suggests that [teacher-centered instruction] may not be the best way to help the students understand what we’re trying to get them to understand” (Alex).
- Lecture hinders understanding of the process of science. The instructors also expressed a desire to get students to learn the process of science early in their education, rather than to passively receive information. “We are being asked as science professors more and more to try and get our students to understand that science is a process, earlier and earlier in their career, and to model what real science is like in their education” (Alex).
- Lecture promotes overreliance on memorization. The instructors discussed a goal to modify the course so as to decrease focus on memorization and increase emphasis on problem solving and conceptual understanding. Julie described: “BSCI207 is the biology majors’ class, and it’s a lot of what the pre meds are taking, and so, critical thinking I think [is important], we’re constantly trying to get them to not just memorize and regurgitate but to put the ideas together.”
We also rearranged the material. So they [the lectures] used to be in a taxonomic orientation, I would give a whole lecture titled the biology of fungi, and the students complained that this taxonomic focus seemed to resemble the structure of BSCI106 [the prerequisite course]. I decided to explode those taxonomic lectures, and take the bits of content that I still thought were valuable, and spread them into other parts. So for example the stuff on mating types, which is wacky and interesting to me, and I hope to the students, is now in a lecture on sex. And they don’t realize half the lecture is on fungi. So they’re susceptible to packaging I think, and we don’t get the complaint any more that the course is redundant to BSCI106 (Alex).
Organisms don’t care about our disciplinary boundaries of research. The organism doesn’t understand that there’s biophysics, and biochemistry, and evolutionary biology, and ecology, and genetics. All these attributes of their biology have to function simultaneously on several different spatial and temporal scales … if we think they do, then we continually miss things that otherwise would fall out naturally if we were a little less wedded to our disciplines.
Relatedly, the instructors noted that most students enrolled in BSCI207 without having taken introductory physics or chemistry, which they thought was preventing students from drawing upon highly relevant concepts (e.g., thermodynamics) from these courses for biology.
- Underrepresented groups do poorly in traditional classes. The instructors quantitatively examined student performance for specific student subgroups (i.e., underrepresented minority students, female students) in previous BSCI207 semesters. They observed that there were disproportionate D/F/W grades for underrepresented students. Coupled with the science education literature documenting the ability of active learning to help underrepresented groups ( Preszler, 2009 ; Haak et al ., 2011 ; Eddy and Hogan, 2014 ), the instructors speculated that adding active learning to the traditional class might help underrepresented students.
In Fall 2014, the instructors went through the process of course revision that follows the adapted model by Rogers (2003) and Andrews and Lemons (2015 ; see Figure 1 ). In the following sections, we discuss their progression through the innovation-decision model. Table 1 shows a summary of the change process for the Fall 2014 semester.
First Iteration of the instructors’ change process
Before the Fall 2014 semester, the instructors engaged in several efforts to increase knowledge about evidence-based teaching approaches to modify the course. The knowledge sources were as follows:
I will go ask [physics education professional] questions. When something doesn’t go well I’ll meet with the postdocs [from physics education research group (PERG)] over there and say, what are they not getting here, how can we make this better, so I’m always trying to get resources to help.
- Reading the science education literature. As a new instructor, Julie participated in the college workshop for new instructors. The workshop was led by the director of the teaching and learning center, who provided several resources for using evidence-based teaching approaches, including an article giving an overview of learning styles ( Felder, 1993 ), a book on teaching tips ( McKeachie and Svinicki, 2006 ), and the book Scientific Teaching ( Handelsman et al ., 2007 ). In her interview, Julie commented, “So I read a lot of books,… I think it was getting students to think about math, I read one of the books [that the director of the college teaching and learning center] had given me [ Scientific Teaching ].”
- Observing other instructors teaching. The instructors had observed another instructor who implemented evidence-based teaching approaches in a small class of BSCI207 (<40 students). This pilot implementation was successful, and the instructors were interested in investigating whether the learner-centered teaching model used could be scaled up to a large-enrollment class.
Persuasion/Decision.
Following the knowledge-generation phase, the instructors felt prepared to change their teaching to a more learner-centered teaching style. They decided to conduct a comparative experiment during the first implementation of the GAEs (i.e., traditional vs. GAE classes; see Marbach-Ad et al ., in press, 2017 ). Although the instructors were aware of the literature documenting the effectiveness of learner-centered teaching, they had several reasons to execute the experiment:
- Obtain evidence for overall effectiveness. The instructors were unsure whether their activities were the best way to change the course (e.g., they were unsure of the challenges that would emerge, how the intervention would impact students). The instructors also wished to explore cost-effectiveness, since they knew that changing the course would require a high instructor time commitment.
- Convince colleagues to adopt learner-centered teaching approaches. The instructors noted that faculty in the department were unconvinced of the superiority of learner-centered teaching approaches, and they thought that a comparison study bringing empirical evidence might demonstrate that changing one’s teaching style is worthwhile. Alex stated, “[A] lot of my motivation for this experiment was to try to provide some evidence that these approaches were worth the effort, and because there is resistance clearly, from some of our colleagues who have been teaching the course for a long time.”
- Respond to grant award requirements. As mentioned earlier, the institution announced a call for proposals for instructors to revise their teaching. The instructions required applicants to propose comparative experiments during course revision to document effectiveness.
Implementation.
As proposed in the provost grant application, the instructors executed the comparison study. In the traditional class, instructors delivered a 50-minute lecture three times per week. In the GAE class, one lecture was replaced with a GAE. The GAE consisted of a brief 20-minute introductory lecture (a short version of the lecture presented to traditional class students) and a 30-minute group activity. As scientists, the instructors wished to manipulate the addition of the GAE day only and to keep remaining variables constant across classes. Therefore, homework assignments, examinations, optional computer tutorials, and office hours availability were consistent in both classes (see Table 2 ).
Fall 2014 class comparison
In the GAE class, on the day of the GAEs, students were instructed to sit with groups of three to five students (of their own choosing) and to leave empty rows between groups. Students were asked to have at least one laptop per group. As discussed previously, the GAEs were designed to be more learner centered relative to traditional lecture classes. To illustrate this here, we give Alex’s description of the membrane transport GAE: “The students had a little computer simulation, and they used that to generate data that they then entered into a Google docs spreadsheet in real time in the class, and there were enough students in the class that their responses produced this beautiful textbook plot of transport rate versus concentration. They built that relationship in a way that otherwise I would have just told them.”
Reflection.
Following the Fall 2014 semester, the instructors reflected on the various pros and cons of the learner-centered teaching intervention in the interviews. Observers and students also provided feedback that was used by the instructors to reflect on both sections of the course and on the comparative experiment. Several themes emerged from these data:
It’s much less about my spouting facts, it’s about my thinking ahead of time to get them to draw conclusions and get them to cement ideas. My role was partly just to control the chaos sometimes, and to control that the TAs had the information they needed so they could provide guidance to the students.
Importantly, observers noted that the instructors were very actively engaged with student groups throughout the GAEs, helping students to work through problems and understand concepts. Julie also commented that teaching with GAEs requires greater proficiency with material than lecturing: “To use these activities, you have to know the material better than if you’re going to straight lecture. And I think some instructors are maybe still learning BSCI207, what is all the material in it. And until you teach it straight a couple of times you probably don’t have the background to really understand.”
We spent less time talking about dating the origins of life using various methods (fossil record, carbon dating); we got rid of a lecture on prokaryotes and had to shrink some of the nutrient assimilation information from two lectures to one.
The instructors explained that, in order to minimize loss of content coverage, they decided to have a GAE class only once per week and to pick GAEs corresponding to lecture topics for which “there was the least amount of lost material by focusing on a particular exercise” (Alex). An additional solution was to move in-class lectures to online, preclass lectures. Julie described this change: “We also ask students to review some of the material that is lost during lecture time into the prep slides they review ahead of time.” However, Julie wondered whether students would benefit from online lectures to the same degree as in-person lectures: “I am still worried they don’t get so much out of those [online lectures] and so miss much of that information.”
- Engagement in learning. Overall, the instructors reflected that most GAEs provided a space for students to interact with one another, TAs, and instructors: Julie added, “I think it was nice to see the energy in the class and the way the students took to the activities, it was different for them.” Observers noted that the GAE class treatment condition was usually associated with increased student interactivity. Specifically, they noted that students in the GAE class were not only more engaged in the GAEs, but that they also tended to raise more questions during the PowerPoint presentations relative to students in the traditional class. Students reflected on their note cards following GAEs, and in the end-of-semester survey, noting that they felt that many of the GAEs were engaging (see Marbach-Ad et al ., in press, 2017 ).
- Giving students control over learning. The instructors noted, “The GAEs represented a chance to turn the class over to the students for some part of the time, where they could do something actively, instead of just sitting there listening to us” (Alex).
It’s actually a bit more how real science works, right, even as somebody who runs a lab, I don’t go into my lab and sit there and talk to my graduate students for four hours, I mean we have a brief conversation about how they should tackle something, and then they go off and work more on it. So it’s more of a checking in and then separating again. That’s kind of how this class works, the GAEs do give the students a little more of a feel of how collaborative real science works, and how no one person is sort of dictating everything, everyone needs to be a bit independent. … I think that this active model gives the students, for the first time, a real taste of how a real scientist would approach a problem.
Students commented on the opportunity afforded by GAEs to take an active role in their learning: “I learned how to apply what we learn in lecture class to actual problems”; “I kind of felt like a real scientist since I was put in a situation in which I had to make a hypothesis myself.”
- Disengagement. The instructors noted that, for some GAEs, students were disengaged. For example, in the GAE on stress and strain, two students were doing measurements in front of the class for 10–15 minutes, and the remaining students were instructed to input data into Excel files. These data were then used to make calculations. Students also expressed their dissatisfaction with this activity on the note cards that they handed in to the instructors: “I feel I understood the concept well once Dr. Julie wrote the plots on the board. This activity was more tedious and like busy work”; “ We could have easily compared values without experimentally finding them. I didn’t feel this deepened my understanding of concepts.”
- Insufficient time for reflection. The instructors noted that most exercises were too long, which did not leave sufficient time for reflection. Alex noted, “Well I think also making sure that if we get the exercise done in the right, short amount of time, then that does give us time to add a reflection at the end. Connecting the results of our exercise back to some larger idea.”
- Student preparation. The instructors felt that students would gain more from the exercise, if they were to come to GAE classes with better understanding of concepts relevant to the GAE. Then, more time could also be allotted for summary and reflection on important concepts. Alex commented, “We probably will need the students to do a bit of preparation before they come in to these active exercises, so that we can spend less time setting it up, and more time summing it up.”
- Assessments and grading misaligned with GAEs. In this implementation, instructors kept the same assessment plan for both the traditional class and the GAE class in order to compare achievement across classes. This resulted in a mismatch between the course activities and the assessments in the GAE section. For example, there were no final examination questions specifically covering GAE material. Of note, the instructors analyzed their final examination questions before conducting the experiment and saw that the questions required students to demonstrate high levels of thinking ( Bloom and Krathwohl, 1956 ; e.g., knowledge application, quantitative analysis), and they believed the GAEs would improve students’ abilities in these areas. Further, the instructors did not count GAE participation toward final grades, which instructors and observers believed had a detrimental effect on GAE attendance. Julie noted that “on the GAE days, only 60% of the students would come. That was partly because they wouldn’t get any credit for it, and they weren’t seeing that it was helping them learn the material better.” Analyses showed that students with higher grade point averages (GPAs) were those who chose to attend on the GAE days (see Marbach-Ad et al ., in press, 2017 ). Given this, the instructors felt that attendance should be incentivized in future implementations of the learner-centered teaching intervention to motivate and benefit a wider range of students.
- Resistance to learner-centered activities. The instructors felt that students’ low attendance specifically on GAE days may also have been because the students did not perceive the benefit of GAEs for their learning. “I feel sort of parental here, maybe the GAEs are like broccoli and brussels sprouts, they need them, they just don’t know it yet” (Alex).
- Group dysfunction. The instructors and observers noted several issues with the groups. Some groups were not engaged, and some students were not participating within their groups (e.g., one student would be left out). In some activities, some groups would finish the activity very quickly and would subsequently appear bored and waiting for further summary or instruction. Julie was frustrated with these occurrences and noted, “People would be sitting there on their phones.” One reason for student disengagement could be that students groups were unassigned and could include different students each week: students “would sit and associate with whoever was around them” (Julie).
- Auditorium-setting challenges. The instructors commented on the difficulty of doing GAEs in the large auditorium: “It’s still tricky to think about how you actually stage all of this, there is a bit of theater to running a large class with 200 students, how you move from one aspect of the process to another [lecture to group activities] quickly, without losing people, without too much noise and disturbance” (Alex).
- Little impact on grade distributions. Alex and Julie were hopeful that the GAEs would lead to large improvements in students’ grades as compared with traditional learning. However, the effect of GAEs was very small. Alex commented, “This was the biggest outcome from my perspective, and it drove much of the revisions for 2015. This is interesting, as it shows that even though we were unable to realize a big payoff in the first year, we nevertheless saw something that we thought was worth keeping and hopefully improving upon.”
- TA training required. The instructors reflected that they did not provide adequate TA preparation for the GAEs: “We hadn’t really prepared the GAEs enough ahead of time so that we could talk about them with the TAs. The TAs at times were really clueless about what was supposed to be happening” (Julie). TAs, although instructed to guide and facilitate groups, apparently lacked the skills to engage students, as observers noted that most of them passively waited for students to ask questions rather than actively approaching students with questions, instructions, etc.
On the basis of their reflection, Julie and Alex decided to continue teaching with GAEs and to seek new knowledge to improve GAEs. In the following sections, we discuss their continued progression through the innovation-decision model (see Figure 1 ). A summary of the change process in Fall 2015 is shown in Table 3 .
Second iteration of the instructors’ change process
- Learn about methods to form successful groups. The instructors reviewed the literature and consulted with the director of the teaching and learning center and other faculty members in the department to form new strategies on building effective groups in auditorium settings. The literature shows that groups work best when they are permanent and students are held accountable to other group members ( Michaelsen and Black, 1994 ; Michaelsen et al ., 2004 , 2008). The literature also shows that taking student diversity into account is important in creating successful groups ( Watson et al ., 1993 ). For example, Watson and colleagues (1993) reported that, although it takes time, heterogeneous groups outperformed homogeneous groups on several performance measures, including generating perspectives and alternative solutions. The instructors also learned from the director of the teaching and learning center about the Pogil method ( pogil.org ), in which students are assigned different roles during group work (e.g., recorder, facilitator). They weighed the pros and cons of implementing this method in the classroom.
- Learn about methods to flip courses. The instructors learned from models of flipped classes ( Hamdan et al ., 2013 ; Jensen et al ., 2015 ), which highlight how to capitalize on out-of-class time to cover material to prepare for face-to-face active learning. In this regard, instructors sought assistance from the information technology office about presentation software (i.e., Camtasia) that can deliver automated lectures effectively.
- Seek expert guidance. During the summer, the instructors again consulted with science education experts to enhance the GAEs. For example, they consulted with a science education expert on how to revise the concept map assignment. Julie described how this guidance helped her “leave the activity a bit more free form and get the students to make a graphic organizer of their own design rather than trying to fill in some pre-designed boxes.” As another example, the science educator recommended strategies about how to streamline GAEs to maximize time spent on developing conceptual understanding and minimize time spent on the mechanics of exercises.
- Learn about strategies to enhance TA support. The instructors wished to decrease student to TA ratio. However, GTAs require departmental funding, which was unavailable. The teaching and learning center director and the biological sciences administration offered to involve ULAs who are unpaid but receive alternative benefits, such as leadership and teaching experience and undergraduate course credit. This model was reported to be successful in our university ( Schalk et al ., 2009 ) and in other institutions ( Otero et al ., 2010 ).
Following reflection on the comparative experiment, instructors sought to keep improving the course and decided to make several changes:
- Teach all sections with learner-centered teaching. Although the instructors reported that keeping the GAE class format requires more time to prepare relative to lecturing and takes time from their research (“fine tuning the GAEs—that took weeks” [Julie]), they decided to implement the GAEs in all sections and to work to improve them.
[Last semester] I had a couple of students up front doing the experiment, and everyone else was kind of twiddling their thumbs while we gathered the data. We talked about the data but we didn’t really have time [to do data analysis and summarize concepts]. I think this year I’m just going to give them last year’s data, and have each group do some analysis.
As another way to modify GAEs, instructors decided to utilize more outside resources such as published, case-based activities. Julie described, “I’d love to come up with some more case studies that we could do. You know the Buffalo site [ http://sciencecases.lib.buffalo.edu/cs/collection ] has all the case studies for all the science classes. So I’m constantly perusing that. A couple of the GAEs that I developed actually come from there.”
I haven’t figured out what the best prep work is. What Alex has been doing is taking the slides he showed last year and just posting them online. I’m not sure that’s the best, or really enough.… But, then they just read. I mean he tries to put more words on them. I tried to find some videos that I thought were appropriate, and I’m not sure that’s any better. I was going to do some of these with Camtasia. In this way you can actually have the slides and actually talk over them and record. But I couldn’t make the software work. I haven’t really gone there yet, I will have to figure that out.
To encourage students to prepare for the GAEs, the instructors decided to give a preclass quiz covering the out-of-class preparatory materials. Julie described,
We’re also doing a quiz this time, we’re giving that preparatory information, they have to have done it by the morning before, they have to take a little 2-point quiz [before class] to show that they’ve covered that material. Then we have the whole class time [for the GAE] so that we’re not so rushed in trying to do to many things at one time.
- Train the TAs better, add ULAs, and involve both teams in the process of GAE development. The instructors decided to expand the team of assistants to decrease the ratio between students and TAs. Julie described the change from Fall 2014 to Fall 2015: “We have a bigger team. We have two of these ULAs, and then we have three UTAs, and two GTAs. So a team of seven helpers, and each person has a different job. The ULAs are specifically supposed to be trying out the GAEs ahead of time. So we kind of run things past them. And then we meet with all the TAs, and then talk through the GAEs beforehand. They have an assigned part of the class, where each of them is hopefully seeing the same students over and over, and hopefully getting to work with them to develop a rapport, and they go in the middle of the activity, so kind of checking in, so what do you think, kind of getting students to verbalize.” The benefit of this new format, where each TA was responsible for a subsection of the large class, was that it approximated a smaller class discussion session in which students could get to know their TAs more personally.
- Revise group structure. On the basis of the literature and their previous experiences, the instructors decided to assign permanent, diverse groups of four at the beginning of the semester. They also decided to instruct students on how to sit in the auditorium with their groups (in two rows rather than in a single line, to enhance group communication) and to award points for completing group work exercises.
In the Fall 2015 implementation, there were several changes to the course (for a comparison of 2014 and 2015 GAE classes, see Table 4 ).
GAE class comparison between Fall 2014 and Fall 2015
- Modify the activities. The instructors devoted a full weekly class period to the GAE instead of 30 minutes. On the basis of their experiences in the previous semester, they revised some GAEs and adapted them to the time frame. Although they had more time for the GAEs, they wished to make them more efficient and interactive: “I think we had to cut some, with the GAEs, because they were taking way too long, but I think in a few cases we simplified them, took out 1/3 of them or something” (Alex). Instead of the 20-minute pre-GAE lecture that was presented in the Fall 2014 implementation, students were asked to prepare for activities at home by watching videos, reviewing lecture slides, and reading textbook materials. In contrast with Fall 2014, the students were awarded three points for participating in the GAE activity and two points for completing a quiz covering preparatory materials that was due before the GAE class. The instructors wished to assign points to these activities in order to “really give them weight” (Julie). “[The activities] formed a large part of the exams as well. So making the activities more integral to the class was a big change” (Julie).
So the next time I drew a map in the room, [which showed] two students in the front, and two students in the back. When you have a very formal auditorium, you have to try and help them assort with each other and talk with each other. The other thing we did was, we were giving each group two copies of the assignment, so they didn’t each have one. So that kind of helped, that kind of had them sharing things.
Finally, TAs and ULAs were assigned to stay with one section of the lecture hall throughout the semester. Thus, TAs and ULAs developed a rapport with a large group of students throughout the semester and were able to learn their names, which facilitated communication.
- Add more and better-trained TAs. Before Fall 2015, the instructors trained the TAs to better engage with student groups in class. Class observation data showed that, in Fall 2014, some TAs were lacking in their ability to engage actively with students. One observer described, “When I observed the classes last year [Fall 2014], they [TAs] were standing in the side [of the auditorium], and sometimes they got to students, but just students that raised their hands. They weren’t active. They were very passive, most of them, because they didn’t know what to do.” Following the implementation in Fall 2015, the observer noticed a change in TA involvement: “Now, it’s more about instruction, they circulate between groups and encourage them to ask questions, they encourage students that aren’t participating … it’s not enough to throw them [the TAs] in the classroom.”
Overall, instructors noticed improvements in the areas that they targeted to improve, and they also felt there were areas that they wished to continue improving.
- Student preparation. Julie described that although new techniques were put in place to increase student preparation, students often seemed unprepared for the activities: “And my data for that is essentially for the first 20 minutes of the GAE they would spend saying, what are we doing? There was a lot of flailing. It took them a lot longer to get going with the GAE than I thought, and I’m not sure if that’s because the preparatory material is not really preparing them, or that they just took the online quiz and didn’t really go through the preparatory material.” Julie thought about changing the nature of the preparatory lectures, “I would still like to explore turning those into little online lectures rather than having them read the slides.”
- Student attendance . Alex commented that the strategy of assigning points to participating in GAEs “made a big difference in attendance […] by incentivizing their attendance, at least on GAE days, they were coming.” The instructors commented that incentivizing participation in the GAEs and the preactivity quizzes increased the amount of student–instructor interaction regarding point grabbing. Alex stated, “The downside of associating points with everything is that I think we spent the largest fraction of our student interaction time dealing with the points related to the GAEs, excused absences, non-excused absences, anxiety about the points, I mean these are tiny amounts of points, but the students took it very seriously. But I think it was one of the top 3 issues that students came up with this semester.”
- Mechanics of exercises. Julie was very frustrated with how students could not effectively operate Excel software: “And they still don’t know Excel. My biggest frustration was that I thought Excel would make their lives easier, and it made their lives harder. I’m almost ready to go back to pencil and paper, just to get them to plot things and think about things, because they’re not getting back to the scientific inquiry and hard thinking, they’re just so stuck in which box do I click.” Alex added, “My issue is that the preparatory materials do a good job preparing them intellectually for what’s the point, but then they do get stuck on the mechanics, what they’re doing with their hands.”
- Allocating time for reflection. The instructors described that they improved substantially in the area of summarizing major concepts and timing activities: “I think we did a pretty good job of every 15 or 20 minutes bringing them back together and saying ok, you would have done this by now. There were a couple that worked really well, and a couple where we were still pressed for time. I think that generally it was far improved” (Julie). Because the instructors had the full class period to devote to the GAE and did not need to compare learner-centered teaching with teacher-centered instruction, they felt that the timing of the activities was much improved. Julie noted, however, “I always overestimate what students can do. I’m still adjusting.”
- Technical issues. There were difficulties with connecting to the wireless Internet in the lecture hall, particularly among students who failed to download the appropriate tools before coming to class. Further, students have different types of computers and software programs and knowledge of software programs required for the course.
The organized approach helped students see the material as well as make a few friends, in fact, I remember coming onto my dorm floor and seeing four people from my class working together, and they were actually in that GAE group, they had made a study group because they were used to working together. One of the aims of this project gets students communicating instead of competing.
Julie commented that there is still room for improvement in the student groups: “I saw a number of groups where at least one person would be left out. I don’t know if that’s a physical orientation, if we could point them toward each other it would be better. Next year one thing we talked about is going to groups of 3, because with 3 you can always get across each other and be more … everybody can talk to each other.” The instructors considered the benefits of the Pogil. Julie explained that they tried to appoint a different group member to act as the scribe each week during GAE activities as a way to increase student participation in groups. The instructors did not strictly enforce this policy, as they were not sure it was beneficial.
And we also had some undergraduates this year, … and I think they were really helpful because they understand what the students are capable of, more than we do … a lot of times they can give you some insight into what’s going on or what classes undergraduates are most likely taking at the same time. It was very helpful.
Finally, Lisa felt that the level of engagement among the teaching staff was higher than for a standard lecture course: “Everyone was very engaged, it’s a unique class to TA for, because I feel like the TAs and the professors are far more engaged than in a standard lecture course, so it was kind of nice.” Alex reflected that, in the future, “It would be even better,” since they will have “a whole floor of ULAs that had us for 207,” and they “will be well-positioned” to assist in the redesigned course.
This case study examines instructor change processes when moving from teacher-centered instruction toward learner-centered teaching. In this study, we examined the change process through the lens of the innovation-decision model ( Rogers, 2003 ; Andrews and Lemons, 2015 ), which recognizes several stages of change: knowledge, decision/persuasion, implementation, and reflection. The model is iterative, recognizing that transforming courses may require multiple revisions as instructors reflect on the inherent challenges and imperfections that arise when changing a course ( Henderson, 2005 ). Consistent with this literature, the first implementation of learner-centered course revision was fraught with imperfections, and the instructors persisted through two rounds of course revision before gaining satisfaction with their teaching approach, although they plan to continue enhancing the course with each semester.
Andrews and Lemons (2015) note that dissatisfaction with one’s current teaching approach is an important motivator leading instructors to change their teaching. Our instructors were dissatisfied with the lecture mode of teaching in their courses due to personal dislike for it, and the sense that it encouraged student reliance on memorization and hindered interdisciplinary thinking. Other motivators for change included 1) awareness of national recommendations to use learner-centered teaching ( AAAS, 2011 ); 2) a hope that underrepresented students would benefit from learner-centered instruction, based on education literature documenting such benefits ( Okebukola, 1986 ; Seymour and Hewitt, 1997 ); and 3) institutional support (i.e., a provost office grant initiative).
These motivations led the instructors to seek new knowledge about learner-centered teaching approaches and how to implement them, which, according to the adapted innovation-decision model ( Andrews and Lemons, 2015 ), is a first step toward changing a biology course. In the present study, knowledge-seeking strategies included consultation with science education experts and information technology experts, reading the empirical literature, observing other faculty members who had adopted evidence-based teaching practices, and involvement with a discipline-based FLC. Following the knowledge stage, the instructors progressed through the decision/persuasion and implementation stages of change. In the reflection stage, the instructors discussed what worked well, challenges, and areas they wished to improve in the subsequent iteration. We present here implications from this study for instructors seeking to change their courses, and also for administrators wishing to promote learner-centered instruction at their institutions.
IMPLICATIONS FOR INSTRUCTORS
Weimer (2013) noted that engaging students in their own learning is messy, unpredictable, and challenging as compared with teacher-centered instruction. The process can be difficult for the faculty members who want to change as well as for the students. First, the instructor must adopt a new role as “instructor-facilitator” ( Weimer, 2013 ), giving up a degree of control to the students to take responsibility for their own learning. Relating to their new role, our instructors reported that, on the one hand, the instructor-facilitator role felt like controlling chaos at times, particularly in the beginning, but that it was markedly beneficial for student learning and for their own teaching. For instance, it gave students an opportunity to be independent learners and to engage with their peers in collaborative problem solving, more closely modeling the process of science. Thus, although it may be intimidating to share control over the learning process with students, it appears that there are benefits for both students and instructors.
Second, learner-centered teaching encourages instructors to cover fewer topics in greater depth, as opposed to more topics in less depth ( Weimer, 2013 ). Despite being uncomfortable with losing content coverage due to the function of BSCI207 as a preparation course for the MCAT and a prerequisite, our instructors decided to remove some course topics and consolidate others into shorter units. Next, they implemented several solutions to the necessary loss of content coverage. First, they moved lecture content to required preclass, online lectures that substituted for in-class content coverage. Second, they were strategic about which course topics they used to redesign as GAEs. Specifically, they selected course topics that were historically conceptually challenging for students (e.g., membrane transport). Our faculty members’ transition process provides an example of how faculty members can identify and implement solutions for concerns about loss of content coverage.
Third, a fundamental principle of learner-centered teaching is to encourage collaboration in the classroom ( Weimer, 2013 ). To this end, our instructors implemented GAEs, a series of group work–based activities. Student collaboration is important, because it promotes sharing of the learning agenda ( Johnson et al ., 1984 ; Weimer, 2013 ), and collaboration is a skill that is essential for the workplace ( Hart Research Associates, 2015 ). Group work is a common and accessible strategy that instructors can use to increase learner-centered teaching in their classrooms. Our instructors experienced various challenges and implemented several revisions to group work activities throughout their change process. The most successful strategies for optimizing group work included 1) increasing the number of TAs and the amount of TA training; 2) creating diverse and permanent student groups to increase accountability ( Michaelsen et al ., 2004 ); 3) assigning grades and preparation assignments for group work activities; and 4) restructuring group work activities to provide more time for whole-class summary and reflection on concepts. Group work is just one type of teaching strategy that can increase learner-centered teaching. Each instructor needs to discover what kinds of approaches are most suitable to increase their level of learner-centered teaching. When selecting and implementing new teaching strategies, it is highly recommended to seek guidance from experts, more experienced faculty members, or from a teaching and learning community.
Transitioning away from lecture-based instruction to learner-centered instruction can be challenging for students as well as instructors. The literature has shown that students resist many learner-centered approaches that require them to engage in the classroom rather than sit anonymously in lecture ( Michaelsen et al ., 2008 ; Shekhar et al ., 2015 ). Our instructors learned about student resistance through several means: 1) student feedback that was collected on note cards at the end of GAE classes, 2) end-of-semester surveys asking students to reflect on each activity, and 3) low attendance on GAE days as compared with lecture class days. It is important for instructors transitioning their courses to monitor student resistance and satisfaction, as our instructors used these data to modify the activities from the first to second iteration.
The instructors used several strategies to reduce student resistance. First, through student feedback, instructors learned that they needed to provide students with better explanations for the purpose of doing GAEs as opposed to sitting in lecture class. Weimer (2013) emphasizes the importance of providing students explicit instruction on how to best learn. Therefore, at the second iteration of the learner-centered implementation, the instructors were explicit about the rationale for the GAEs. At various points throughout the semester, the instructors explained how the GAEs were helpful in enhancing skills (e.g., critical thinking, problem solving, collaboration, understanding the interdisciplinary nature of science, relating course material to everyday life and to scientific research) that are recommended by national organizations ( AAAS, 2011 ) and employers ( Hart Research Associates, 2015 ). Second, instructors awarded class participation points for completing GAE exercises and grades for completing the preclass online quiz. This strategy resulted in better alignment between requirements of students and course assessments, which accords with Wiggins and McTighe’s (2005) backward design theory. This method of GAE grading resulted in much higher student attendance as compared with the first iteration. Third, instructors used evidence-based strategies to reduce resistance within student groups, including creating permanent, diverse groups at the start of the semester. Fourth, instructors took student feedback into account with regard to their satisfaction with specific activities and modified activities with the goal of maximizing student engagement.
IMPLICATIONS FOR ADMINISTRATORS
Given that changing one’s teaching from teacher-centered instruction to learner-centered teaching is challenging, there must be administrative support for these efforts.
First, administrators can play a key role in acknowledging the importance of learner-centered teaching. Historically, universities have failed to encourage faculty members to prioritize teaching to a similar degree as research ( Fairweather et al ., 1996 ). Unfortunately, many tenure-track faculty members at research-intensive universities fear that they may be penalized for investing the time to adopt learner-centered teaching. Research-oriented universities should prioritize teaching in order to support more widespread adoption of evidence-based teaching approaches. Julie reflected on her frustration with the university’s message that teaching is devalued relative to research:
I think for assistant professors, I was actually scolded for putting time into teaching and trying to participate in teaching improvements and so, I think it’s discouraged, perhaps rightly so, because they’re not going to value it, so if that’s going to take away from what’s required to get tenure, to get promoted, they want you to know that. So they’re just being honest perhaps.
As part of a university culture that values learner-centered teaching, administrators (e.g., chairs, promotion committees) should acknowledge instructors who are making the effort to transition their courses and understand if their teaching evaluations are lower during the initial semesters of transition.
Second, as evidenced by our study and by others in the literature, transitioning from lecture-based teaching to learner-centered teaching requires a large time commitment from instructors. Thus, funding and release time are valuable supports that administrators can provide to improve the quality of teaching at their institutions. The provost grant was a fundamental support contributing to our instructors’ success in transitioning a core biology course. Further, the fact that teaching fellowships were awarded from the university provost shows that our research-intensive university is beginning to value faculty members’ adoption of learner-centered teaching. Alex commented on these fellowships:
The message comes through that the university values teaching, otherwise we wouldn’t have these fellowships from the Provost, that’s about as high up as it gets, I mean there is this signal, a voice that says, great, please do this. But then when the rubber meets the road, are you going to get promoted? It is not considered a substitute for quality research productivity as a research-active faculty.
Third, learner-centered instruction requires more human resources relative to teacher-centered instruction (e.g., for grading, facilitating small-group discussions, demonstrations, assisting in revising course activities). Administrators should consider ways to assign more TAs to courses that use learner-centered teaching. TAs and/or ULAs could be compensated through financial means or through other methods like course credit. Our university, for example, has developed a training program for undergraduate TAs, in which they receive training in how to facilitate small groups.
Fourth, in universities where there are state-of-the art facilities for teaching and learning, there should be a priority for courses that adopt innovative teaching approaches. In our university, such facilities are in a state of development, and administrators are planning to incentivize faculty who are using evidence-based teaching approaches by giving them priority to teach in the new, state-of-the art teaching and learning facility, which includes classrooms with round tables, movable seats, and advanced technology.
Finally, universities should provide support for a campus teaching and learning expert and an FLC. These resources were fundamental in the transition process of our faculty members. FLCs may be discipline-based ( Marbach-Ad et al ., 2010 ) or campus-wide ( Cox, 2001 ). FLCs and teaching and learning experts can provide pedagogical and curricular guidance, as well as emotional support for the stressors associated with teaching.
Supplementary Material
Acknowledgments.
This work has been approved by the University of Maryland Institutional Review Board (IRB protocol 601750-2). We thank our teaching team members who participated in the study.
- American Association for the Advancement of Science. Vision and Change in Undergraduate Biology Education: A Call to Action. Washington, DC: 2011. http://visionandchange.org/files/2011/03/Revised-Vision-and-Change-Final-Report.pdf (accessed 8 November 2016) [ Google Scholar ]
- Andrews TC, Lemons PP. It’s personal: biology instructors prioritize personal evidence over empirical evidence in teaching decisions. CBE Life Sci Educ. 2015; 14 :ar7. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Baldwin RG. The climate for undergraduate teaching and learning in STEM fields. New Dir Teach Learn. 2009; 117 :9–17. [ Google Scholar ]
- Bloom BS, Krathwohl DR. Taxonomy of Educational Objectives: The Classification of Educational Goals, Handbook 1: Cognitive Domain. New York: Longmans; 1956. [ Google Scholar ]
- Bourrie DM, Cegielski CG, Jones-Farmer LA, Sankar CS. Identifying characteristics of dissemination success using an expert panel. Decision Sci J Innov Educ. 2014; 12 :357–380. [ Google Scholar ]
- Brownell SE, Tanner KD. Barriers to faculty pedagogical change: lack of training, time, incentives, and… tensions with professional identity. CBE Life Sci Educ. 2012; 11 :339–346. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Carleton KL, Rietschel CH, Marbach-Ad G. Group active engagements using quantitative modeling of physiology concepts in large-enrollment biology classes. J Microbiol Biol Educ. 2017; 17 ( in press ) [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Connolly MR, Seymour E. 2015. Why Theories of Change Matter (WCER Working Paper No. 2015-2). www.wcer.wisc.edu/publications/workingPapers/papers.php (accessed 8 November 2016)
- Cox MD. Faculty learning communities: change agents for transforming institutions into learning organizations. To Improve the Academy. 2001; 19 :69–93. [ Google Scholar ]
- Creswell JW. Research Design: Qualitative, Quantitative, and Mixed Methods Approaches, 2nd ed. Thousand Oaks, CA: Sage; 2003. [ Google Scholar ]
- Dancy M, Henderson C. Pedagogical practices and instructional change of physics faculty. Am J Phys. 2010; 78 :1056–1063. [ Google Scholar ]
- Ebert-May D, Derting TL, Hodder J, Momsen JL, Long TM, Jardeleza SE. What we say is not what we do: effective evaluation of faculty professional development programs. BioScience. 2011; 61 :550–558. [ Google Scholar ]
- Eddy S, Hogan K. Getting under the hood: how and for whom does increasing course structure work. CBE Life Sci Educ. 2014; 13 :453–468. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ellsworth JB. Surviving Change: A Survey of Educational Change Models. Washington, DC: Office of Educational Research and Improvement; 2000. [ Google Scholar ]
- Fairweather J, Colbeck C, Paulson K, Campbell C, Bjorklund S, Malewski E. Engineering Coalition of Schools for Excellence and Leadership (ECSEL): Year 6. University Park: Center for the Study of Higher Education, Penn State University; 1996. [ Google Scholar ]
- Felder RM. Reaching the second tier. J Coll Sci Teach. 1993; 23 :286–290. [ Google Scholar ]
- Fink LD. Creating Significant Learning Experiences: An Integrated Approach to Designing College Courses. Hoboken, NJ: Wiley; 2013. [ Google Scholar ]
- Freeman S, Eddy S, McDonough M, Smith MK, Okoroafor N, Jordt H, Wenderoth MP. Active learning increases student performance in science, engineering, and mathematics. Proc Natl Acad Sci USA. 2014; 111 :8410–8415. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Haag ES, Marbach-Ad G. Quantitative modeling of membrane transport and anisogamy by small groups within a large-enrollment organismal biology course. J Microbiol Biol Educ. 2017; 17 ( in press ) [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Haak DC, HilleRisLambers J, Pitre E, Freeman S. Increased structure and active learning reduce the achievement gap in introductory biology. Science. 2011; 332 :1213–1216. [ PubMed ] [ Google Scholar ]
- Hamdan N, McKnight P, McKnight K, Arfstrom KM. A review of flipped learning. 2013. Flipped Learning Network. flippedlearning.org/wp-content/uploads/2016/07/LitReview_FlippedLearning.pdf (accessed 8 November 2016)
- Hammer D. Epistemological beliefs in introductory physics. Cogn Instr. 1994; 12 :151–183. [ Google Scholar ]
- Handelsman J, Miller S, Pfund C. Scientific Teaching. New York: Freeman; 2007. [ PubMed ] [ Google Scholar ]
- Hart Research Associates. Falling Short? College Learning and Career Success. Washington, DC: 2015. [ Google Scholar ]
- Henderson C. The challenges of instructional change under the best of circumstances: a case study of one college physics instructor. Am J Phys. 2005; 73 :778–786. [ Google Scholar ]
- Henderson C, Dancy MH. Barriers to the use of research-based instructional strategies: the influence of both individual and situational characteristics. Phys Rev Spec Top Phys Educ Res. 2007; 3 :020102. [ Google Scholar ]
- Jensen JL, Kummer TA, Godoy PD. Improvements from a flipped classroom may simply be the fruits of active learning. CBE Life Sci Educ. 2015; 14 :ar5. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Johnson DW, Johnson RT, Holubec EJ, Roy P. Circles of Learning: Cooperation in the Classroom. Alexandria, VA: Association for Supervision and Curriculum Development; 1984. [ Google Scholar ]
- Kezar A, Gehrke S, Elrod S. Implicit theories of change as a barrier to change on college campuses: an examination of STEM reform. Rev High Educ. 2015; 38 :479–506. [ Google Scholar ]
- Marbach-Ad G, McAdams KC, Benson S, Briken V, Cathcart L, Chase M, El-Sayed NM, Frauwirth K, Fredericksen B, Joseph SW, Lee V. A model for using a concept inventory as a tool for students’ assessment and faculty professional development. CBE Life Sci Educ. 2010; 9 :408–416. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Marbach-Ad G, Rietschel C, Saluja N, Carleton KL, Haag E. The use of group activities in introductory biology supports learning gains and uniquely benefits high-achieving students. J Microbiol Biol Educ. 2017; 17 ( in press ) [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Maykut P, Morehouse R. Beginning Qualitative Research: A Philosophic and Practical Approach. Bristol, PA: Falmer; 1994. [ Google Scholar ]
- McKeachie W, Svinicki M. McKeachie’s Teaching Tips: Strategies, Research, and Theory for College and University Teachers. Boston. MA: Houghton Mifflin; 2006. [ Google Scholar ]
- Merriam SB. Qualitative Research: A Guide to Design and Implementation. San Francisco, CA: Jossey-Bass; 2009. [ Google Scholar ]
- Michaelsen LK, Black RH. In: Collaborative Learning: A Sourcebook for Higher Education, vol. 2. University Park: National Center on Postsecondary Teaching, Learning, and Assessment, Pennsylvania State University; 1994. Building learning teams: the key to harnessing the power of small groups in education; pp. 65–81. [ Google Scholar ]
- Michaelsen LK, Knight AB, Fink LD. Team-Based Learning: A Transformative Use of Small Groups in College Teaching. Sterling, VA: Stylus; 2004. [ Google Scholar ]
- Michaelsen LK, Sweet M, Parmelee DX (eds.), editors. Team-Based Learning: Small Group Learning’s Next Big Step, New Directions for Teaching and Learning, Number 116. Hoboken, NJ: Wiley; 2008. [ Google Scholar ]
- Okebukola PA. Cooperative learning and students’ attitudes to laboratory work. School Sci Math. 1986; 86 :582–590. [ Google Scholar ]
- Otero V, Pollock S, Finkelstein N. A physics department’s role in preparing physics teachers: the Colorado learning assistant model. Am J Phys. 2010; 78 :1218–1224. [ Google Scholar ]
- Preszler RW. Replacing lecture with peer-led workshops improves student learning. CBE Life Sci Educ. 2009; 8 :182–192. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Rogers EM. Diffusion of Innovations, 5th ed. New York: The Free Press; 2003. [ Google Scholar ]
- Schalk KA, McGinnis JR, Harring JR, Hendrickson A, Smith AC. The undergraduate teaching assistant experience offers opportunities similar to the undergraduate research experience. J Microbiol Educ. 2009; 10 :32–42. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Seidel SB, Tanner KD. What if students revolt?—Considering student resistance: origins, options, and opportunities for investigation. CBE Life Sci Educ. 2013; 12 :586–595. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Seymour E, Hewitt NM. Talking about Leaving: Why Undergraduates Leave the Sciences. Boulder, CO: Westview; 1997. [ Google Scholar ]
- Shekhar P, Demonbrun M, Borrego M, Finelli C, Prince M, Henderson C, Waters C. Development of an observation protocol to study undergraduate engineering student resistance to active learning. Int J Eng Educ. 2015; 31 :597–609. [ Google Scholar ]
- Watson WE, Kumar K, Michaelsen LK. Cultural diversity’s impact on interaction process and performance: comparing homogeneous and diverse task groups. Acad Manage J. 1993; 36 :590–602. [ Google Scholar ]
- Weimer M. Learner-Centered Teaching. San Francisco: Jossey-Bass; 2013. [ Google Scholar ]
- Wieman C, Perkins K, Gilbert S. Transforming science education at large research universities: a case study in progress. Change. 2010; 42 :6–14. [ Google Scholar ]
- Wiggins GP, McTighe J. Understanding by Design. Alexandria, VA: Association for Supervision and Curriculum Development; 2005. [ Google Scholar ]
- Yin RK. Case Study Research: Design and Methods, 3rd ed. Thousand Oaks, CA: Sage; 2003. [ Google Scholar ]

- school Campus Bookshelves
- menu_book Bookshelves
- perm_media Learning Objects
- login Login
- how_to_reg Request Instructor Account
- hub Instructor Commons
Margin Size
- Download Page (PDF)
- Download Full Book (PDF)
- Periodic Table
- Physics Constants
- Scientific Calculator
- Reference & Cite
- Tools expand_more
- Readability
selected template will load here
This action is not available.

8.1: Case Study: Genes and Inheritance
- Last updated
- Save as PDF
- Page ID 16760

- Suzanne Wakim & Mandeep Grewal
- Butte College
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
Case Study: Cancer in the Family
People tend to look similar to their biological parents, as illustrated by the family tree in Figure \(\PageIndex{1}\). But, you can also inherit traits from your parents that you can’t see. Rebecca becomes very aware of this fact when she visits her new doctor for a physical exam. Her doctor asks several questions about her family's medical history, including whether Rebecca has or had relatives with cancer. Rebecca tells her that her grandmother, aunt, and uncle, who have all passed away, all had cancer. They all had breast cancer, including her uncle, and her aunt additionally had ovarian cancer. Her doctor asks how old they were when they were diagnosed with cancer. Rebecca is not sure exactly, but she knows that her grandmother was fairly young at the time, probably in her forties.
Rebecca’s doctor explains that while the vast majority of cancers are not due to inherited factors, a cluster of cancers within a family may indicate that there are mutations in certain genes that increase the risk of getting certain types of cancer, particularly breast and ovarian cancer. Some signs that cancers may be due to these genetic factors are present in Rebecca’s family, such as cancer with an early age of onset (e.g. breast cancer before age 50), breast cancer in men, and breast cancer and ovarian cancer within the same person or family.
Based on her family medical history, Rebecca’s doctor recommends that she see a genetic counselor because these professionals can help determine whether the high incidence of cancers in her family could be due to inherited mutations in their genes. If so, they can test Rebecca to find out whether she has the particular variations of these genes that would increase her risk of getting cancer.
When Rebecca sees the genetic counselor, he asks how her grandmother, aunt, and uncle with cancer are related to her. She says that these relatives are all on her mother’s side — they are her mother’s mother and siblings. The genetic counselor records this information in the form of a specific type of family tree, called a pedigree, indicating which relatives had which type of cancer and how they are related to each other and to Rebecca. He also asks her ethnicity. Rebecca says that her family, on both sides, are Ashkenazi Jews, meaning Jews whose ancestors came from central and eastern Europe. “But what does that have to do with anything?” she asks. The counselor tells Rebecca that mutations in two tumor-suppressor genes called BRCA1 and BRCA2, located on chromosome 17 and 13, respectively, are particularly prevalent in people of Ashkenazi Jewish descent and greatly increase the risk of getting cancer. About 1 in 40 Ashkenazi Jewish people have one of these mutations, compared to about 1 in 800 in the general population. Her ethnicity, along with the types of cancer, age of onset, and the specific relationships between her family members who had cancer indicate to the counselor that she is a good candidate for genetic testing for the presence of these mutations.
Rebecca says that her 72-year-old mother never had cancer, and nor had many other relatives on that side of the family, so how could the cancers be genetic? The genetic counselor explains that the mutations in the BRCA1 and BRCA2 genes, although dominant, are not inherited by everyone in a family. Also, even people with mutations in these genes do not necessarily get cancer — the mutations simply increase their risk of getting cancer. For instance, 55 to 65% of women with a harmful mutation in the BRCA1 gene will get breast cancer before age 70, compared to 12% of women in the general population who will get breast cancer sometime over the course of their lives.
Rebecca is not sure she wants to know whether she has a higher risk of cancer. The genetic counselor understands her apprehension but explains that if she knows that she has harmful mutations in either of these genes, her doctor will screen her for cancer more often and at earlier ages. Therefore, any cancers she may develop are likely to be caught earlier when they are often much more treatable. Rebecca decides to go through with the testing, which involves taking a blood sample, and nervously waits for her results.
Chapter Overview: Genetics
At the end of this chapter, you will find out Rebecca ’s test results. By then, you will have learned how mutations in genes such as BRCA1 and BRCA2 can be passed down and cause disease. Especially, you will learn about:
- How Gregor Mendel discovered the laws of inheritance for certain types of traits.
- The science of heredity, known as genetics, and the relationship between genes and traits.
- Simple and more complex inheritance of some human traits.
- Genetic Disorders.
As you read this chapter, keep Rebecca’s situation in mind and think about the following questions:
- What do the BRCA1 and BRCA2 genes normally do? How can they cause cancer?
- Are BRCA1 and BRCA2 considered linked genes? And are they on autosomes or sex chromosomes?
- After learning more about pedigrees, draw the pedigree for cancer in Rebecca’s family. Use the pedigree to help you think about why it is possible that her mother does not have one of the BRCA gene mutations, even if her grandmother, aunt, and uncle did have it.
- Why do you think certain gene mutations are prevalent in certain ethnic groups?
Attributions
- Caelius and Valerius family tree by Ann Martin , licensed CC BY 2.0 via Flickr
- Text adapted from Human Biology by CK-12 licensed CC BY-NC 3.0

- GPU Computing
- Cloud Computing
- Lattice Microbes
- Atomic Resolution Brownian Dynamics
- VND (Neuronal)
Publications
Lectures and talks, computational facility, case studies, publications database.
The case studies provided on this page exploit the molecular graphics program VMD for teaching molecular cell biology. The case studies start out like a conventional textbook chapter, but utilize VMD molecular graphics to offer a much more detailed view of the subjects than commonly possible in textbooks. The case studies induce the reader to inspect by means of VMD the systems introduced. For this purpose the case studies show the molecular systems through graphical images, but provide also the files that permit the reader to regenerate the images with few mouse clicks and then explore in depth the structures shown using the program VMD. Students can rotate the images, enlarge them, alter the views in many ways, and analyze structures and sequences. The readers, undergraduate or graduate students, are expected to have a basic familiarity with the molecular graphics program VMD that is achieved best by completing the " Using VMD " tutorial.
The case studies below introduce various, simple, yet relevant and interesting subjects also found in textbooks. Readers will be amazed how much more they learn reading the case studies not in a passive manner, but in an active manner by using VMD.

- Viruses are the smallest life form in existence. Satellite Tobacco Mosaic Virus (STMV) is one of the simplest viruses. STMV has a protective outer coat consisting of 60 identical proteins. The coat surrounds the virus's genetic material, in this case a ribonucleic acid (RNA) molecule. STMV will be explored in this case study to illustrate the principles of virus structure . . .
- STMV Case Study [ pdf , 6.2M]
- Required case study files: [ .tgz , 29.4M]

- Water is essential for sustaining life on Earth. Almost 75% of the Earth's surface is covered by it. It composes roughly 70% of the human body by mass. It is the medium associated with nearly all microscopic life processes. Much of the reason that water can sustain life is due to its unique properties...
- Water Case Study [ pdf , 1.1M]
- Required case study files: [ .tgz , 65.3M]

- With the 50th aniversary of the discovery of the DNA structure by James Watson and Francis Crick and the race for the human genome, with all its controversies and awe-inspiring medical implications, we have all heard a lot about DNA, the molecule of life...
- DNA Case Study [ pdf , 2.0M]
- Required case study files: [ .tgz , 81.3M]

- Membranes are essential to cellular organisms. They are like fortresses in that they provide a barrier between the inside and outside with guarded drawbridges in the form of proteins that regulate the influx and efflux of material. Unlike the rigid walls of a fortress, membranes are fluid and are able to bend and move. The bricks forming a membrane, called lipids, freely move. Plasma membranes enclose and define the boundaries of a cell, maintaining a barrier between the interior of a cell, the cytosol, and the extracellular environment...
- Bilayer Case Study [ pdf , 1003k]
- Required case study files: [ .tgz , 39.2M]

- Bovine pancreatic trypsin inhibitor (BPTI) is one of the smallest and simplest globular proteins that inhibits protein digestive action (digesting protein to peptides) of the enzyme trypsin in cow (bovine) pancreas. BPTI is a member of the family of serine protease inhibitors. These enzymes have many conserved cysteines that form disulfide bonds that stabilize protein three-dimensional structures. BPTI has a relatively broad specificity in that it can inhibit several kinds of digestive enzymes...
- BPTI Case Study [ pdf , 4.9M]
- Required case study files: [ .tgz , 27.6M]

- Without a doubt, the most organized and coordinated machine known is the biological cell. Inside its micrometer-scale diameter, a wide variety of macromolecules (DNA, proteins, sugars, lipids, etc.) work together in a cooperative way, balancing energy and matter to keep the cell alive. To maintain harmony and efficiency between various functions, most processes have to be turned on or off according to different cellular stages and changes within the environment. To this end, together with the mechanisms to assemble functional proteins and to turn on their functions, there should be counterparts to suppress and disassemble proteins when they are no longer needed. Ubiquitin is a key player in eukaryotic intracellular protein degradation...
- Ubiquitin Case Study [ pdf , 1.7M] Required case study files: [ .tgz , 40.2M])

- Myoglobin is a small, monomeric protein which serves as an intracellular oxygen storage site. It is found in abundance in the skeletal muscle of vertebrates, and is responsible for the characteristic red color of muscle tissue. Myoglobin is closely related to hemoglobin, which consists of four myoglobin-like subunits that form a tetramer and are responsible for carrying oxygen in blood. In humans, blood-borne cardiac myoglobin can serve as a biomarker of heart attack, since blood myoglobin levels rise in two to three hours following muscle injury...
- Myoglobin Case Study [ pdf , 2.8M]
- Required case study files: [ .tgz , 63.5M])

- The organization of water is critical to most biological processes. Although cell membranes are to some extent water-permeable, they cannot facilitate the rapid exchange of large volumes of water, as required by the kidneys in the human body. Aquaporins (AQPs) are a family of specialized integral membrane proteins which function mainly as water channels. These highly efficient water channels can explain how we secrete tears, saliva and sweat, how our kidneys concentrate urine, and how our brains maintain spinal fluids...
- Aquaporin Case Study [ pdf , 3.2M]
- Required case study files: [ .tgz , 16.9M]

- The physiological function of ion channels had been observed long before they were known to exist. Many cells such as nerve and muscle cells, were known to have excitable cell membranes that would respond to variations in their electrical membrane potential through an all-or-nothing response. It was soon discovered that the regulators of ion passage across biological membranes were specialized proteins called ion channels. Ion channels are transmembrane proteins that selectively allow a given species of ions to pass through them...
- Ion Channels Case Study [ pdf , 3.0M]
- Required case study files: [ .tgz , 13.4M]

- In the extreme sport of bungee jumping, a daring athlete leaps from a great height and free-falls while a tethered cord tightens and stretches to absorb the energy from the descent. The bungee cord protects the jumper from serious injury, because its elasticity allows it to extend and provide a cushioning force that opposes gravity during the fall. Amazingly, nature also uses elasticity to dampen biological forces at the molecular level, such as during extension of a muscle fiber under stress. The molecular bungee cord that serves this purpose in the human muscle fiber is the protein titin, which functions to protect muscle fibers from damage due to overstretching...
- Titin Case Study [ pdf , 992k]
- Required case study files: [ .tgz , 52.6M])

- Sunlight is ultimately the energy source for nearly all life on Earth. Many organisms, such as plants, algae, and some bacteria, have developed a means to harvest sunlight and turn it into chemical energy, a process known as photosynthesis. Photosynthesis occurs with an amazingly high efficiency, not surprising given the more than 3.5 billion years of evolution...
- LH2 Case Study [ pdf , 13.0M]
- Required case study files: [ .tgz , 1.7M])

- The ribosome, one of the largest molecular machines in living cells, is in charge of protein synthesis. The ribosome translates genetic code into amino acid sequences that fold into protein structures. Ribosomes are the birth place of proteins in living cells. The bacterial ribosome plays also a key role in medicine as a drug target for antibiotics. Antibiotics are employed by biological cells, for example those arising in molds, to fight bacteria...
- Ribosome Case Study [ pdf , 10.0M]
- Required case study files: [ .tgz , 48.0M])

Systems biology of persistent infection: tuberculosis as a case study
Affiliation.
- 1 Centre for Integrative Systems Biology at Imperial College, Imperial College London, London SW7 2AZ, UK. [email protected]
- PMID: 18536727
- DOI: 10.1038/nrmicro1919
The human immune response does an excellent job of clearing most of the pathogens that we encounter throughout our lives. However, some pathogens persist for the lifetime of the host. Despite many years of research, scientists have yet to determine the basis of persistence of most pathogens, and have therefore struggled to develop reliable prevention and treatment strategies. Systems biology provides a new and integrative tool that will help to achieve these goals. In this article, we use Mycobacterium tuberculosis as an example of how systems-biology approaches have begun to make strides in uncovering important facets of the host-pathogen interaction.
Publication types
- Research Support, N.I.H., Extramural
- Research Support, Non-U.S. Gov't
- Bacteriology / trends*
- Molecular Epidemiology
- Mycobacterium tuberculosis / genetics*
- Proteome / physiology*
- Systems Biology*
- Tuberculosis / immunology
- Tuberculosis / physiopathology*
- Tuberculosis / prevention & control
- Tuberculosis / therapy
Grants and funding
- MC_U117581288/MRC_/Medical Research Council/United Kingdom
- R01 LM009027/LM/NLM NIH HHS/United States
- R01 HL 072682/HL/NHLBI NIH HHS/United States
- BB/C519670/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- Search Menu
- Sign in through your institution
- Advance articles
- Regular articles
- Spotlight articles
- Points of view
- Software for systematics and evolution
- Announcements
- Why Publish
- Author Guidelines
- Submission Site
- Open Access
- About Systematic Biology
- About the Society of Systematic Biologists
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Self-Archiving Policy
- Dispatch Dates
- Journals on Oxford Academic
- Books on Oxford Academic

Article Contents
Supplementary material, conflict of interest, acknowledgments, data availability.
- < Previous
Geogenomic Predictors of Genetree Heterogeneity Explain Phylogeographic and Introgression History: A Case Study in an Amazonian Bird ( Thamnophilus aethiops )
- Article contents
- Figures & tables
- Supplementary Data
Lukas J Musher, Glaucia Del-Rio, Rafael S Marcondes, Robb T Brumfield, Gustavo A Bravo, Gregory Thom, Geogenomic Predictors of Genetree Heterogeneity Explain Phylogeographic and Introgression History: A Case Study in an Amazonian Bird ( Thamnophilus aethiops ), Systematic Biology , Volume 73, Issue 1, January 2024, Pages 36–52, https://doi.org/10.1093/sysbio/syad061
- Permissions Icon Permissions
Can knowledge about genome architecture inform biogeographic and phylogenetic inference? Selection, drift, recombination, and gene flow interact to produce a genomic landscape of divergence wherein patterns of differentiation and genealogy vary nonrandomly across the genomes of diverging populations. For instance, genealogical patterns that arise due to gene flow should be more likely to occur on smaller chromosomes, which experience high recombination, whereas those tracking histories of geographic isolation (reduced gene flow caused by a barrier) and divergence should be more likely to occur on larger and sex chromosomes. In Amazonia, populations of many bird species diverge and introgress across rivers, resulting in reticulated genomic signals. Herein, we used reduced representation genomic data to disentangle the evolutionary history of 4 populations of an Amazonian antbird, Thamnophilus aethiops , whose biogeographic history was associated with the dynamic evolution of the Madeira River Basin. Specifically, we evaluate whether a large river capture event ca. 200 Ka, gave rise to reticulated genealogies in the genome by making spatially explicit predictions about isolation and gene flow based on knowledge about genomic processes. We first estimated chromosome-level phylogenies and recovered 2 primary topologies across the genome. The first topology (T1) was most consistent with predictions about population divergence and was recovered for the Z-chromosome. The second (T2), was consistent with predictions about gene flow upon secondary contact. To evaluate support for these topologies, we trained a convolutional neural network to classify our data into alternative diversification models and estimate demographic parameters. The best-fit model was concordant with T1 and included gene flow between non-sister taxa. Finally, we modeled levels of divergence and introgression as functions of chromosome length and found that smaller chromosomes experienced higher gene flow. Given that (1) genetrees supporting T2 were more likely to occur on smaller chromosomes and (2) we found lower levels of introgression on larger chromosomes (and especially the Z-chromosome), we argue that T1 represents the history of population divergence across rivers and T2 the history of secondary contact due to barrier loss. Our results suggest that a significant portion of genomic heterogeneity arises due to extrinsic biogeographic processes such as river capture interacting with intrinsic processes associated with genome architecture. Future phylogeographic studies would benefit from accounting for genomic processes, as different parts of the genome reveal contrasting, albeit complementary histories, all of which are relevant for disentangling the intricate geogenomic mechanisms of biotic diversification. [Amazonia; biogeography; demographic modeling; gene flow; gene tree; genome architecture; geogenomics; introgression; linked selection; neural network; phylogenomic; phylogeography; reproductive isolation; speciation; species tree.]
A key goal of speciation research is to understand the biogeographic mechanisms associated with population divergence and homogenization ( Endler 1977 ). Although the reduction of gene flow between populations caused by biogeographic barriers (henceforth geographic isolation) typically plays a crucial role in speciation, populations may diverge despite high gene flow ( Nosil 2008 ). Such gene flow, especially between non-sister populations, can result in heterogeneous levels of differentiation across the genome ( Keller et al. 2013 ; Gompert et al. 2014 ; Mallet et al. 2016 ; Meier et al. 2017 ; Pulido-Santacruz et al. 2020 ), a pattern further intensified by interactions with selection and genome architecture ( Feder et al. 2012 ; Cruickshank and Hahn 2014 ; Irwin et al. 2018 ; Manthey et al. 2021 ). For example, genomic regions experiencing strong disruptive selection or low recombination (e.g., large chromosomes; Haenel et al. 2018 ) may resist the homogenizing effects of gene flow, resulting in elevated peaks of divergence and contrasting genealogies when compared with other parts of the genome. In these cases, recombination rate and introgression should covary when there is baseline selection against gene flow due to divergent selection or cohesion among co-evolved genes that reduce gene flow. Thus, the genomic landscape of divergence not only reflects disparate levels of differentiation between populations but also a profusion of genealogical relationships ( Fontaine et al. 2015 ; Mallet et al. 2016 ; Wen et al. 2016 ). Dealing with and modeling evolutionary history in light of this heterogeneous landscape is crucial for obtaining a detailed understanding of biogeographic processes ( Provost et al. 2022 ).
Although the genomic landscape can be highly heterogeneous, the signals of distinct processes, such as divergence and gene flow, are nonrandomly distributed across the genome ( Van Doren et al. 2017 ). For instance, variation in recombination rates directly impacts levels of gene flow and divergence ( Wang et al. 2022 ). One widely recognized mechanism by which this operates is the breakdown of blocks of linked loci affected by selection. Specifically, linked selection (selection in the genome impacting nearby sites) can significantly diminish genetic variation in blocks of linked loci through genetic hitchhiking of nearby sites ( Lohmueller et al. 2011 ; Feder et al. 2012 ). When linked selection within populations is strong, measures of relative divergence, such as F ST , which contain a term for within population variation, are expected to increase, but absolute divergence, d xy , may be reduced (barring strong genomic island effects) by the corresponding depletion of allelic diversity ( Charlesworth 1998 ; Nachman and Payseur 2012 ; Cruickshank and Hahn 2014 ; Van Doren et al. 2017 ; Irwin et al. 2018 ). In regions of the genome where recombination is high, blocks of linked loci are more frequently broken down, lessening the effects of linked selection ( Tigano et al. 2022 ). Where recombination is low, however, the effects of selection are elevated as longer blocks of linked loci are able to persist. Recombination rate varies considerably across the genome and is particularly associated with chromosome size. Because each chromosome must undergo at least one crossing-over event during Meiosis ( Mather 1938 ), smaller chromosomes experience more recombination per base than large chromosomes ( Haenel et al. 2018 ; Tigano et al. 2022 ). Thus, regions of the genome with higher rates of recombination, such as smaller chromosomes, are also expected to have higher rates of introgression, when population divergence occurs with gene flow ( Martin et al.2019 ; Manthey et al. 2021 ). Quantifying the variation and predictability of these genomic processes that are intrinsic to organisms can help shed light on the reticulated history of recent radiations.
In contrast to intrinsic genomic architecture, biogeographic history is an important extrinsic factor influencing reticulation and the genomic landscape ( Burbrink and Gehara 2018 ; Thom et al. 2021 ; Provost et al. 2022 ). As levels of isolation associated with physiographic barriers vary through space and time, so too do rates of selection, gene flow, and divergence ( Endler 1977 ; Aguilée et al. 2013 ; Delmore et al. 2018 ; He et al. 2019 ). In many parts of the world, population isolation and connectivity vary, in part, as functions of spatiotemporal variation in the environment ( Flantua et al. 2019 ; He et al. 2019 ; Musher et al. 2019 ). For example, rates of isolation and gene flow among populations that diverged across the Isthmus of Panama may have been affected by the wax and wane of humid and dry forests across that region ( David Webb 1991 ; Vrba 1992 ; Smith et al. 2012 ; Musher et al. 2020 ). Likewise, sea-level fluctuations and rainfall patterns have directly affected the distribution and amount of flooded forest habitat in Amazonia, which also affected levels of gene flow between populations of organisms that occur there ( Thom et al. 2020 ; Sawakuchi et al. 2022 ; Luna et al. 2023 ). In Amazonian lowlands, differentiated populations often experience pervasive gene flow, sometimes from multiple non-sister lineages, a factor that complicates inference about their historical relationships, biogeography, and systematics ( Pulido-Santacruz et al. 2018 ; Del-Rio et al. 2022 ; Musher et al. 2022 ). This is because gene flow between non-sister taxa results in a network of interpopulation relationships that violates the assumptions of a bifurcating model of evolutionary history ( Mallet et al. 2016 ; Thom et al. 2018 ). Thus, if biogeography drives opportunities for isolation and contact between non-sister taxa, it should result in distinct predictable signatures of genealogy in the genomes of diverging populations.
Many lowland terra-firme (non-flooded forest) Amazonian birds have geographically isolated populations across rivers yet experience high levels of gene flow ( Barrera-Guzmán et al. 2022 ; Del-Rio et al. 2022 ; Musher et al. 2022 ). Rivers are key biogeographic barriers for many lowland Amazonian birds, driving population isolation and genetic structure across the landscape ( Sick 1967 ; Capparella 1991 ; Ribas et al. 2012 ; Smith et al. 2014 ; Ferreira et al. 2017 ). However, 3 well-known features of Amazonian lowlands add complexity to this system. First, the Amazon Basin, especially its southern portion, is characterized by several large tributaries running in quasi-parallel, forming isolated blocks of habitat (interfluves) wherein a given taxon may be surrounded by 2 or more closely related taxa that occur on opposite river margins. Second, Amazonian rivers get narrower toward their headwaters, which is associated with increased gene flow across their upper portions ( Weir et al. 2015 ). Finally, lowland river basins continuously rearrange via tributary capture (the movement of a tributary from one basin to another) and avulsion (the erosion of channel boundaries, leading to channel migration) ( Gascon et al. 2000 ; Albert et al. 2018 ). In this geographic configuration, there are opportunities for multiple non-sister taxa to interact, and partially isolated populations can experience gene flow across rivers, leading to highly reticulated patterns of diversification across species’ genomes ( Musher et al. 2022 ).
Inferring the history of population isolation and gene flow under these conditions is a major challenge for researchers studying Amazonia because limited knowledge about the historical relationships of taxa also hampers an understanding of the mechanisms that contribute to the region’s high biodiversity ( Cracraft et al. 2020 ). Previous studies of Amazonian birds have greatly advanced our knowledge of the history of the region’s taxa, but in many cases, limited data or sparse spatial sampling has resulted in weak resolution of species’ complex histories of isolation and gene flow. Genomic approaches, however, are revealing many of the evolutionary and biogeographic mechanisms driving species accumulation in the Neotropics ( Thom et al. 2018 , 2020 ; Pulido-Santacruz et al. 2020 ; Schley et al. 2020 ). This is especially important given geologists’ growing understanding that the Amazonian landscape has been highly dynamic ( Bicudo et al. 2019 ; Pupim et al. 2019 ; Ruokolainen et al. 2019 ). Thus, important questions for Amazonian biogeography include, (1) how do we infer population history under conditions of high gene flow among non-sister taxa, especially in the context of a dynamic landscape? (2) What are the consequences of these complex histories of isolation and gene flow for Amazonian organisms at the genomic level? (3) do genealogical patterns vary predictably across the genome in a way that is informative for biogeographic inference? and (4) to what extent can reduced representation genomic data shed light on these questions?
In this study, we address these questions by testing competing hypotheses for the biogeographic history of a passerine bird, the White-shouldered antshrike Thamnophilus aethiops (Thamnophilidae), in southern Amazonia. The southern Amazonian lowlands are particularly dynamic and experienced a major riverine restructuring, wherein large tributaries moved among watersheds and across the landscape during the Quaternary ( Latrubesse 2002 ; Rossetti 2014 ; Ruokolainen et al. 2019 ; Rossetti et al. 2021 ). Some of these past movements occurred near the headwaters of the modern Madeira River, a large tributary of the Amazon that is a well-known biogeographic barrier for many bird species ( Fernandes 2013 ; Smith et al. 2014 ; Silva et al. 2019 ). For example, paleochannels between the modern upper Madeira and Purus Rivers indicate that the Madeira extended its basin by capturing a large tributary of the Purus ca. 200 Ka or less ( Fig. 1 ) ( Ruokolainen et al. 2019 ). This suggests that the upper portion of the Madeira formed more recently than the lower, likely becoming a barrier for many terrestrial organisms over 2 stages.

Spatially explicit predictions of the river capture scenario (top row). The stippled lines show the movement of a paleo-river, historically draining water via the Purus watershed (time = t0) and its subsequent capture by the modern Madeira watershed (time = t1). Phylogenetic predictions assume a set of historical relationships (t0); the predicted topology (t1) matches that inferred from mitochondrial DNA in a previous study ( Thom and Aleixo 2015 ). Red arrows indicate opportunities for gene flow between non-sister populations. The maps in the top row are colored by topography, with dark pinks representing lower elevations (e.g., river channels) and darker greens representing uplands. The bottom row shows the study region, sampling (white circles), and key interfluvial areas (from west to east): W Inambari (purple), E Inambari (teal), S Rondônia (pale green), and N Rondônia (red).
The Madeira River capture scenario provides predictions about the history of population divergence and gene flow, wherein populations east of the Upper Madeira (Southern Rondônia; Fig. 1 ) are expected to be more closely related to populations west of the Madeira (Inambari) than to other populations within Rondônia ( Fernandes 2013 ; Ferreira et al. 2017 ). Such a scenario also suggests that there will be more genetic differentiation, and therefore potential incompatibilities across the genomes of more deeply diverged populations (e.g., those in Rondônia) than between more recently diverged populations (e.g., populations within Inambari). Previous studies of T. aethiops have been equivocal with respect to these predictions ( Thom and Aleixo 2015 ; Musher et al. 2022 ). For example, the mitochondrial (mtDNA) phylogeny is consistent with the expectations of river capture, recovering a sister relationship between populations on opposite sides of the Madeira River (but not its downstream portions). Genomic data were more ambiguous, however, recovering relatively strong support for the monophyly of Rondônia and Inambari populations, which conflicts with the expectations of river capture. Thus, if the population in S Rondônia is historically related to Inambari populations, as the river capture scenario predicts, then we expect regions of the genome supporting monophyly of Rondônia to be driven by introgressive hybridization after secondary contact. Herein, we test these alternative genealogical predictions while also utilizing and modeling information about genome architecture. For instance, if gene flow increases on smaller autosomes, genealogies resulting from secondary contact should be less likely on larger autosomes and sex chromosomes. Rather, regions of the genome inferred to have low gene flow should better track phylogeny sensu stricto (i.e., the history of population divergences).
Study Taxon
Thamnophilus aethiops is well-suited for testing our hypotheses as it exhibits both subspecific variation and genetic structure across southern Amazonian rivers ( Thom and Aleixo 2015 ; Musher et al. 2022 ). Populations east of the Madeira River belong to a single subspecies, T. a. punctuliger . Populations west of the Madeira fall into one of 3 subspecies: T. a. injunctus occurs between the Purus and Madeira Rivers, T. a. juruanus occurs between the Jurua and Purus Rivers, and T. a. kapouni occurs west of the Juruá. Superficially, the phenotypes of T. a. injunctus and T. a. punctuliger are most similar, with individuals of both taxa being lighter gray overall and marked with white spots on the wing coverts, unlike populations farther west. Moreover, T. aehiops is a fairly sedentary understory species, so gene flow expectations across stable rivers are relatively low.

Sampling and Genotyping-by-sequencing Data Assembly
We downloaded genotyping-by-sequencing (GBS) data from a previous study ( Musher et al. 2022 ) and re-assembled it using iPyrad version 0.9.81 ( Elshire et al. 2011 ; Eaton and Overcast 2020 ). These data are available on the NIH Sequence Read Archive under project ID PRJNA966941. Raw reads were deposited in NCBI Sequence Read Archive under ID SRR26669395. We were interested in the history and relationships of populations west of the Tapajós River and south of the Amazon River (southwestern Amazon Basin), so we excluded samples falling outside of this region. This dataset totaled 51 samples ( Supplementary Tables S1 and S2 ). First, we demultiplexed and cleaned raw reads. Specifically, we used TGCA as our restriction overhang (Pstl-I, which cleaves DNA at 5ʹ-CTGCA/G-3ʹ sites was the restriction enzyme used in library prep), allowed a maximum of 5 low-quality ( Q < 20) base calls in each read with a Phred Q-score offset of 33, and discarded any cleaned reads with fewer than 35 bp. We then mapped cleaned reads to a newly assembled reference genome of a closely related species Thamnophilus caerulescens (divergence time: ~4 Ma; Harvey et al. (2020) ; see Supplementary Materials ). iPyrad uses vsearch ( Rognes et al. 2016 ) to merge overlapping paired reads and then bwa ( Li and Durbin 2009 ) to map those paired reads to a reference genome and determine locus homology, discarding any unmapped or replicated reads. In sum, paired-end reads are mapped to the reference genome based on sequence similarity, and reads that cluster at the same genomic loci are then aligned using muscle ( Edgar 2022 ). Each of these “aligned clusters” represents a single GBS locus. We applied a minimum statistical coverage of six, which is considered the minimum value for accurate base calling that is widely used in RADSeq studies ( Eaton and Overcast 2020 ; Barreto et al. 2022 ; Donoghue et al. 2022 ; Hanes et al. 2022 ). When estimating consensus allele sequences from clustered reads, we allowed default settings of a maximum of 5% ambiguous base calls and 5% heterozygous sites. These last 2 filters reduce the risk of incorporating erroneous alignments into the dataset that are likely to have increased heterozygosity and may be caused when many ambiguous bases are included in a consensus.
Population Structure and Ancestry
Characterization of the population structure of T. aethiops is crucial for delineating populations for downstream analysis and characterization of patterns of admixture. We first investigated genetic structure among populations using principal components analysis (PCA), implemented in the iPyrad API ( Eaton and Overcast 2020 ). Specifically, we used k- means iterative clustering to assign individuals to populations under a 4-population model ( k = 4). We iteratively clustered sample single-nucleotide polymorphisms (SNPs) present across 90% of individuals and clustered individuals based on an assumed value for the number of populations. We repeated this clustering 5 times, allowing more missing data at each successive iteration until reaching a minimum coverage of 75% of individuals for a given SNP. Next, missing genotypes for all samples were imputed by choosing one random genotype from the assigned population based on the k- means assignments. This method allowed imputation without a priori geographic bias. We then performed PCA on unlinked genotype calls (one randomly sampled genotype per locus) in this imputed dataset.
We then used sparse non-negative matrix factorization (sNMF), implemented in the R-package LEA3 ( Frichot et al. 2014 ; Gain and François 2021 ), to calculate admixture coefficients for each population and infer the best-fit number of ancestral populations ( k ) in the dataset. We tested values of k from one through 5 to infer the optimal number of ancestries. As sNMF results are sometimes sensitive to the regularization parameter, α , we explored our results under multiple values of α (10, 100, 1000, 10,000) using only 10 replicates. However, given our relatively large dataset, our results were stable across all α -values, so we performed 1000 iterations of each k value using α = 10 to obtain our final results. The best-fit value of k was determined by choosing the k with the lowest cross-entropy value ( Frichot et al. 2014 ). In addition to the best-fit value of k , we also visualized the results from other values of k , to better characterize additional, biologically relevant population structure present in the data. To assess the stability of population assignments in sNMF, we ran this procedure multiple times, replicating the 1000 iterations of sNMF over multiple runs.
Genetrees, Chromosome Trees, and Species Tree Topologies
We evaluated the phylogenetic relationships for the populations recovered in sNMF and PCA analyses ( K = 4), which were also consistent with previously published mitochondrial clades ( Thom and Aleixo 2015 ). We estimated genetrees for sliding windows across the genome using TreeSlider, a python program available within the iPyrad API ( Eaton and Overcast 2020 ). TreeSlider keeps the most common allele for each locus in each population and estimates the maximum likelihood phylogeny at each locus using RAxML ( Stamatakis 2014 ). We specifically compared the results from TreeSlider for 10 kb, 50 kb, 100 kb, and 200 kb sliding windows, first requiring a minimum of 5 SNPs, and then a minimum of 10 SNPs to retain each window. We found that 50 kb windows with a minimum of 5 SNPs retained the most (4858; Supplementary Table S2 ) loci, so we used the genetrees from this dataset for all downstream phylogenetic analyses.
To understand how phylogenetic history varied across the genome and among chromosomes, we analyzed genetree results at multiple scales using ASTRAL v5.7.8 ( Zhang et al. 2018 ). ASTRAL estimates an unrooted species tree given a set of genetrees by identifying the maximum number of shared induced quartets within the provided genetrees. We used ASTRAL to estimate the interrelationships of four populations of interest, corresponding to those identified using sNMF and PCA. To understand how these relationships varied among parts of the genome, we first ran ASTRAL for all 4858 genetrees together (genome-wide species tree), second for only autosomal genetrees (autosomal tree; that is, excluding genetrees from sex chromosomes), and finally for genetrees from each chromosome independently (chromosomal trees). As no loci on Chromosome 33 met our criteria (50 kb minimum of 5 SNPs), we excluded this chromosome from all ASTRAL analyses.
To quantify variation in support for competing phylogenetic hypotheses, we calculated unrooted topology weights for alternative topologies across windows of the genome using TWISST ( Martin and Van Belleghem 2017 ). This approach provides an assessment of the relative likelihood of alternative topologies for individual genomic windows. Here, we estimated genetrees from SNPs on sliding windows using PHYML v3.0 ( Guindon et al. 2010 ) following Martin and Van Belleghem (2017) . Due to the patchiness of GBS datasets, we analyzed windows of varying lengths with exactly 100 SNPs each. The larger window size and stricter filtering applied here enabled the estimation of relatively high-resolution genetrees, reducing noise in this analysis. We performed 2 independent runs testing the relationships between 4 taxa. First, we calculated topology weights for 3 unrooted topologies using the 4 ingroup populations. This configuration allowed us to test the sister relationship between E and W Inambari populations. Next, we added the reference genome ( T. caerulescens ) to the dataset and removed the W Inambari population, allowing us to test S Rondônia as sister to Inambari populations. Because TWISST quantifies the relative weights of unrooted topologies, running our analyses with 4 taxa enabled us to evaluate support for alternative quartets identified in ASTRAL as well as the mitochondrial topology, without the noise of more complicated 5-taxon statements. Initial tests with our data set revealed that 5-taxon runs had poor resolution given the larger set of alternative topologies.
Given the overall low support for the mtDNA tree across the genome (see “Results” section), we explored whether the few genomic windows with high support for mtDNA topology were linked to nuclear genes associated with mitochondrial activity. We selected genes associated with mitochondrial activity according to Morales et al. (2018) (734 N-mt genes; Gene Ontology term: 0005739 and 0006119) using our annotated reference genome, located within 100 kb of windows with topology weights above 3 standard deviations from the mean for the mtDNA tree.
Demographic Modeling
To estimate genome-wide topology for the 4 populations while accounting for gene flow, we used a combination of coalescent simulations performed with PipeMaster ( Gehara et al. 2017 ), and supervised machine learning implemented in Keras v2.3 (Arnold 2017; https://github.com/rstudio/keras ). We first simulated data under 3 demographic models matching alternative phylogenetic hypotheses under the effect of gene flow between selected populations. For the purposes of this section, we abbreviate N Rondônia as “NR,” S Rondônia as “SR,” W Inambari as “WI,” and E Inambari as “EI.” The simulated models were: Model 1) (NR,(SR(EI, WI))) with gene flow between SR and both NR and EI (M SR<->NR and M SR<->EI ); Model 2) ((NR, SR),(EI, WI)) with gene flow between SR and EI (M SR<->EI ); and Model 3) (NR,(WI,(EI, SR))) with gene flow between populations on both sides of the Madeira River (M SR<->NR and M WI<->EI ). Each of the 3 models matches the results obtained with independent phylogenetic analysis: Models 1 and 2 match the alternative topologies obtained with ASTRAL (T1 and T2; see “Results” section); whereas Model 3 matches the mtDNA topology of Thom and Aleixo (2015) (T3; see “Results” section). Symmetric gene flow was allowed in the model between pairs of non-sister populations potentially in contact (e.g., occurring on opposite sides of a river) that could explain the discordance observed in phylogenetic estimates. We only allowed symmetric gene flow between populations to reduce the number of parameters of our models and because our primary goal was to test for alternative phylogenetic topologies while accounting for gene flow; our intent was not to estimate the absolute values of demographic parameters. For all models we set relatively large and uniform priors based on reasonable values for lowland Amazonian species: Ne 100,000–1,000,000 diploid individuals (for all populations); M pop1<->pop2 0.1–3.0 migrants per generation; Tdiv (Divergence time in generations) Model 1: WI/EI 50,000–800,000, SR/WI+EI 500,000–1,200,000, NR/WI+EI+SR 750,000–1,500,000; Model 2: WI/EI 50,000–800,000, NR/SR 50,000–800,000, NR+SR/WI+EI 750,000–1,500,000; and Model 3: EI/SR 50,000–800,000, WI/EI+SR 500,000–1,200,000, NR/WI+EI+SR 750,000–1,500,000. We assumed a fixed mutation rate of 2.42 × 10 −9 mutations per generation and a 1-year generation time ( Jarvis et al. 2014 ; Zhang et al. 2014 ). Although an assumed 1-year generation time may underestimate the true generation time, limited data about survivorship and reproduction in Amazonian birds precludes a more precise estimate ( Saether et al. 2005 ).
To obtain the observed data on which simulations were based, we initially converted the alleles file produced by iPyrad into individual fasta alignments with the iPyrad.alleles.loci2fasta function of PipeMaster. Since PipeMaster is sensitive to missing data, we also applied multiple filters to the sequence data. First, we removed individuals missing more than 50% of the loci from alignments using Alignment_Refiner_v2.py ( Portik et al. 2016 ) and excluded alignment positions not recovered for at least 80% of the individuals using trimAL ( Capella-Gutierrez et al. 2009 ). Finally, we excluded loci with fewer than 51 individuals, shorter than 100 bp, and missing more than 50% of the sites using AMAS and custom R scripts ( Borowiec 2016 ). The genetic data for each model was simulated on PipeMaster based on the number of retained loci, matching their length and number of individuals using msABC ( Pavlidis et al. 2010 ). To summarize genetic variation of observed and simulated data, we calculated multiple population genetics summary statistics, including mean and variance across loci: (1) the number of segregating sites, both per population and summed across populations; (2) nucleotide diversity ( π ), both per population and for all populations combined; (3) Watterson’s theta per population and for all populations combined; (4) pairwise F ST between populations; the number of shared alleles between pairs of populations; (5) the number of private alleles per population and between pairs of populations; and (6) the number of fixed alleles per population and between pairs of populations. These summary statistics were used as feature vectors on a neural network (nnet) approach implemented in Keras designed to estimate the classification probability of the 3 simulated models given our data and associated demographic parameters.
After careful parameter exploration, the final architecture of our neural network had three hidden layers with 32 internal nodes and a “relu” activation function. For model classification, our output layer was composed of 3 nodes and a “softmax” activation function. Three-quarters of the simulations were used as training data, and the remaining 25% were used to test the accuracy of our approach in assigning simulations to the correct model. The neural network training step was run for 1000 epochs using the “adam” optimizer and a batch size of 20,000 using 5% of the data for validation. To track improvements in model classification during training, we calculated the overall accuracy and the sparse_categorical_crossentropy for each epoch. After identifying the most probable model for our observed data, we estimated demographic parameters with a neural network designed to predict continuous variables. Here, we used a similar architecture to the one described above but set an output layer with a single node and a “relu” activation. We used this approach to estimate the effective size of each population, the amount of gene flow between populations, and divergence times. To assess improvements in accuracy during training, we used the mean absolute percentage error (MAE) as an optimizer. We trained the neural network for 3000 epochs with a batch size of 10,000 and a validation split of 0.1. To account for variation in parameter estimation, we ran 10 replicates and summarized the results calculating mean values for each demographic parameter. We also assessed the accuracy of parameter estimation by calculating the coefficient of correlation between estimated and true simulated values of the testing data set.
Testing the Link Between Chromosome Length, Linked Selection, and Introgression
To test for predicted patterns associated with linked selection across the genome, we calculated nucleotide diversity ( π ), relative divergence ( F ST ), and absolute divergence ( d xy ) across the genome. To calculate π , d xy , and F ST , we used pixy, a command line utility that handles missing data by adjusting the site-level denominators and sequence length ( Korunes and Samuk 2021 ). Summary statistics were estimated based on vcf files with invariant sites included. To examine the robustness of these estimates given the incomplete nature of GBS datasets, we employed pixy for multiple sliding window widths and data filters (see Supplemental Materials ). We specifically examined pixy results for (1) 50 kb windows with an unfiltered vcf; (2) 50 kb windows with a vcf filtered for sites with a minimum depth of 10×, and (3) 250 kb windows with a vcf filtered for sites with a minimum depth of 10×. As we found that pixy results were robust to these depth filters and window sizes (see Supplemental Materials ), we opted to use the dataset that retained the most variants, the 50 kb windows with a minimum depth of 6×.
To better understand how topology and rates of gene flow vary among chromosomes, we fit data to generalized linear models (glms) in R ( R Core Team 2019 ) to test for an association with chromosome length and bootstrapped these models to examine their sensitivity (see Supplemental Materials ). In so doing, we make the assumption that genome structure is relatively conserved across New World suboscine birds (Passeriformes, Tyrannides) since the chromosome lengths of the reference genome are derived from those in Chiroxiphia lanceolata , a manakin. This assumption is supported given the high synteny within passerine birds ( Dawson et al. 2007 ; Ellegren 2010 ; Delmore et al. 2018 ; Coelho et al. 2019 ; Peñalba et al. 2020 ). We first modeled π , d xy , and F ST (unfiltered 50 kb-window dataset) as functions of chromosome length (glm family = “gaussian”) based on the results from the pixy analyses outlined above. To do this, we calculated the average value for each statistic across each chromosome by taking the mean value across all windows after eliminating empty windows (i.e., NA values). As the distribution of chromosome lengths was right-skewed, we evaluated these models after log 10 transforming the data. This allowed us to track general patterns associated with linked selection and recombination in the data. For example, since prior studies have shown that gene flow is expected to increase on smaller chromosomes ( Martin et al. 2019 ), understanding how patterns of divergence, genealogy, and of introgression change with chromosome length enables us to evaluate whether evidence for certain phylogenetic topologies might be driven by gene flow between non-sister populations. For all Gaussian regressions, we also estimated Pearson’s correlation coefficient ( r ).
Next, we used logistic regression (family = ”binomial”) to test for an association between chromosome length and phylogenetic topology, scoring genetrees (i.e., RAxML trees from the sliding window analysis above) as monophyletic (1) or non-monophyletic (0) for populations in Rondônia, Inambari, and S Rondônia+E Inambari. These logistic regression analyses enabled us to test for associations between genealogical relationships across the genome and chromosome length. ASTRAL recovered 2 alternative topologies across the genome corresponding to non-monophyletic (T1) and monophyletic (T2) Rondônia populations, and mtDNA recovered a sister relationship between E Inambari and S Rondônia (T3). Thus, these analyses were aimed at identifying how support for these three topologies varied across the genome to better evaluate whether certain topologies are associated with introgression. For logistic regression, we estimated the coefficient ( β ) and odds-ratio ( ψ ) for each model. We ran these logistic regression models on all 4858 genetrees as well as a subset of 1727 genetrees reconstructed from windows with a minimum of 10 SNPs. The latter allowed us to examine the robustness of these models to potential gene tree error. We also replicated this analysis on the chromosome-level ASTRAL phylogenies instead of genetrees.
To obtain direct measures of introgression, we then estimated the introgression proportion ( f dM ) across the genome using window-based ABBA–BABA tests in non-overlapping windows with exactly 100 SNPs using ABBABABAwindows.py ( https://github.com/simonhmartin/genomics_general ( Martin et al. 2014 ). An initial analysis using windows of fixed width (50 kb and 250 kb) produced highly biased f dM estimates, with high variance in f dM estimates for larger chromosomes. This was due to the incomplete nature of GBS datasets and amplification bias that led to denser sequence coverage on smaller chromosomes and sparser sequence coverage on larger chromosomes ( DaCosta and Sorenson 2014 ) (see Supplementary materials ). Thus, we chose to define windows by the number of SNPs to help reduce this bias and generate windows with equivalent information content. We specifically explored introgression between N and S Rondônia, which were defined by positive values of f dM assuming T1 as the species tree (P1 = WI, P2 = SR, P3 = NR, out = reference; see “Results” section).
These introgression results were then used to model f dM and the proportion of derived alleles shared by N and S Rondônia per window (variants shared by P2 and P3 but not P1 or P4; i.e., ABBA’s) as functions of chromosome length to understand how introgression varied across the genome. As positive values of f dM indicate introgression between P2 and P3 (N and S Rondônia), we modeled positive f dM values as a function of chromosome length after excluding negative values from the data frame. To evaluate whether introgression on the Z-chromosome was lower than mean autosomal introgression, we also used an unpaired Bayesian t- test for distributions with unequal variances implemented in the R-package Bolstad ( Curran 2013 ) to statistically test differences in mean values of f dM (p2<->p3) across windows on the Z-chromosome versus the mean f dM (p2<->p3) for windows on all autosomes combined.
Finally, previous work has shown that recombination not only increases on small chromosomes but also on the periphery of large chromosomes ( Haenel et al. 2018 ). To test for a potential effect of increased gene flow on the periphery of large chromosomes, we modeled f dM (p2<->p3) as a function of percent distance from a chromosome’s center (defined as the distance in base pairs of a window from the center of a chromosome divided by half the chromosome’s length in base pairs). All statistical analyses were performed in R version 4.1.2 ( R Core Team 2019 ). From all analyses, we excluded the W-chromosome, which does not undergo crossing-over and is subject to error given many males in the dataset. We also removed Chromosomes 31, 32, and 33, for which we recovered relatively little data.
GBS Assembly
Our final GBS assembly included a total of 118,127 loci with a mean and standard deviation of 62,882.65 and 17,506.31 loci assembled per sample, respectively (range = 11,955–85,123 loci). Within this dataset, we recovered a total of 1,277,143 SNPs.
Our results point to between 2 and 4 populations across the region corresponding to W Inambari, E Inambari, S Rondônia, and N Rondônia, including individuals with a shared coefficient of ancestrality between W Inambari and S Rondônia, as well as between N and S Rondônia ( Fig. 2 ). Although the best-fit value for the number of ancestries was k = 2 ( Supplementary Fig. S1 ), corresponding to populations east and west of the Madeira River, higher values of k were consistent with the PCA results, recovering additional distinct ancestries across the Aripuanã ( k = 3) and Purus ( k = 4) Rivers. Contrastingly, PCA consistently recovered 4 clusters of samples corresponding to areas delimited by the 3 focal rivers ( Fig. 2 ). These PCA clusters match the spatial boundaries of mitochondrial clades from a previous study ( Thom and Aleixo 2015 ). Still, in consistence with the sNMF results, PC1 (explaining 8.5% of the genomic variation) separated populations from across the Madeira river, PC2 (explaining 6.8% of the genomic variation) separated populations across the Aripuanã river, and PC3 (explaining 2.9% of the genomic variation) separated populations across the Purus river.

Results of PCA and ancestry on the full GBS dataset from samples spanning 4 populations. Colors correspond to shaded regions on the map ( Fig. 1 ): W Inambari (purple), E Inambari (teal), S Rondônia (pale green), and N Rondônia (red). (Left) Results of PCA on all samples. (Right) sNMF results for the optimal K value of K = 2 (top), as well as plots for K = 3 (center) and K = 4 (bottom).
Genetrees, Chromosome Trees, Species Tree, and Demographic History
We detected 3 competing phylogenetic topologies across the genome, 2 of which predominated among chromosome trees ( Fig. 3 ): Topology 1 (T1) matched the genome-wide species tree and was recovered for 9 of the 34 chromosomes, including the Z-chromosome; T2 matched the autosomal tree and was recovered for 22 of the autosomes. There were also 3 unique topologies recovered for Chromosome 29, Chromosome 31, and Chromosome 32 ( Supplementary Fig. S2 ). None of these were concordant with the mitochondrial topology recovered in a previous study (henceforth, T3) ( Thom and Aleixo 2015 ). Overall, populations from Rondônia were recovered as monophyletic for 22 chromosome trees and Inambari populations were monophyletic for all but 3 chromosomes.

Chromosome-level topologies inferred in astral for 50 kb sliding windows across the genome. Two chromosome tree topologies predominated across the genome: T1 (red chromosomes/red box), which is consistent with the genome-wide tree, and T2 (blue chromosomes/blue box), which is consistent with the autosomal tree. The mitochondrial topology is also shown (T3) with bootstrap support, based on the results of a previous study ( Thom and Aleixo 2015 ). The tree at the lower left shows the best-fit demographic model inferred using a convolutional neural network. Bars in the left column are proportional to chromosome length.
Demographic modeling similarly recovered T1 as the population history (model classification probability: model 2 = 0.99), finding high gene flow between S Rondônia and E Inambari as well as between N and S Rondônia ( Fig. 3 ). Model classification yielded high accuracy, with training data being correctly assigned to the simulated model 99% of the time (accuracy = 0.99). The neural network regression approach designed for demographic parameter estimations produced accurate estimates for most parameters ( R 2 simulated ~ estimated > 0.90), except for the 2 oldest divergence times, likely due to the high amount of gene flow between populations ( R 2 ~ 0.61; Supplementary Table S4 ). Effective population sizes were in general consistent with the size of the geographic distribution of the species, with the most restricted E Inambari populations having the smallest size (198,314 individuals; R 2 = 0.93; MAE = 72,272). However, because we only modeled symmetric gene flow between non-sister populations (and not all possible gene flow scenarios), we suggest these Ne be interpreted with caution. The divergence between E and W Inambari populations occurred around 150 Ka (149,546ya; R 2 = 0.95; MAE = 48,492), followed by the split between the ancestor of the Inambari populations from S Rondônia at about 600 Ka (616,206ya; R 2 = 0.61; MAE = 117,941), and the deepest divergence at around 820 Ka (824,957ya; R 2 = 0.61; MAE = 125,875). Gene flow estimates were high between both migration edges of the model suggesting considerable introgression between E Inambari and S Rondônia, and between Rondônia populations. Parameter estimates were in general contained within simulated priors except for gene flow estimates. Additional runs adjusting priors for gene flow drastically affected the accuracy of model classification, thus we assumed these constrained and conservative estimates.
The support for the 3 alternative topologies varied across the genome ( Supplementary Figs. S3 and S4 ; Table 1 ). The unrooted T1/T2 topology (T1 and T2 are the same when the ingroup quartet is unrooted) had the highest support across the genome in our analysis for the ingroup populations. However, when including the reference to differentiate between T1 and T2 topologies, both T1/T3 (T1 and T3 are the same when this quartet is unrooted) and T2 had similar support across the genome. There were relatively few outlier loci supporting T3 compared with the other topologies, and we did not find any association between n-mt genes and outlier peaks supporting the mitochondrial topology.
Number of outlier windows supporting 3 alternative topologies (T1/T2, T3, and a third unrooted topology Tx), and the overall number of genes, and number of genes linked to mitochondrial activity less than 100 kb from outlier peaks (N-mt genes) on Supplementary Fig. S4 [ingroup TWISST analysis].
Associations with Chromosome Length
We detected significant associations between chromosome length, genetree topologies, and genome statistics ( Fig. 4 ). First, we found a significant association with F ST ( n = 32, 50 kb windows: r = 0.818, P < 0.0001; 250 kb windows: n = 32, r = 0.666, P < 0.0001; Fig. 4a ), d xy (50 kb windows: n = 32, r = −0.856, P < 0.0001; 250 kb windows: n = 32, r = −0.931, P < 0.0001; Fig. 4b ), and π (S Rondônia: 50 kb windows, n = 32, r = −0.885, P < 0.0001, 250 kb windows, n = 34, r = −0.868, P < 0.0001; 50 kb windows N Rondônia: n = 32, r = −0.883, P < 0.0001; 250 kb windows: n = 34, r = −0.846, P < 0.0001; Fig. 4c , d ). Bootstrapping ( Supplementary Fig. S5 ) and pairwise correlations based on alternate VCF filtering and window sizes showed that the results of our linear regression models were not affected by unequal sequence coverage across windows and chromosomes ( Fig. 5 , Supplementary Figs. S6 and S7 ). We recovered a negative association between chromosome length and genetree topology, wherein genetrees on larger chromosomes have a reduced probability of recovering monophyly of Rondônia ( β = −0.231, ψ = 0.794, P = 0.0005; Fig. 4f ) and recapitulated this result at the level of chromosomes, though the latter was only weakly supported ( β = −0.0003, ψ = 0.999, P = 0.054; Fig. 4e ).

Logistic and linear regression tests for associations between genomic characteristics and chromosome length. Y -axis values in the top row reflect chromosome-wide means of 50-kb windows for (a) FST, (b) dxy, (c) pi for N Rondônia, and (d) pi for S Rondônia. Plots in the bottom row examine the influence of introgression on genetree variation across the genome showing (e) a nonsignificant association between chromosome topologies and chromosome length despite (f) a significant negative association in genetree topologies consistent with T2 and chromosome size, and (g and h) significant negative associations between introgression and chromosome length. Models for each association are shown as the solid line with the shaded gray area representing the model standard error. Bullseye points in plots g and h represent values for the Z-chromosome. Derived variants shared by Rondônia were defined as SNPs showing an ABBA pattern assuming topology T1, as indicated using the tree at the top right of plot g. The topology tips, from left to right are E Inambari, S Rondônia, N Rondônia, and T. caerulescens (outgroup), with A’s and B’s representing alternative alleles.

Heatmap of Pearson’s coefficients for pairwise correlations showing pairwise correlations of within-chromosome means of genetic summary statistics for values of dxy, FST, and pi. Empty (white) boxes represent correlations with nonsignificant P -values ( P > 0.05). Correlation coefficients are printed on each square.
We also confirmed that introgression varied across the genome and was negatively correlated with chromosome length ( Fig. 4g , h ). Specifically, we found significant negative correlations between f dM ( n = 31, r = −0.476, P = 0.007) and the proportion of derived variants shared by N and S Rondônia (ABBA’s; n = 31, r = −0.420, P = 0.019) with chromosome length. The Bayesian t -test confirmed ( df = 237.74, t = 3.566, P = 0.0005) that the Z-chromosome had lower levels of introgression (mean f dM = 0.178) than autosomes (mean f dM = 0.212). Overall, f dM values averaged positive across the genome, implying stronger gene flow between N and S Rondônia than between N Rondônia and E Inambari (positive values indicate an excess of gene flow between P2 and P3, whereas negative values indicate an excess between P1 and P3). We found no relationship between f dM and distance from chromosome center ( Supplementary Fig. S9 ).
Nonrandom Variation in Genealogical History Across the Genome
We combine phylogenomics and population genetics to investigate the interplay between genomic architecture and biogeographic processes in generating predictable patterns of genetree variation across the genome of an Amazonian antbird, T. aethiops . We found that accounting for chromosome length informs phylogenetic and biogeographic inference in cases of high gene flow among non-sister taxa; it may be necessary to examine genome architecture to properly interpret phylogeographic signal when divergence occurs with gene flow. Our results also suggest that reduced representation genomic data such as genotype-by-sequencing can be used with genomic-architecture-aware approaches, recapitulating expected associations between genomic processes and the signal for ancestry and introgression.
We tested 3 competing hypotheses for the relationships of 4 spatially adjacent and genetically differentiated populations that are semi-isolated across Amazonian tributaries. Two of these topologies, T1 and T2, were equally supported across genome-wide sliding windows ( Supplementary Fig. S3 ), which made inferring T. aethiops ’ evolutionary history challenging. We found that genealogical signal for these competing hypotheses was nonrandomly distributed across the genome; areas of low gene flow such as the Z-chromosome and larger autosomes tend to support T1, whereas areas with elevated gene flow such as smaller autosomes tend to recover genetrees consistent with T2 ( Fig. 4e–f ). The third topology, T3, was not well supported across the nuclear genome ( Supplementary Fig. S4 ) but was recovered for mitochondrial DNA. Importantly, introgression was negatively correlated with chromosome length and the Z-chromosome exhibited especially lower introgression than autosomes, consistent with expectations ( Supplementary Fig. S10 ). Thus, we suggest that T1 may be seen as representing the initial branching pattern among the 4 taxa, whereas the prevalence of T2 on smaller autosomes probably resulted from introgression between N and S Rondônia. These results suggest that the genome-wide diversification history of T. aethiops might be better explained by a complex network of differentiation and introgression between multiple interacting populations.
The idea that the prevalence of T2 is driven by gene flow is supported by theoretical predictions about genome architecture. Smaller chromosomes are expected to exhibit higher levels of gene flow than larger chromosomes due to their higher recombination rates that more effectively break the linkage between introgressed variants ( Martin et al. 2019 ; Tigano et al. 2022 ). During Meiosis, each chromosome must undergo at least one crossing-over event resulting in fewer recombination events per base on longer chromosomes than shorter ones. This becomes complicated in birds, which may also experience increased selection on smaller chromosomes due to denser gene content in those regions ( Henderson and Brelsford 2020 ). However, in birds, recombination is also thought to be tied to gene promoter regions ( Singhal et al. 2015 ). Thus, although there is an increase in the density of targets of selection along smaller chromosomes, there is also additional recombination acting to reduce the effects of such linked selection. To test this assumption, we modeled multiple population genetic statistics as functions of chromosome length. We found that F ST decreased on smaller chromosomes, but π and d xy were negatively correlated with chromosome length. These patterns are consistent with expectations associated with a reduction in overall genetic diversity due to linked selection on larger chromosomes, thereby increasing F ST and decreasing d xy . We also found that introgression statistics such as f dM and the proportion of derived alleles shared by Rondônia populations (i.e., ABBA’s given T1) were negatively correlated with chromosome length. Overall, these results confirm our assumption that rates of gene flow between N and S Rondônia are higher on smaller chromosomes and likely driven by increased recombination.
Biogeographic and Geogenomic Implications
Understanding how variation in genealogical history is associated with genomic processes enables a closer look at the processes driving population divergence and speciation in T. aethiops and can illuminate our understanding of Amazonian biogeography in general. Amazonian biogeography has been at the center of discussions on how landscape evolution leads to allopatric speciation ( Haffer 1997 ; Ribas et al. 2012 ), but increasingly, researchers are discovering that the histories of taxa across this landscape are marked by high gene flow ( Barrera-Guzmán et al. 2022 ). Recent genomic studies have reported introgression across rivers and considerable phylogenetic conflict, often despite strong genetic and phenotypic structuring ( Thom et al. 2021 ; Del-Rio et al. 2022 ; Musher et al. 2022 ). This includes examples of hybrid speciation ( Barrera-Guzmán et al. 2018 ), mitonuclear discordance ( Del-Rio et al. 2022 ), mitochondrial capture ( Ferreira et al. 2018 ), and extensive introgression across river headwaters ( Weir et al. 2015 ). Phylogenetic relationships among interfluvial populations have been used to inform paleogeographic models of landscape evolution and have also helped to generate and test multiple biogeographic hypotheses ( Cracraft and Prum 1988 ; Ribas et al. 2012 ). However, the unique configuration of the Amazon Basin with massive, unstable tributaries flowing in parallel facilitates episodic or continuous gene flow, which can result in reticulate patterns of differentiation ( Barrera-Guzmán et al. 2018 ; Thom et al. 2018 ). This process directly affects phylogenetic inference and, if not fully understood, hampers researchers from obtaining a detailed understanding of the region’s biogeographic history. Our study highlights how genealogical patterns vary predictably across the genome and inform biogeographic inference ( Martin et al. 2019 ).
We suggest that large-scale river capture events can result in historical signatures of discordant genealogy across the genomes of species that respond to rivers as barriers ( Musher et al. 2022 ). The river capture scenario postulated in prior studies ( Fernandes 2013 ; Weeks et al. 2016 ; Ruokolainen et al. 2019 ) predicts a sister relationship between Inambari and S Rondônia with gene flow between E and W Inambari as well as between N and S Rondônia. Thus, either T1 or T3 might match the spatial-phylogenetic expectations under a river-vicariance model. However, our results are consistent with a more nuanced set of expectations associated with barrier change wherein genomic heterogeneity is associated with multiple distinct genealogical histories. Assuming that T1 reflects the history of population isolation (or at least reduced introgression) across the Aripuanã, then T2 results from secondary contact and lineage fusion within the traditionally recognized area of endemism, Rondônia ( Cracraft 1985 ). This gene flow among N and S Rondônia appears to be resulting in autosomal homogenization; sNMF preferred a model of k = 2, recovering 2 ancestral populations corresponding to Inambari and Rondônia ( Fig. 2 and Supplementary Fig. S1 ). Within Rondônia, the lack of any apparent plumage variation in T. aethiops also supports the notion that there is homogenizing gene flow between the N and S Rondônia populations.
A key objective of this study was to dissect the interplay between genomic and biogeographic processes in generating genomic heterogeneity, which requires some knowledge about landscape history. Recently, researchers have proposed a field of study, called “geogenomics,” wherein patterns of genomic differentiation and genomically inferred timings of divergence and gene flow can be used to help test paleogeographic models ( Dawson et al. 2022 ; Ribas et al. 2022 ). As shown here and elsewhere, spatial diversification patterns within Amazonia are reticulated, and unraveling the evolutionary history of taxa in this system is nontrivial ( Dagosta and Pinna 2017 ; Dagosta and De Pinna 2019 ). Understanding the geological context of river capture while accounting for intrinsic genomic processes, however, aids in the interpretation of alternative phylogenetic histories. For instance, if the Madeira headwaters were captured within the past few hundred thousand years, the Aripuanã river must predate the upper Madeira, as implied by both T1 and T3. Given that T1 is probably less impacted by introgression and populations within Rondônia are now homogenizing, the Aripuanã probably represented a more important barrier for T. aethiops prior to the capture event that is now weakened. In this way, we have a window into the process driving the formation of areas of endemism. As new barriers form on the landscape, old barriers erode; if differentiated taxa are not reproductively isolated, as appears the case for T. aethiops populations in Rondônia, they may fuse into a single taxon whose distribution conforms to the boundaries of the new river-barriers, leaving behind only reciprocally monophyletic mitochondrial groups that potentially match the ancestral landscape configuration.
Extensive paleochannels between the Jiparaná and Aripuanã tributaries of the middle Madeira Basin suggest that these rivers have historically been larger and behaved as a dynamic megafan, a hypothesis also supported by biological data ( Latrubesse 2002 ; Wilkinson et al. 2010 ; Ferreira et al. 2017 ). We thus postulate a historical river somewhere in the vicinity of these 2 tributaries that could have acted as a historical barrier to dispersal. Under this scenario, the paleo-Madeira River would have been flowing via the Jiparaná or Aripuanã basins or somewhere in between ( Hayakawa and Rossetti 2015 ), acting as a historical barrier for taxa located on either side. Likewise, the Tapajós basin to the east was probably drained via this paleo-Madeira River ( Rossetti 2014 ). If so, the formation of the modern Tapajós would have drawn water away from this basin, reducing the barrier’s strength. Once the Purus tributary was captured, the modern Madeira formed by extending its headwaters, which, in turn, generated a new barrier on the landscape. If taken at face value, our best-fit demographic model suggests that this river capture occurred roughly 600 Ka, at least 400 ky prior to the current geological estimate ( Ruokolainen et al. 2019 ), but in line with divergence times of some other taxa in the region ( Silva et al. 2019 ; Musher et al. 2022 ).
Interpreting the Mitochondrial Topology
Given that T1 and T2 are the primary genealogical signals across the genome, we are left to evaluate the mechanisms that gave rise to the mitochondrial tree (T3). The dispersal ecology of resident Amazonian bird species is poorly understood, but male birds are generally considered philopatric, which means it is unlikely that male-biased dispersal drives mtDNA patterns. T3 was estimated based on 2 loci, cytochrome- b and NADH-dehydrogenase subunit 2, and is well supported ( Thom and Aleixo 2015 ). Mitochondrial DNA is known to have faster coalescent times due to its reduced (one quarter) effective population size and has thus traditionally been viewed as an efficacious phylogenetic marker for detecting divergences that occur over short timescales ( Zink and Barrowclough 2008 ). However, mtDNA is a single locus and, therefore, might be expected to disagree with the species tree under certain conditions ( Maddison 1997 ). For example, if we assume T1 to be the “true” species tree, then T3 may have resulted from mitochondrial capture during hybridization between E Inambari and S Rondônia, as documented in other groups ( Ferreira et al. 2018 ; Myers et al. 2022 ). If this were the case, we should find portions of the genome that are associated with the cell respiratory system to be introgressed due to the same event ( Morales et al. 2018 ). Yet, there is limited support for T3 across the genome ( Supplementary Fig. S4 ), and the windows that do support T3 are not clearly linked to genes associated with mitochondrial activity ( Table 1 ). However, because we used GBS data for this analysis, we could be missing crucial mitonuclear gene clusters in our dataset.
Alternatively, incomplete lineage sorting or biogeographic history could have given rise to the mtDNA topology. For example, it could have resulted from a deep coalescent event wherein W Inambari haplotypes failed to coalesce with E inambari haplotypes. This is especially likely in instances of very rapid divergence ( Degnan and Rosenberg 2006 ). Given that divergences in our ingroup occurred in under 1 million years, this is certainly plausible, despite the rapid fixation rate of mtDNA. However, it is also possible that T3 represents the history of population isolation across rivers ( Fig. 1 ); it is striking that the distribution of reciprocally monophyletic populations based on mtDNA ( Thom and Aleixo 2015 ) matches the spatial patterns recovered in the PCA and that T. a. injunctus (E Inambari) is most phenotypically similar to T. a. punctuliger (Rondônia). Therefore, if the T3 topology indicates the signal of population isolation across rivers then, introgression between multiple non-sister lineages has nearly erased the signal of that isolation from the genome since relatively few nuclear loci support T3 ( Table 1 ). In this case, T1 does not exactly show the history of isolation across rivers, but might just reflect fewer genetic incompatibilities between E and W Inambari, which are more recently diverged, than between N and S Rondônia. In other words, T1 may not reflect a lack of gene flow altogether, but instead easier gene flow between E and W Inambari, which lack as many incompatibilities.
The genomes of populations that diversify on dynamic landscapes contain the signatures of multiple histories; that is, they are reticulated. To dissect these histories of isolation and secondary contact, we argue that it is important to understand the biogeographic mechanisms that give rise to predictable genomic patterns. If one’s objective is to model the relationships of taxa—that is, reconstruct phylogeny—T1 is the best-supported topology given our phylogenetic and demographic modeling results. However, if the goal is to decipher the complex biogeographic history of these taxa across space and time, we may want to know whether the mtDNA tree resulted from mitochondrial capture, deep coalescence, historical isolation, or even simply phylogenetic error. We suggest that T3 could represent the historical population divergences across rivers as they formed. The divergence time based on mtDNA between E Inambari and S Rondônia occurred at roughly 200 Ka ( Thom and Aleixo 2015 ), which lines up well with geological estimates for the Madeira River capture event ( Ruokolainen et al. 2019 ). However, future studies based on whole-genome resequencing data may be necessary to fully understand the complicated patterns of isolation, reticulation, and homogenization in T. aethiops , and the relationships between these processes and genome architecture. It would also enable more detailed models of historical demography and selection not possible with GBS data.
Concluding Remarks
Gene flow may be a creative or destructive force with regard to divergence and speciation. Introgression among divergent populations, wherein standing genetic variation among populations is episodically reshuffled into novel combinations might be an underappreciated speciation mechanism ( Marques et al. 2019 ). We showed that introgression remains high among taxa of a common understory antbird, T. aethiops . Specifically, taxa within T. aethiops seem to remain differentiated, despite ongoing and apparently intense introgression. This has been shown in many other Neotropical taxa ( Martin et al. 2013 ; Ebersbach et al. 2020 ; Musher et al. 2022 ). Given the potential for high rates of episodic isolation and reconnection due to the movement of large rivers, conditions in Amazonian lowlands, like those in some African lakes ( Aguilée et al. 2013 ; Meier et al. 2017 ), are potentially ideal for this combinatorial mechanism of adaptation to promote diversification in lowland birds. Still, the process of high introgression can also result in higher rates of extinction (i.e., homogenization) for these young, weakly differentiated taxa, as they fuse with other lineages ( Harvey et al. 2017 ; Barrera-Guzmán et al. 2022 ). Nonetheless, our results suggest that a nontrivial portion of genealogical heterogeneity across the genome arises due to extrinsic processes—such as river-course rearrangement—interacting with intrinsic processes associated with genome architecture.
Data available from the Dryad Digital Repository: https://doi.org/10.5281/zenodo.7460835 .
L.J.M. was funded in part by NSF grant DEB-1855812 to J.D. Weckstein.
The authors declare no conflict of interest.
We are grateful to E. Griffith, A. Del Grosso, J. Weckstein, and J. Merwin for comments on an earlier draft of this manuscript. We also thank B. Faircloth for advice and assistance, as well as the Bauer Core Facility and Cannon/Odyssey HPC system staff at the Harvard University Faculty of Arts and Sciences for support and computational resources needed for sequencing and assembly of the T. caerulescens reference genome. We thank S.V. Edwards and the Department of Organismic and Evolutionary Biology at Harvard University for providing financial support to generate the reference genome of Thamnophilus caerulescens . L.J.M. was funded in part by NSF grant DEB-1855812 to J.D. Weckstein. We also thank curators and staff at the Instituto Nacional de Pesquisas Amazonica, Museu Paraense Emilio Goeldi, American Museum of Natural History, and Louisiana Museum of Natural Science for loaning invaluable genetic resources. Reviews from F. Burbrink and two anonymous referees greatly improved the quality of this manuscript.
Supplemental material and scripts are available https://doi.org/10.5061/dryad.tht76hf3b . The T. caerulescens assembly and raw reads have been submitted to NCBI under project number PRJNA1016420.
Aguilée R. , Claessen D. , Lambert A. 2013 . Adaptive radiation driven by the interplay of eco-evolutionary and landscape dynamics . Evolution 67 : 1291 – 1306 .
Google Scholar
Albert J.S. , Craig J.M. , Tagliacollo V.A. , Petry P. 2018 . Upland and lowland fishes: a test of the river capture hypothesis . In: Hoorn C. , Perrigo A. , Antonelli A. , editors. Mountains, climate and biodiversity . New York (NY) : Wiley-Blackwell . p. 273 – 294 .
Google Preview
Barrera-Guzmán A.O. , Aleixo A. , Faccio M. , Dantas S. de M , Weir J.T. 2022 . Gene flow, genomic homogenization and the timeline to speciation in Amazonian manakins . Mol. Ecol . 31 : 4050 – 4066 .
Barrera-Guzmán A.O. , Aleixo A. , Shawkey M.D. , Weir J.T. 2018 . Hybrid speciation leads to novel male secondary sexual ornamentation of an Amazonian bird . Proc. Natl. Acad. Sci. U.S.A . 115 : E218 – E225 .
Barreto S.B. , Knowles L.L. , Mascarenhas R. , Affonso P.R.A. de M. , Batalha-Filho H. 2022 . Drainage rearrangements and in situ diversification of an endemic freshwater fish genus from north-eastern Brazilian rivers . Freshw. Biol . 67 : 759 – 773 .
Bicudo T.C. , Sacek V. , de Almeida R.P. , Bates J.M. , Ribas C.C. 2019 . Andean tectonics and mantle dynamics as a pervasive influence on Amazonian ecosystem . Sci. Rep . 9 : 16879 .
Borowiec M.L. 2016 . AMAS: a fast tool for alignment manipulation and computing of summary statistics . PeerJ 4 : e1660 .
Burbrink F.T. , Gehara M. 2018 . The biogeography of deep time phylogenetic reticulation . Syst. Biol . 67 : 743 – 744 .
Capella-Gutierrez S. , Silla-Martinez J.M. , Gabaldon T. 2009 . trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses . Bioinformatics 25 : 1972 – 1973 .
Capparella A.P. 1991 . Neotropical avian diversity and riverine barriers . Acta Congressus Internationalis Ornithologici 20 : 307 – 316 .
Charlesworth B. 1998 . Measures of divergence between populations and the effect of forces that reduce variability . Mol. Biol. Evol . 15 : 538 – 543 .
Coelho L.A. , Musher L.J. , Cracraft J. 2019 . A multireference-based whole genome assembly for the obligate ant-following antbird, rhegmatorhina melanosticta (Thamnophilidae) . Diversity 11 : 144 .
Cracraft J. 1985 . Historical biogeography and patterns of differentiation within the South American avifauna: areas of endemism . Ornithol. Monogr 36 : 49 – 84 .
Cracraft J. , Prum R.O. 1988 . Patterns and processes of diversification: speciation and historical congruence in some neotropical birds . Evolution 42 : 603 – 620 .
Cracraft J. , Ribas C.C. , d’Horta F.M. , Bates J. , Almeida R.P. , Boubli J.P. , Campbell K.E. , Cruz F.W. , Ferreira M. , Fritz S.C. , Grohmann C.H. , Latrubesse E.M. , Lohmann L.G. , Musher L.J. , Nogueira A. , Sawakuchi A.O. , Baker P. 2020 . The origin and evolution of Amazonian species diversity . In: Rull V. , Carnaval A.C. , editors. Neotropical diversification: patterns and processes . Springer . p. 225 – 244 .
Cruickshank T.E. , Hahn M.W. 2014 . Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow . Mol. Ecol . 23 : 3133 – 3157 .
Curran J.M. 2013 . Bolstad2: Bolstad functions . R package version 1.0-28 . In: Bolstad W.M. , editor. Understanding computational Bayesian statistics . Vol. 644 . John Wiley & Sons .
DaCosta J.M. , Sorenson M.D. 2014 . Amplification biases and consistent recovery of loci in a double-digest RAD-seq protocol . PLoS One 9 : e106713 .
Dagosta F.C.P. , De Pinna M. 2019 . The fishes of the Amazon: distribution and biogeographical patterns, with a comprehensive list of species . Bull. Am. Mus. Nat. Hist . 2019 : 1 .
Dagosta F.C.P. , de Pinna M. 2017 . Biogeography of Amazonian fishes: deconstructing river basins as biogeographic units . Neotrop. Ichthyol 15 : e170034 .
David Webb S. 1991 . Ecogeography and the great American interchange . Paleobiology 17 : 266 – 280 .
Dawson D.A. , Akesson M. , Burke T. , Pemberton J.M. , Slate J. , Hansson B. 2007 . Gene order and recombination rate in homologous chromosome regions of the chicken and a passerine bird . Mol. Biol. Evol . 24 : 1537 – 1552 .
Dawson M.N. , Ribas C.C. , Dolby G.A. , Fritz S.C. 2022 . Geogenomics: toward synthesis . J. Biogeogr . 49 : 1657 – 1661 .
Degnan J.H. , Rosenberg N.A. 2006 . Discordance of species trees with their most likely gene trees . PLoS Genet . 2 : e68 .
Delmore K.E. , Lugo Ramos J.S. , Van Doren B.M. , Lundberg M. , Bensch S. , Irwin D.E. , Liedvogel M. 2018 . Comparative analysis examining patterns of genomic differentiation across multiple episodes of population divergence in birds . Evol. Lett . 2 : 76 – 87 .
Del-Rio G. , Rego M.A. , Whitney B.M. , Schunck F. , Silveira L.F. , Faircloth B.C. , Brumfield R.T. 2022 . Displaced clines in an avian hybrid zone (Thamnophilidae: Rhegmatorhina) within an Amazonian interfluve . Evolution 76 : 455 – 475 .
Donoghue M.J. , Eaton D.A.R. , Maya-Lastra C.A. , Landis M.J. , Sweeney P.W. , Olson M.E. , Cacho N.I. , Moeglein M.K. , Gardner J.R. , Heaphy N.M. , Castorena M. , Rivas A.S. , Clement W.L. , Edwards E.J. 2022 . Replicated radiation of a plant clade along a cloud forest archipelago . Nat. Ecol. Evol . 6 : 1318 – 1329 .
Eaton D.A.R. , Overcast I. 2020 . ipyrad: interactive assembly and analysis of RADseq datasets . Bioinformatics 36 : 2592 – 2594 .
Ebersbach J. , Posso-Terranova A. , Bogdanowicz S. , Gómez-Díaz M. , García-González M.X. , Bolívar-García W. , Andrés J. 2020 . Complex patterns of differentiation and gene flow underly the divergence of aposematic phenotypes in Oophaga poison frogs . Mol. Ecol . 29 : 1944 – 1956 .
Edgar R.C. 2022 . Muscle5: high-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny . Nat. Commun . 13 : 6968 .
Ellegren H. 2010 . Evolutionary stasis: the stable chromosomes of birds . Trends Ecol. Evol . 25 : 283 – 291 .
Elshire R.J. , Glaubitz J.C. , Sun Q. , Poland J.A. , Kawamoto K. , Buckler E.S. , Mitchell S.E. 2011 . A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species . PLoS One 6 : e19379 .
Endler J.A. 1977 . Geographic variation, speciation, and clines . Monogr. Popul. Biol . 10 : 1 – 246 .
Feder J.L. , Egan S.P. , Nosil P. 2012 . The genomics of speciation-with-gene-flow . Trends Genet . 28 : 342 – 350 .
Fernandes A.M. 2013 . Fine-scale endemism of Amazonian birds in a threatened landscape . Biodivers. Conserv . 22 : 2683 – 2694 .
Ferreira M. , Aleixo A. , Ribas C.C. , Santos M.P.D. 2017 . Biogeography of the Neotropical genus Malacoptila (Aves: Bucconidae): the influence of the Andean orogeny, Amazonian drainage evolution and palaeoclimate . J. Biogeogr . 44 : 748 – 759 .
Ferreira M. , Fernandes A.M. , Aleixo A. , Antonelli A. , Olsson U. , Bates J.M. , Cracraft J. , Ribas C.C. 2018 . Evidence for mtDNA capture in the jacamar Galbula leucogastra/chalcothorax species-complex and insights on the evolution of white-sand ecosystems in the Amazon basin . Mol. Phylogenet. Evol . 129 : 149 – 157 .
Flantua S.G.A. , O’Dea A. , Onstein R.E. , Giraldo C. , Hooghiemstra H. 2019 . The flickering connectivity system of the north Andean páramos . J. Biogeogr . 41 : 1227 .
Fontaine M.C. , Pease J.B. , Steele A. , Waterhouse R.M. , Neafsey D.E. , Sharakhov I.V. , Jiang X. , Hall A.B. , Catteruccia F. , Kakani E. , Mitchell S.N. , Wu Y.-C. , Smith H.A. , Love R.R. , Lawniczak M.K. , Slotman M.A. , Emrich S.J. , Hahn M.W. , Besansky N.J. 2015 . Extensive introgression in a malaria vector species complex revealed by phylogenomics . Science 347 : 1258524 .
Frichot E. , Mathieu F. , Trouillon T. , Bouchard G. , François O. 2014 . Fast and efficient estimation of individual ancestry coefficients . Genetics 196 : 973 – 983 .
Gain C. , François O. 2021 . LEA 3: factor models in population genetics and ecological genomics with R . Mol. Ecol. Resour . 21 : 2738 – 2748 .
Gascon C. , Malcolm J.R. , Patton J.L. , da Silva M.N. , Bogart J.P. , Lougheed S.C. , Peres C.A. , Neckel S. , Boag P.T. 2000 . Riverine barriers and the geographic distribution of Amazonian species . Proc. Natl. Acad. Sci. U.S.A . 97 : 13672 – 13677 .
Gehara M. , Garda A.A. , Werneck F.P. , Oliveira E.F. , da Fonseca E.M. , Camurugi F. , Magalhães F.M. , Lanna F.M. , Sites J.W. Jr , Marques R. , Silveira-Filho R. , São Pedro V.A. , Colli G.R. , Costa G.C. , Burbrink F.T. 2017 . Estimating synchronous demographic changes across populations using hABC and its application for a herpetological community from northeastern Brazil . Mol. Ecol . 26 : 4756 – 4771 .
Gompert Z. , Lucas L.K. , Buerkle C.A. , Forister M.L. , Fordyce J.A. , Nice C.C. 2014 . Admixture and the organization of genetic diversity in a butterfly species complex revealed through common and rare genetic variants . Mol. Ecol . 23 : 4555 – 4573 .
Guindon S. , Dufayard J.-F. , Lefort V. , Anisimova M. , Hordijk W. , Gascuel O. 2010 . New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 30 . Syst. Biol . 59 : 307 – 321 .
Haenel Q. , Laurentino T.G. , Roesti M. , Berner D. 2018 . Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics . Mol. Ecol . 27 : 2477 – 2497 .
Haffer J. 1997 . Alternative models of vertebrate speciation in Amazonia: an overview . Biodivers. Conserv . 6 : 451 – 476 .
Hanes M.M. , Shell S. , Shimu T. , Crist C. , Machkour-M’Rabet S. 2022 . The phylogeographic history of Megistostegium (Malvaceae) in the dry, spiny thickets of southwestern Madagascar using RAD-seq data and ecological niche modeling . Ecol. Evol . 12 : e8632 .
Harvey M.G. , Seeholzer G.F. , Smith B.T. , Rabosky D.L. , Cuervo A.M. , Brumfield R.T. 2017 . Positive association between population genetic differentiation and speciation rates in New World birds . Proc. Natl. Acad. Sci. U.S.A . 114 : 6328 – 6333 .
Harvey M.G. , Bravo G.A. , Claramunt S. , Cuervo A.M. , Derryberry G.E. , Battilana J. , Seeholzer G.F. , McKay J.S. , O’Meara B.C. , Faircloth B.C. , Edwards S.V. , Perez-Emanm J. , Moyle R.G. , Sheldon F.H. , Aleixo A. , Smith B.T. , Chesser R.T. , Silveira L.F. , Cracraft J. , Brumfield R.T. , Derryberry E. 2020 . The evolution of a tropical biodiversity hotspot . Science . 370 : 1343 – 1348 .
Hayakawa E.H. , Rossetti D.F. 2015 . Late quaternary dynamics in the Madeira river basin, southern Amazonia (Brazil), as revealed by paleomorphological analysis . An. Acad. Bras. Cienc . 87 : 29 – 49 .
Henderson E.C. , Brelsford A. 2020 . Genomic differentiation across the speciation continuum in three hummingbird species pairs . BMC Evol. Biol . 20 : 113 .
He Z. , Li X. , Yang M. , Wang X. , Zhong C. , Duke N.C. , Wu C.-I. , Shi S. 2019 . Speciation with gene flow via cycles of isolation and migration: insights from multiple mangrove taxa . Natl. Sci. Rev . 6 : 275 – 288 .
Irwin D.E. , Milá B. , Toews D.P.L. , Brelsford A. , Kenyon H.L. , Porter A.N. , Grossen C. , Delmore K.E. , Alcaide M. , Irwin J.H. 2018 . A comparison of genomic islands of differentiation across three young avian species pairs . Mol. Ecol . 27 : 4839 – 4855 .
Jarvis E.D. , Mirarab S. , Aberer A.J. , Li B. , Houde P. , Li C. , Ho S.Y.W. , Faircloth B.C. , Nabholz B. , Howard J.T. , Suh A. , Weber C.C. , da Fonseca R.R. , Li J. , Zhang F. , Li H. , Zhou L. , Narula N. , Liu L. , Ganapathy G. , Boussau B. , Bayzid M.S. , Zavidovych V. , Subramanian S. , Gabaldón T. , Capella-Gutiérrez S. , Huerta-Cepas J. , Rekepalli B. , Munch K. , Schierup M. , Lindow B. , Warren W.C. , Ray D. , Green R.E. , Bruford M.W. , Zhan X. , Dixon A. , Li S. , Li N. , Huang Y. , Derryberry E.P. , Bertelsen M.F. , Sheldon F.H. , Brumfield R.T. , Mello C.V. , Lovell P.V. , Wirthlin M. , Schneider M.P.C. , Prosdocimi F. , Samaniego J.A. , Vargas Velazquez A.M. , Alfaro-Núñez A. , Campos P.F. , Petersen B. , Sicheritz-Ponten T. , Pas A. , Bailey T. , Scofield P. , Bunce M. , Lambert D.M. , Zhou Q. , Perelman P. , Driskell A.C. , Shapiro B. , Xiong Z. , Zeng Y. , Liu S. , Li Z. , Liu B. , Wu K. , Xiao J. , Yinqi X. , Zheng Q. , Zhang Y. , Yang H. , Wang J. , Smeds L. , Rheindt F.E. , Braun M. , Fjeldsa J. , Orlando L. , Barker F.K. , Jønsson K.A. , Johnson W. , Koepfli K.-P. , O’Brien S. , Haussler D. , Ryder O.A. , Rahbek C. , Willerslev E. , Graves G.R. , Glenn T.C. , McCormack J. , Burt D. , Ellegren H. , Alström P. , Edwards S.V. , Stamatakis A. , Mindell D.P. , Cracraft J. , Braun E.L. , Warnow T. , Jun W. , Gilbert M.T.P. , Zhang G. 2014 . Whole-genome analyses resolve early branches in the tree of life of modern birds . Science 346 : 1320 – 1331 .
Keller I. , Wagner C.E. , Greuter L. , Mwaiko S. , Selz O.M. , Sivasundar A. , Wittwer S. , Seehausen O. 2013 . Population genomic signatures of divergent adaptation, gene flow and hybrid speciation in the rapid radiation of Lake Victoria cichlid fishes . Mol. Ecol . 22 : 2848 – 2863 .
Korunes K.L. , Samuk K. 2021 . pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data . Mol. Ecol. Resour . 21 : 1359 – 1368 .
Latrubesse E.M. 2002 . Evidence of Quaternary palaeohydrological changes in middle Amazônia: the Aripuanã-Roosevelt and Jiparaná“ fans” . Z. Geomorphol. Suppl . 129 : 61 – 72 .
Li H. , Durbin R. 2009 . Fast and accurate short read alignment with Burrows-Wheeler transform . Bioinformatics 25 : 1754 – 1760 .
Lohmueller K.E. , Albrechtsen A. , Li Y. , Kim S.Y. , Korneliussen T. , Vinckenbosch N. , Tian G. , Huerta-Sanchez E. , Feder A.F. , Grarup N. , Jørgensen T. , Jiang T. , Witte D.R. , Sandbæk A. , Hellmann I. , Lauritzen T. , Hansen T. , Pedersen O. , Wang J. , Nielsen R. 2011 . Natural selection affects multiple aspects of genetic variation at putatively neutral sites across the human genome . PLoS Genet . 7 : e1002326 .
Luna L.W. , Naka L.N. , Thom G. , Knowles L.L. , Sawakuchi A.O. , Aleixo A. , Ribas C.C. 2023 . Late Pleistocene landscape changes and habitat specialization as promoters of population genomic divergence in Amazonian floodplain birds . Mol. Ecol . 32 : 214 – 228 .
Maddison W.P. 1997 . Gene trees in species trees . Syst. Biol . 46 : 523 – 536 .
Mallet J. , Besansky N. , Hahn M.W. 2016 . How reticulated are species ? Bioessays 38 : 140 – 149 .
Manthey J.D. , Klicka J. , Spellman G.M. 2021 . The genomic signature of allopatric speciation in a songbird is shaped by genome architecture (Aves: Certhia Americana) . Genome Biol. Evol . 13 : evab120 .
Marques D.A. , Meier J.I. , Seehausen O. 2019 . A combinatorial view on speciation and adaptive radiation . Trends Ecol. Evol . 34 : 531 – 544 .
Martin S.H. , Dasmahapatra K.K. , Nadeau N.J. , Salazar C. , Walters J.R. , Simpson F. , Blaxter M. , Manica A. , Mallet J. , Jiggins C.D. 2013 . Genome-wide evidence for speciation with gene flow in Heliconius butterflies . Genome Res . 23 : 1817 – 1828 .
Martin S.H. , Davey J.W. , Jiggins C.D. 2014 . Evaluating the use of ABBA–BABA statistics to locate introgressed loci . Mol. Biol. Evol . 32 : 244 – 257 .
Martin S.H. , Davey J.W. , Salazar C. , Jiggins C.D. 2019 . Recombination rate variation shapes barriers to introgression across butterfly genomes . PLoS Biol . 17 : e2006288 .
Martin S.H. , Van Belleghem S.M. 2017 . Exploring evolutionary relationships across the genome using topology weighting . Genetics 206 : 429 – 438 .
Mather K. 1938 . Crossing-over . Biol. Rev. Camb. Philos. Soc . 13 : 252 – 292 .
Meier J.I. , Marques D.A. , Mwaiko S. , Wagner C.E. , Excoffier L. , Seehausen O. 2017 . Ancient hybridization fuels rapid cichlid fish adaptive radiations . Nat. Commun . 8 : 14363 .
Morales H.E. , Pavlova A. , Amos N. , Major R. , Kilian A. , Greening C. , Sunnucks P. 2018 . Concordant divergence of mitogenomes and a mitonuclear gene cluster in bird lineages inhabiting different climates . Nat. Ecol. Evol . 2 : 1258 – 1267 .
Musher , Galante P.J. , Thom G. , Huntley J.W. , Blair M.E. 2020 . Shifting ecosystem connectivity during the Pleistocene drove diversification and gene-flow in a species complex of Neotropical birds (Tityridae: Pachyramphus) . J. Biogeogr . 66 : 167 .
Musher L.J. , Ferreira M. , Auerbach A.L. , Cracraft J. 2019 . Why is Amazonia a “source” of biodiversity? Climate-mediated dispersal and synchronous speciation across the Andes in an avian group (Tityrinae) . Proc. Roy. Soc. B . 286 : 20182343 .
Musher L.J. , Giakoumis M. , Albert J. , Del-Rio G. , Rego M. , Thom G. , Aleixo A. , Ribas C.C. , Brumfield R.T. , Smith B.T. , Cracraft J. 2022 . River network rearrangements promote speciation in lowland Amazonian birds . Sci. Adv . 8 : eabn1099 .
Myers E.A. , Mulcahy D.G. , Falk B. , Johnson K. , Carbi M. , de Queiroz K. 2022 . Interspecific gene flow and mitochondrial genome capture during the radiation of Jamaican Anolis Lizards (Squamata; Iguanidae) . Syst. Biol . 71 : 501 – 511 .
Nachman M.W. , Payseur B.A. 2012 . Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice . Philos. Trans. R. Soc. Lond. B Biol. Sci 367 : 409 – 421 .
Nosil P. 2008 . Speciation with gene flow could be common . Mol. Ecol . 17 : 2103 – 2106 .
Pavlidis P. , Laurent S. , Stephan W. 2010 . msABC: a modification of Hudson’s ms to facilitate multi-locus ABC analysis . Mol. Ecol. Resour . 10 : 723 – 727 .
Peñalba J.V. , Deng Y. , Fang Q. , Joseph L. , Moritz C. , Cockburn A. 2020 . Genome of an iconic Australian bird: high-quality assembly and linkage map of the superb fairy-wren ( Malurus cyaneus ) . Mol. Ecol. Resour . 20 : 560 – 578 .
Portik D.M. , Smith L.L. , Bi K. 2016 . An evaluation of transcriptome-based exon capture for frog phylogenomics across multiple scales of divergence (Class: Amphibia, Order: Anura) . Mol. Ecol. Resour . 16 : 1069 – 1083 .
Provost K. , Shue S.Y. , Forcellati M. , Smith B.T. 2022 . The genomic landscapes of desert birds form over multiple time scales . Mol. Biol. Evol . 39 : msac200 .
Pulido-Santacruz P. , Aleixo A. , Weir J.T. 2018 . Morphologically cryptic Amazonian bird species pairs exhibit strong postzygotic reproductive isolation . Proc. Biol. Sci . 285 : 20172081 .
Pulido-Santacruz P. , Aleixo A. , Weir J.T. 2020 . Genomic data reveal a protracted window of introgression during the diversification of a neotropical woodcreeper radiation . Evolution 74 : 842 – 858 .
Pupim F.N. , Sawakuchi A.O. , Almeida R.P. , Ribas C.C. , Kern A.K. , Hartmann G.A. , Chiessi C.M. , Tamura L.N. , Mineli T.D. , Savian J.F. , Grohmann C.H. , Bertassoli D.J. , Stern A.G. , Cruz F.W. , Cracraft J. 2019 . Chronology of Terra Firme formation in Amazonian lowlands reveals a dynamic Quaternary landscape . Quat. Sci. Rev . 210 : 154 – 163 .
R Core Team . 2019 . R: a language and environment for statistical computing .
Ribas C.C. , Aleixo A. , Nogueira A.C.R. , Miyaki C.Y. , Cracraft J. 2012 . A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years . Proc. Biol. Sci . 279 : 681 – 689 .
Ribas C.C. , Fritz S.C. , Baker P.A. 2022 . The challenges and potential of geogenomics for biogeography and conservation in Amazonia . J. Biogeogr . 49 : 1839 – 1847 .
Rognes T. , Flouri T. , Nichols B. , Quince C. , Mahé F. 2016 . VSEARCH: a versatile open source tool for metagenomics . PeerJ 4 : e2584 .
Rossetti D.F. 2014 . The role of tectonics in the late Quaternary evolution of Brazil’s Amazonian landscape . Earth-Sci. Rev . 139 : 362 – 389 .
Rossetti D.F. , Vasconcelos D.L. , Valeriano M.M. , Bezerra F.H.R. 2021 . Tectonics and drainage development in central Amazonia: the Juruá River . Catena 206 : 105560 .
Ruokolainen K. , Moulatlet G.M. , Zuquim G. , Hoorn C. , Tuomisto H. 2019 . Geologically recent rearrangements in central Amazonian river network and their importance for the riverine barrier hypothesis . Front Biogeogr 11 : e45046 .
Saether B.-E. , Lande R. , Engen S. , Weimerskirch H. , Lillegård M. , Altwegg R. , Becker P.H. , Bregnballe T. , Brommer J.E. , McCleery R.H. , Merilä J. , Nyholm E. , Rendell W. , Robertson R.R. , Tryjanowski P. , Visser M.E. 2005 . Generation time and temporal scaling of bird population dynamics . Nature 436 : 99 – 102 .
Sawakuchi A.O. , Schultz E.D. , Pupim F.N. , Bertassoli D.J. Jr , Souza D.F. , Cunha D.F. , Mazoca C.E. , Ferreira M.P. , Grohmann C.H. , Wahnfried I.D. , Chiessi C.M. , Cruz F.W. , Almeida R.P. , Ribas C.C. 2022 . Rainfall and sea level drove the expansion of seasonally flooded habitats and associated bird populations across Amazonia . Nat. Commun . 13 : 4945 .
Schley R.J. , Pennington R.T. , Pérez-Escobar O.A. , Helmstetter A.J. , de la Estrella M. , Larridon I. , Sabino Kikuchi I.A.B. , Barraclough T.G. , Forest F. , Klitgård B. 2020 . Introgression across evolutionary scales suggests reticulation contributes to Amazonian tree diversity . Mol. Ecol . 29 : 4170 – 4185 .
Sick H. 1967 . Rios e enchentes na Amazônia como obstáculo para a avifauna . Atas do simpósio sobre a biota amazônica 5 : 495 – 520 .
Silva S.M. , Peterson A.T. , Carneiro L. , Burlamaqui T.C.T. , Ribas C.C. , Sousa-Neves T. , Miranda L.S. , Fernandes A.M. , d’Horta F.M. , Araújo-Silva L.E. , Batista R. , Bandeira C.H.M.M. , Dantas S.M. , Ferreira M. , Martins D.M. , Oliveira J. , Rocha T.C. , Sardelli C.H. , Thom G. , Rêgo P.S. , Santos M.P. , Sequeira F. , Vallinoto M. , Aleixo A. 2019 . A dynamic continental moisture gradient drove Amazonian bird diversification . Sci. Adv . 5 : eaat5752 .
Singhal S. , Leffler E.M. , Sannareddy K. , Turner I. , Venn O. , Hooper D.M. , Strand A.I. , Li Q. , Raney B. , Balakrishnan C.N. , Griffith S.C. , McVean G. , Przeworski M. 2015 . Stable recombination hotspots in birds . Science 350 : 928 – 932 .
Smith B.T. , Amei A. , Klicka J. 2012 . Evaluating the role of contracting and expanding rainforest in initiating cycles of speciation across the Isthmus of Panama . Proc. Biol. Sci . 279 : 3520 – 3526 .
Smith B.T. , McCormack J.E. , Cuervo A.M. , Hickerson M.J. , Aleixo A. , Cadena C.D. , Pérez-Emán J. , Burney C.W. , Xie X. , Harvey M.G. , Faircloth B.C. , Glenn T.C. , Derryberry E.P. , Prejean J. , Fields S. , Brumfield R.T. 2014 . The drivers of tropical speciation . Nature 515 : 406 – 409 .
Stamatakis A. 2014 . RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies . Bioinformatics 30 : 1312 – 1313 .
Thom G. , Aleixo A. 2015 . Cryptic speciation in the white-shouldered antshrike ( Thamnophilus aethiops , Aves–Thamnophilidae): the tale of a transcontinental radiation across rivers in lowland … . Mol. Phylogenet. Evol . 82 Pt A : 95 – 110 .
Thom G. , Amaral F.R.D. , Hickerson M.J. , Aleixo A. , Araujo-Silva L.E. , Ribas C.C. , Choueri E. , Miyaki C.Y. 2018 . Phenotypic and genetic structure support gene flow generating gene tree discordances in an Amazonian Floodplain endemic species . Syst. Biol . 67 : 700 – 718 .
Thom G. , Moreira L.R. , Batista R. , Gehara M. , Aleixo A. , Smith B.T. 2021 . Genomic architecture controls spatial structuring in Amazonian birds . bioRxiv . 2021.12.01.470789 . doi: 10.1101/2021.12.01.470789
Thom G. , Xue A.T. , Sawakuchi A.O. , Ribas C.C. , Hickerson M.J. , Aleixo A. , Miyaki C. 2020 . Quaternary climate changes as speciation drivers in the Amazon floodplains . Sci. Adv . 6 : eaax4718 .
Tigano A. , Khan R. , Omer A.D. , Weisz D. , Dudchenko O. , Multani A.S. , Pathak S. , Behringer R.R. , Aiden E.L. , Fisher H. , MacManes M.D. 2022 . Chromosome size affects sequence divergence between species through the interplay of recombination and selection . Evolution 76 : 782 – 798 .
Van Doren B.M. , Campagna L. , Helm B. , Illera J.C. , Lovette I.J. , Liedvogel M. 2017 . Correlated patterns of genetic diversity and differentiation across an avian family . Mol. Ecol . 26 : 3982 – 3997 .
Vrba E.S. 1992 . Mammals as a key to evolutionary theory . J. Mammal . 73 : 1 – 28 .
Wang L. , Liu S. , Yang Y. , Meng Z. , Zhuang Z. 2022 . Linked selection, differential introgression and recombination rate variation promote heterogeneous divergence in a pair of yellow croakers . Mol. Ecol . 31 : 5729 – 5744 .
Weeks B.C. , Claramunt S. , Cracraft J. 2016 . Integrating systematics and biogeography to disentangle the roles of history and ecology in biotic assembly . J. Biogeogr . 43 : 1546 – 1559 .
Weir J.T. , Faccio M.S. , Pulido-Santacruz P. , Barrera-Guzmán A.O. , Aleixo A. 2015 . Hybridization in headwater regions, and the role of rivers as drivers of speciation in Amazonian birds . Evolution 69 : 1823 – 1834 .
Wen D. , Yu Y. , Hahn M.W. , Nakhleh L. 2016 . Reticulate evolutionary history and extensive introgression in mosquito species revealed by phylogenetic network analysis . Mol. Ecol . 25 : 2361 – 2372 .
Wilkinson M.J. , Marshall L.G. , Lundberg J.G. , Kreslavsky M.H. 2010 . Megafan environments in northern South America and their impact on Amazon Neogene aquatic ecosystems . In: Hoorn C. , Wesselingh F.P. , editors. Amazonia, landscape and species evolution: a look into the past . 1st edition. pp. 162 – 184 .
Zhang C. , Rabiee M. , Sayyari E. , Mirarab S. 2018 . ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees . BMC Bioinf . 19 : 153 .
Zhang G. , Li C. , Li Q. , Li B. , Larkin D.M. , Lee C. , Storz J.F. , Antunes A. , Greenwold M.J. , Meredith R.W. , Ödeen A. , Cui J. , Zhou Q. , Xu L. , Pan H. , Wang Z. , Jin L. , Zhang P. , Hu H. , Yang W. , Hu J. , Xiao J. , Yang Z. , Liu Y. , Xie Q. , Yu H. , Lian J. , Wen P. , Zhang F. , Li H. , Zeng Y. , Xiong Z. , Liu S. , Zhou L. , Huang Z. , An N. , Wang J. , Zheng Q. , Xiong Y. , Wang G. , Wang B. , Wang J. , Fan Y. , da Fonseca R.R. , Alfaro-Núñez A. , Schubert M. , Orlando L. , Mourier T. , Howard J.T. , Ganapathy G. , Pfenning A. , Whitney O. , Rivas M.V. , Hara E. , Smith J. , Farré M. , Narayan J. , Slavov G. , Romanov M.N. , Borges R. , Machado J.P. , Khan I. , Springer M.S. , Gatesy J. , Hoffmann F.G. , Opazo J.C. , Håstad O. , Sawyer R.H. , Kim H. , Kim K.-W. , Kim H.J. , Cho S. , Li N. , Huang Y. , Bruford M.W. , Zhan X. , Dixon A. , Bertelsen M.F. , Derryberry E. , Warren W. , Wilson R.K. , Li S. , Ray D.A. , Green R.E. , O’Brien S.J. , Griffin D. , Johnson W.E. , Haussler D. , Ryder O.A. , Willerslev E. , Graves G.R. , Alström P. , Fjeldså J. , Mindell D.P. , Edwards S.V. , Braun E.L. , Rahbek C. , Burt D.W. , Houde P. , Zhang Y. , Yang H. , Wang J. , Jarvis E.D. , Gilbert M.T.P. , Wang J. ; Avian Genome Consortium . 2014 . Comparative genomics reveals insights into avian genome evolution and adaptation . Science 346 : 1311 – 1320 .
Zink R.M. , Barrowclough G.F. 2008 . Mitochondrial DNA under siege in avian phylogeography . Mol. Ecol . 17 : 2107 – 2121 .
Email alerts
Citing articles via.
- Recommend to your Library
Affiliations
- Online ISSN 1076-836X
- Print ISSN 1063-5157
- Copyright © 2024 Society of Systematic Biologists
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.

The Biology Corner
Biology Teaching Resources

Case Study – A Tiny Heart (old version)

This case study was revised in 2023, get the NEW VERSION !
This case study focuses on a baby boy who was born with a problem with his heart. The story is based on a real scenario, though some of the names have been changed, and the parents gave permission to include photos of the infant.
Students will read about symptoms that occur when a baby is born with stenosis, or a narrowing of the artery. Students consider treatment options and compare the circulation of a fetus to that of an adult. Finally, the Ross Procedure is described where a valve from the pulmonary artery is moved to the aorta.
This case study was made for a high school anatomy class, and may not be appropriate for younger audiences. Students should have already completed the chapter on the circulatory system and have a strong foundation in how the circulatory system works. Case studies are designed to be completed in small groups so that students can have discussions and help each other with difficult vocabulary.
HS-LS1-2 Develop and use a model to illustrate the hierarchical organization of interacting systems that provide specific functions within multicellular organisms
Shannan Muskopf

- Book Solutions
- State Boards
Case Study Questions Class 9 Science The Fundamental Unit of Life
Case study questions class 9 science chapter 5 the fundamental unit of life.
CBSE Class 9 Case Study Questions Science The Fundamental Unit of Life. Important Case Study Questions for Class 9 Exam. Here we have arranged some Important Case Base Questions for students who are searching for Paragraph Based Questions The Fundamental Unit of Life.
At Case Study Questions there will given a Paragraph. In where some Questions will made on that respective Case Based Study. There will various types of marks will given 1 marks, 2 marks, 3 marks or 4 marks.
CBSE Case Study Questions Class 9 Science – The Fundamental Unit of Life
All living Organisms are made up of cells and these cells perform all the functions essential for the survival of the Organism eg. Respiration, digestion, excretion etc. In Unicellular organisms, a single cell carries out all these functions and in multicellular organisms different group of cells carry out different functions.
Cells were first discovered by Robert Hooke in 1665. He observed the cells in a cork slice with the help of a primitive microscope. Leeuwenhoek (1674), with the improved microscope, discovered the free living cells in pond water for the first time. It was Robert Brown in 1831 who discovered the nucleus in the cell. Purkinje in 1839 coined the term ‘protoplasm’ for the fluid substance of the cell.
(1) Who discovered the cell?
(a) Robert Hooke
(b) Leeuwenhoek
(c) Robert Brown
(d) T. Schwann
(2) Who discovered the nucleus in the cell?
(3) Who coined the term ‘Protoplasm’?
(d) Purkinje
(4) What is protoplasm?
(a) Unit of life
(b) Cell organelle
(c) Fluid substance of the cell.
(d) Cytoplasm
(5) Which of these statement is true about the cell?
(a) All organism are made up of cell
(b) Cell is the basic unit of life
(c) Cell is responsible for different metabolic functions
(d) All above
The cell theory, that all the plants and animals are composed of cells and that the cell is the basic unit of life, was presented by two biologists, German zoologist Schleiden (1838) and British zoologist Schwann (1839). The cell theory was further expanded by Virchow (1855) by suggesting that all cells arise from pre-existing cells. With the discovery of the electron microscope in 1940, it was possible to observe and understand the complex structure of the cell and its various organelles.
(1) Theodore Schwann was a _____
(a) British zoologist
(b) German zoologist
(c) British botanist
(d) German botanist
(2) Matthias Schleiden was a _____
(a) German zoologist
(b) British zoologist
(c) German botanist
(d) British botanist
(3) Which of these scientists formulated the cell theory?
(a) Schleiden and Schwann
(b) Rudolf Virchow
(c) Robert Koch
(d) Antony Von Leeuwenhoek
(4) Which scientist was the first to explain that new cells arise from pre-existing cells?
(a) Antony Von Leeuwenhoek
(b) Matthias Schleiden
(c) Rudolph Virchow
(d) Theodore Schwann
(5) Which of these scientists did not contribute to the cell theory?
(a) Robert Koch
(c) Theodore Schwann
(d) Rudolph Virchow
Plasma membrane or Cell membrane is the outermost covering of the cell that separates the contents of the cell from its external environment. The plasma membrane is flexible and is made up of organic molecules called lipids and proteins. The flexibility of the cell membrane also enables the cell to engulf in food and other material from its external environment. Such processes are known as endocytosis.The plasma membrane allows or permits the entry and exit of some materials in and out of the cell. It also prevents movement of some other materials. The cell membrane, therefore, is called a selectively permeable membrane.
Some substances like carbon dioxide or oxygen can move across the cell membrane by a process called diffusion. There is spontaneous movement of a substance from a region of high concentration to a region where its concentration is low. Similar thing happens in cells – some substance like CO2 (carbon dioxide is cellular waste and requires to be excreted out by the cell) accumulates in high concentrations inside the cell. In the cell’s external environment, the concentration of CO2 is low as compared to that inside the cell. As soon as there is a difference of concentration of CO2 inside and outside a cell, CO2 moves out of the cell, from a region of high concentration, to a region of low concentration outside the cell by the process of diffusion.
Water obeys the law of diffusion. The movement of water molecules through such a selectively permeable membrane is called osmosis. The movement of water across the plasma membrane is also affected by the amount of substance dissolved in water. Thus, osmosis is the net diffusion of water across a selectively permeable membrane toward a higher solute concentration.
(1) The plasma membrane is made up of ___________
(a) Proteins
(c) Proteins and Lipids (Lipoproteins)
(d) none of above
(2) Which of the following substance is known as cellular waste?
(b) Nitrogen
(c) Carbon dioxide
(d) None of above
(3) The movement of a substance from the region of higher concentration to the region where its concentration is lower is called as _____________
(a) Osmosis
(b) Diffusion
(c) Excretion of CO2 (carbon dioxide)
(4) Why cell membrane is known as selectively permeable membrane?
(5) What is mean by diffusion?
(6) Define Osmosis.
(d) Cell membrane allows or permits the entry and exit of some materials in and out of the cell. It also prevents movement of some other materials. Hence it is called as selectively permeable membrane.
(e) It is spontaneous movement of a substance from a region of high concentration to a region where its concentration is low. For example, some substances like carbon dioxide or oxygen can move across the cell membrane by a process called diffusion.
Plant cells, in addition to the plasma membrane, have another rigid outer covering called the cell wall. The cell wall lies outside the plasma membrane. The plant cell wall is mainly composed of cellulose. Cellulose is a complex substance and provides structural strength to plants. When a living plant cell loses water through osmosis there is shrinkage or contraction of the contents of the cell away from the cell wall. This phenomenon is known as plasmolysis.
(1) Which of the following is the main constituent of cell wall?
(c) Lipoproteins
(d) Cellulose
(2) Which of the following is outer most covering of the plant cell?
(a) Cell membrane
(b) Plasma membrane
(c) Cell wall
(3) Choose the correct set of statements from the following.
Statement 1 – Cell wall lies outside the plasma membrane.
Statement 2 – Cell wall is mainly composed of cellulose.
Statement 3 – Cellulose is a complex substance and provides structural strength to plants.
Statement 4 – Cell wall lies outside the plasma membrane.
(a) Statement 1 & 3
(b) Statement 1 & 2
(c) Statement 3 & 4
(d) All statement are correct
(4) What is mean by plasmolysis?
(5) What is the reason behind structural strength of plant cell?
(4) When living plant cell loses water through osmosis there is contraction of the contents of the cell away from the cell wall. This phenomenon is called as plasmolysis.
(5) The plant cell wall is mainly composed of cellulose. Cellulose is a complex substance and provides structural strength to plants.
Every cell has a membrane around it to keep its own contents separate from the external environment. Large and complex cells, including cells from multicellular organisms, need a lot of chemical activities to support their complicated structure and function. To keep these activities of different kinds separate from each other, these cells use membrane-bound little structures within themselves. The cytoplasm is the jelly like fluid content inside the plasma membrane which contains many specialised cell organelles. Such as Endoplasmic Reticulum, Golgi apparatus, Lysosomes, Ribosomes, Nucleus, Chloroplast, Mitochondria and Plastids.Each of these organelles performs a specific function for the cell. Some of these organelles are visible only with an electron microscope. They are important because they carry out some very crucial functions in cells.
(1) Identify the statement which is true for cells.
Statement 1 – Some cell organelles are visible only with an electron microscope.
Statement 2 – Cytoplasm is jelly like fluid present inside the cell.
Statement 3 – Cell organelles perform all the functions in cell.
Statement 4 – Every cell has a membrane around it to keep its own contents separate from the external environment.
(b) Statement 2 & 4
(c) Statement 1 & 4
(d) All statement are true.
(2) A suitable term for the various components of cells is ________
(b) cell organelles
(c) chromosomes
(3) The jelly-like fluid substance present in cells is called __________
(a) Protoplasm
(b) Chromosome
(c) Chloroplast
(4) What is cell organelles?
(5) Enlist the any five cell organelles.
(4) Cell organelles are the specialized organelles present with in the cells these organelles are involved in carrying out essential functions.
(5) Endoplasmic Reticulum, Golgi apparatus, Lysosomes, Ribosomes, Nucleus, Chloroplast, Mitochondria and Plastids.
The endoplasmic reticulum is a large network of membrane-bound tubes and sheets. It looks like long tubules or round or oblong bags (vesicles). It is discovered by Porter and Thompson. The ER membrane is similar in structure to the plasma membrane. There are two types of ER– rough endoplasmic reticulum (RER) and smooth endoplasmic reticulum (SER). RER looks rough under a microscope because it has particles called ribosomes attached to its surface. The ribosomes, which are present in all active cells, are the sites of protein manufacture. The manufactured proteins are then sent to various places in the cell depending on need, using the ER. The SER helps in the manufacture of fat molecules, or lipids, important for cell function. Some of these proteins and lipids help in building the cell membrane. This process is known as membrane biogenesis. Some other proteins and lipids function as enzymes and hormones. Although the ER varies greatly in appearance in different cells, it always forms a network system.
(1) Who discovered endoplasmic reticulum?
(a) Porter and Thompson
(b) Robert Brown
(c) Robert Hooke
(d) Koshland
(2) Which are the components of endoplasmic reticulum__________
(a) Cisternae, tubules and vesicles.
(b) Cisternae, chromatids and vacuoles
(c) Both a and b
(d) None of the above
(3) Endoplasmic reticulum membrane which is associated with ribosomes is called_______
(a) ER lumen
(b) Smooth endoplasmic reticulum
(c) Rough endoplasmic reticulum
(d) Endosome
(4) Enlist the types of Endoplasmic Reticulum.
(5) Define membrane biogenesis?
(4) Smooth endoplasmic reticulum and Rough endoplasmic reticulum
(5) The SER helps in the manufacture of fat molecules, or lipids, important for cell function. Some of these proteins and lipids help in building the cell membrane. This process is known as membrane biogenesis.
The Golgi apparatus, first described by Camilo Golgi, consists of a system of membrane-bound vesicles (flattened sacs) arranged approximately parallel to each other in stacks called cisterns. These membranes often have connections with the membranes of ER and therefore constitute another portion of a complex cellular membrane system. The material synthesised near the ER is packaged and dispatched to various targets inside and outside the cell through the Golgi apparatus. Its functions include the storage, modification and packaging of products in vesicles. In some cases, complex sugars may be made from simple sugars in the Golgi apparatus. The Golgi apparatus is also involved in the formation of lysosomes
(d) Camilo Golgi
(2) A system of membrane-bound flattened sacs arranged approximately parallel to each other in stacks are called as _________
(a) Cisterns
(b) Vesicles
(c) Golgi complex
(d) Vacuoles
(3) Membrane bound flattened sacs is termed as _________
(4) Enlist the function of Golgi apparatus.
(5) Name the cell organelles which is involved in the formation of lysosomes?
(4) Functions of Golgi apparatus:
- Storage, modification and packaging of products
- Involved in formation of lysosomes
(5) Golgi apparatus
It’s a suggestion that tries to give all the questions in mcq types in case/paragraph-based questions or if you are putting question-based kindly put only very short answer types questions like one word or one sentence it’s more helpful to all. Rest it’s really good information and provides ideas to us.
Ok Dibya, We will design this page
tomorrow is my science final papper. this case study is come in my papper than i am very happy.
This very helpful casebased questions Thanku for this
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
We have a strong team of experienced Teachers who are here to solve all your exam preparation doubts
An astrologer’s day by r. k naryan mcq question answers, tripura board class 6 bengali solutions chapter 14 আমের কুসি, as you like it mcq for west bengal class 11 exam.

Othello MCQ for West Bengal Class 11 Exam
Sign in to your account
Username or Email Address
Remember Me

Want to create or adapt books like this? Learn more about how Pressbooks supports open publishing practices.
2.1 Case Study: Why Should You Study Human Biology?
Created by CK-12/Adapted by Christine Miller
Case Study: Our Invisible Inhabitants
Lanying is suffering from a fever, body aches, and a painful sore throat that feels worse when she swallows. She visits her doctor, who examines her and performs a throat culture. When the results come back, he tells her that she has strep throat, which is caused by the bacteria Streptococcus pyogenes . He prescribes an antibiotic that will either kill the bacteria or stop it from reproducing, and advises her to take the full course of the treatment even if she is feeling better earlier. Stopping early can cause an increase in bacteria that are resistant to antibiotics.
Lanying takes the antibiotic as prescribed. Toward the end of the course, her throat is feeling much better — but she can’t say the same for other parts of her body! She has developed diarrhea and an itchy vaginal yeast infection. She calls her doctor, who suspects that the antibiotic treatment has caused both the digestive distress and the yeast infection. He explains that our bodies are home to many different kinds of microorganisms, some of which are actually beneficial to us because they help us digest our food and minimize the population of harmful microorganisms. When we take an antibiotic, many of these “good” bacteria are killed along with the “bad,” disease-causing bacteria, which can result in diarrhea and yeast infections.
Lanying’s doctor prescribes an antifungal medication for her yeast infection. He also recommends that she eat yogurt with live cultures, which will help replace the beneficial bacteria in her gut. Our bodies contain a delicate balance of inhabitants that are invisible without a microscope, and changes in that balance can cause unpleasant health effects.
What Is Human Biology?
As you read the rest of this book, you’ll learn more amazing facts about the human organism, and you’ll get a better sense of how biology relates to your health. Human biology is the scientific study of the human species, which includes the fascinating story of human evolution and a detailed account of our genetics, anatomy, physiology, and ecology. In short, the study focuses on how we got here, how we function, and the role we play in the natural world. This helps us to better understand human health, because we can learn how to stay healthy and how diseases and injuries can be treated. Human biology should be of personal interest to you to the extent that it can benefit your own health, as well as the health of your friends and family. This branch of science also has broader implications for society and the human species as a whole.
Chapter Overview: Living Organisms and Human Biology
In the rest of this chapter, you’ll learn about the traits shared by all living things, the basic principles that underlie all of biology, the vast diversity of living organisms, what it means to be human, and our place in the animal kingdom. Specifically, you’ll learn:
- The seven traits shared by all living things: homeostasis , or the maintenance of a more-or-less constant internal environment; multiple levels of organization consisting of one or more cells ; the use of energy and metabolism ; the ability to grow and develop; the ability to evolve adaptations to the environment; the ability to detect and respond to environmental stimuli; and the ability to reproduce .
- The basic principles that unify all fields of biology, including gene theory, homeostasis, and evolutionary theory.
- The diversity of life (including the different kinds of biodiversity), the definition of a species, the classification and naming systems for living organisms, and how evolutionary relationships can be represented through diagrams, such as phylogenetic trees.
- How the human species is classified and how we’ve evolved from our close relatives and ancestors.
- The physical traits and social behaviors that humans share with other primates.
As you read this chapter, consider the following questions about Lanying’s situation:
- What do single-celled organisms (such as the bacteria and yeast living in and on Lanying) have in common with humans?
- How are bacteria, yeast, and humans classified?
- How do the concepts of homeostasis and biodiversity apply to Lanying’s situation?
- Why can stopping antibiotics early cause the development of antibiotic-resistant bacteria?
Attribution
Figure 2.1.1
Photo (face mask) by Michael Amadeus , on Unsplash is used under the Unsplash license (https://unsplash.com/license).
Mayo Clinic Staff (n.d.). Strep throat [online article]. MayoClinic.org. https://www.mayoclinic.org/diseases-conditions/strep-throat/symptoms-causes/syc-20350338
The ability of an organism to maintain constant internal conditions despite external changes.
The smallest unit of life, consisting of at least a membrane, cytoplasm, and genetic material.
The chemical processes that occur in a living organism to sustain life.
The production of offspring by sexual or asexual process.
The variety of life in the world, ecosystem, or in a particular habitat.
Human Biology Copyright © 2020 by Christine Miller is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License , except where otherwise noted.
Share This Book
Case Study Research Method in Psychology
Saul Mcleod, PhD
Editor-in-Chief for Simply Psychology
BSc (Hons) Psychology, MRes, PhD, University of Manchester
Saul Mcleod, PhD., is a qualified psychology teacher with over 18 years of experience in further and higher education. He has been published in peer-reviewed journals, including the Journal of Clinical Psychology.
Learn about our Editorial Process
Olivia Guy-Evans, MSc
Associate Editor for Simply Psychology
BSc (Hons) Psychology, MSc Psychology of Education
Olivia Guy-Evans is a writer and associate editor for Simply Psychology. She has previously worked in healthcare and educational sectors.
On This Page:
Case studies are in-depth investigations of a person, group, event, or community. Typically, data is gathered from various sources using several methods (e.g., observations & interviews).
The case study research method originated in clinical medicine (the case history, i.e., the patient’s personal history). In psychology, case studies are often confined to the study of a particular individual.
The information is mainly biographical and relates to events in the individual’s past (i.e., retrospective), as well as to significant events that are currently occurring in his or her everyday life.
The case study is not a research method, but researchers select methods of data collection and analysis that will generate material suitable for case studies.
Freud (1909a, 1909b) conducted very detailed investigations into the private lives of his patients in an attempt to both understand and help them overcome their illnesses.
This makes it clear that the case study is a method that should only be used by a psychologist, therapist, or psychiatrist, i.e., someone with a professional qualification.
There is an ethical issue of competence. Only someone qualified to diagnose and treat a person can conduct a formal case study relating to atypical (i.e., abnormal) behavior or atypical development.
Famous Case Studies
- Anna O – One of the most famous case studies, documenting psychoanalyst Josef Breuer’s treatment of “Anna O” (real name Bertha Pappenheim) for hysteria in the late 1800s using early psychoanalytic theory.
- Little Hans – A child psychoanalysis case study published by Sigmund Freud in 1909 analyzing his five-year-old patient Herbert Graf’s house phobia as related to the Oedipus complex.
- Bruce/Brenda – Gender identity case of the boy (Bruce) whose botched circumcision led psychologist John Money to advise gender reassignment and raise him as a girl (Brenda) in the 1960s.
- Genie Wiley – Linguistics/psychological development case of the victim of extreme isolation abuse who was studied in 1970s California for effects of early language deprivation on acquiring speech later in life.
- Phineas Gage – One of the most famous neuropsychology case studies analyzes personality changes in railroad worker Phineas Gage after an 1848 brain injury involving a tamping iron piercing his skull.
Clinical Case Studies
- Studying the effectiveness of psychotherapy approaches with an individual patient
- Assessing and treating mental illnesses like depression, anxiety disorders, PTSD
- Neuropsychological cases investigating brain injuries or disorders
Child Psychology Case Studies
- Studying psychological development from birth through adolescence
- Cases of learning disabilities, autism spectrum disorders, ADHD
- Effects of trauma, abuse, deprivation on development
Types of Case Studies
- Explanatory case studies : Used to explore causation in order to find underlying principles. Helpful for doing qualitative analysis to explain presumed causal links.
- Exploratory case studies : Used to explore situations where an intervention being evaluated has no clear set of outcomes. It helps define questions and hypotheses for future research.
- Descriptive case studies : Describe an intervention or phenomenon and the real-life context in which it occurred. It is helpful for illustrating certain topics within an evaluation.
- Multiple-case studies : Used to explore differences between cases and replicate findings across cases. Helpful for comparing and contrasting specific cases.
- Intrinsic : Used to gain a better understanding of a particular case. Helpful for capturing the complexity of a single case.
- Collective : Used to explore a general phenomenon using multiple case studies. Helpful for jointly studying a group of cases in order to inquire into the phenomenon.
Where Do You Find Data for a Case Study?
There are several places to find data for a case study. The key is to gather data from multiple sources to get a complete picture of the case and corroborate facts or findings through triangulation of evidence. Most of this information is likely qualitative (i.e., verbal description rather than measurement), but the psychologist might also collect numerical data.
1. Primary sources
- Interviews – Interviewing key people related to the case to get their perspectives and insights. The interview is an extremely effective procedure for obtaining information about an individual, and it may be used to collect comments from the person’s friends, parents, employer, workmates, and others who have a good knowledge of the person, as well as to obtain facts from the person him or herself.
- Observations – Observing behaviors, interactions, processes, etc., related to the case as they unfold in real-time.
- Documents & Records – Reviewing private documents, diaries, public records, correspondence, meeting minutes, etc., relevant to the case.
2. Secondary sources
- News/Media – News coverage of events related to the case study.
- Academic articles – Journal articles, dissertations etc. that discuss the case.
- Government reports – Official data and records related to the case context.
- Books/films – Books, documentaries or films discussing the case.
3. Archival records
Searching historical archives, museum collections and databases to find relevant documents, visual/audio records related to the case history and context.
Public archives like newspapers, organizational records, photographic collections could all include potentially relevant pieces of information to shed light on attitudes, cultural perspectives, common practices and historical contexts related to psychology.
4. Organizational records
Organizational records offer the advantage of often having large datasets collected over time that can reveal or confirm psychological insights.
Of course, privacy and ethical concerns regarding confidential data must be navigated carefully.
However, with proper protocols, organizational records can provide invaluable context and empirical depth to qualitative case studies exploring the intersection of psychology and organizations.
- Organizational/industrial psychology research : Organizational records like employee surveys, turnover/retention data, policies, incident reports etc. may provide insight into topics like job satisfaction, workplace culture and dynamics, leadership issues, employee behaviors etc.
- Clinical psychology : Therapists/hospitals may grant access to anonymized medical records to study aspects like assessments, diagnoses, treatment plans etc. This could shed light on clinical practices.
- School psychology : Studies could utilize anonymized student records like test scores, grades, disciplinary issues, and counseling referrals to study child development, learning barriers, effectiveness of support programs, and more.
How do I Write a Case Study in Psychology?
Follow specified case study guidelines provided by a journal or your psychology tutor. General components of clinical case studies include: background, symptoms, assessments, diagnosis, treatment, and outcomes. Interpreting the information means the researcher decides what to include or leave out. A good case study should always clarify which information is the factual description and which is an inference or the researcher’s opinion.
1. Introduction
- Provide background on the case context and why it is of interest, presenting background information like demographics, relevant history, and presenting problem.
- Compare briefly to similar published cases if applicable. Clearly state the focus/importance of the case.
2. Case Presentation
- Describe the presenting problem in detail, including symptoms, duration,and impact on daily life.
- Include client demographics like age and gender, information about social relationships, and mental health history.
- Describe all physical, emotional, and/or sensory symptoms reported by the client.
- Use patient quotes to describe the initial complaint verbatim. Follow with full-sentence summaries of relevant history details gathered, including key components that led to a working diagnosis.
- Summarize clinical exam results, namely orthopedic/neurological tests, imaging, lab tests, etc. Note actual results rather than subjective conclusions. Provide images if clearly reproducible/anonymized.
- Clearly state the working diagnosis or clinical impression before transitioning to management.
3. Management and Outcome
- Indicate the total duration of care and number of treatments given over what timeframe. Use specific names/descriptions for any therapies/interventions applied.
- Present the results of the intervention,including any quantitative or qualitative data collected.
- For outcomes, utilize visual analog scales for pain, medication usage logs, etc., if possible. Include patient self-reports of improvement/worsening of symptoms. Note the reason for discharge/end of care.
4. Discussion
- Analyze the case, exploring contributing factors, limitations of the study, and connections to existing research.
- Analyze the effectiveness of the intervention,considering factors like participant adherence, limitations of the study, and potential alternative explanations for the results.
- Identify any questions raised in the case analysis and relate insights to established theories and current research if applicable. Avoid definitive claims about physiological explanations.
- Offer clinical implications, and suggest future research directions.
5. Additional Items
- Thank specific assistants for writing support only. No patient acknowledgments.
- References should directly support any key claims or quotes included.
- Use tables/figures/images only if substantially informative. Include permissions and legends/explanatory notes.
- Provides detailed (rich qualitative) information.
- Provides insight for further research.
- Permitting investigation of otherwise impractical (or unethical) situations.
Case studies allow a researcher to investigate a topic in far more detail than might be possible if they were trying to deal with a large number of research participants (nomothetic approach) with the aim of ‘averaging’.
Because of their in-depth, multi-sided approach, case studies often shed light on aspects of human thinking and behavior that would be unethical or impractical to study in other ways.
Research that only looks into the measurable aspects of human behavior is not likely to give us insights into the subjective dimension of experience, which is important to psychoanalytic and humanistic psychologists.
Case studies are often used in exploratory research. They can help us generate new ideas (that might be tested by other methods). They are an important way of illustrating theories and can help show how different aspects of a person’s life are related to each other.
The method is, therefore, important for psychologists who adopt a holistic point of view (i.e., humanistic psychologists ).
Limitations
- Lacking scientific rigor and providing little basis for generalization of results to the wider population.
- Researchers’ own subjective feelings may influence the case study (researcher bias).
- Difficult to replicate.
- Time-consuming and expensive.
- The volume of data, together with the time restrictions in place, impacted the depth of analysis that was possible within the available resources.
Because a case study deals with only one person/event/group, we can never be sure if the case study investigated is representative of the wider body of “similar” instances. This means the conclusions drawn from a particular case may not be transferable to other settings.
Because case studies are based on the analysis of qualitative (i.e., descriptive) data , a lot depends on the psychologist’s interpretation of the information she has acquired.
This means that there is a lot of scope for Anna O , and it could be that the subjective opinions of the psychologist intrude in the assessment of what the data means.
For example, Freud has been criticized for producing case studies in which the information was sometimes distorted to fit particular behavioral theories (e.g., Little Hans ).
This is also true of Money’s interpretation of the Bruce/Brenda case study (Diamond, 1997) when he ignored evidence that went against his theory.
Breuer, J., & Freud, S. (1895). Studies on hysteria . Standard Edition 2: London.
Curtiss, S. (1981). Genie: The case of a modern wild child .
Diamond, M., & Sigmundson, K. (1997). Sex Reassignment at Birth: Long-term Review and Clinical Implications. Archives of Pediatrics & Adolescent Medicine , 151(3), 298-304
Freud, S. (1909a). Analysis of a phobia of a five year old boy. In The Pelican Freud Library (1977), Vol 8, Case Histories 1, pages 169-306
Freud, S. (1909b). Bemerkungen über einen Fall von Zwangsneurose (Der “Rattenmann”). Jb. psychoanal. psychopathol. Forsch ., I, p. 357-421; GW, VII, p. 379-463; Notes upon a case of obsessional neurosis, SE , 10: 151-318.
Harlow J. M. (1848). Passage of an iron rod through the head. Boston Medical and Surgical Journal, 39 , 389–393.
Harlow, J. M. (1868). Recovery from the Passage of an Iron Bar through the Head . Publications of the Massachusetts Medical Society. 2 (3), 327-347.
Money, J., & Ehrhardt, A. A. (1972). Man & Woman, Boy & Girl : The Differentiation and Dimorphism of Gender Identity from Conception to Maturity. Baltimore, Maryland: Johns Hopkins University Press.
Money, J., & Tucker, P. (1975). Sexual signatures: On being a man or a woman.
Further Information
- Case Study Approach
- Case Study Method
- Enhancing the Quality of Case Studies in Health Services Research
- “We do things together” A case study of “couplehood” in dementia
- Using mixed methods for evaluating an integrative approach to cancer care: a case study
Related Articles
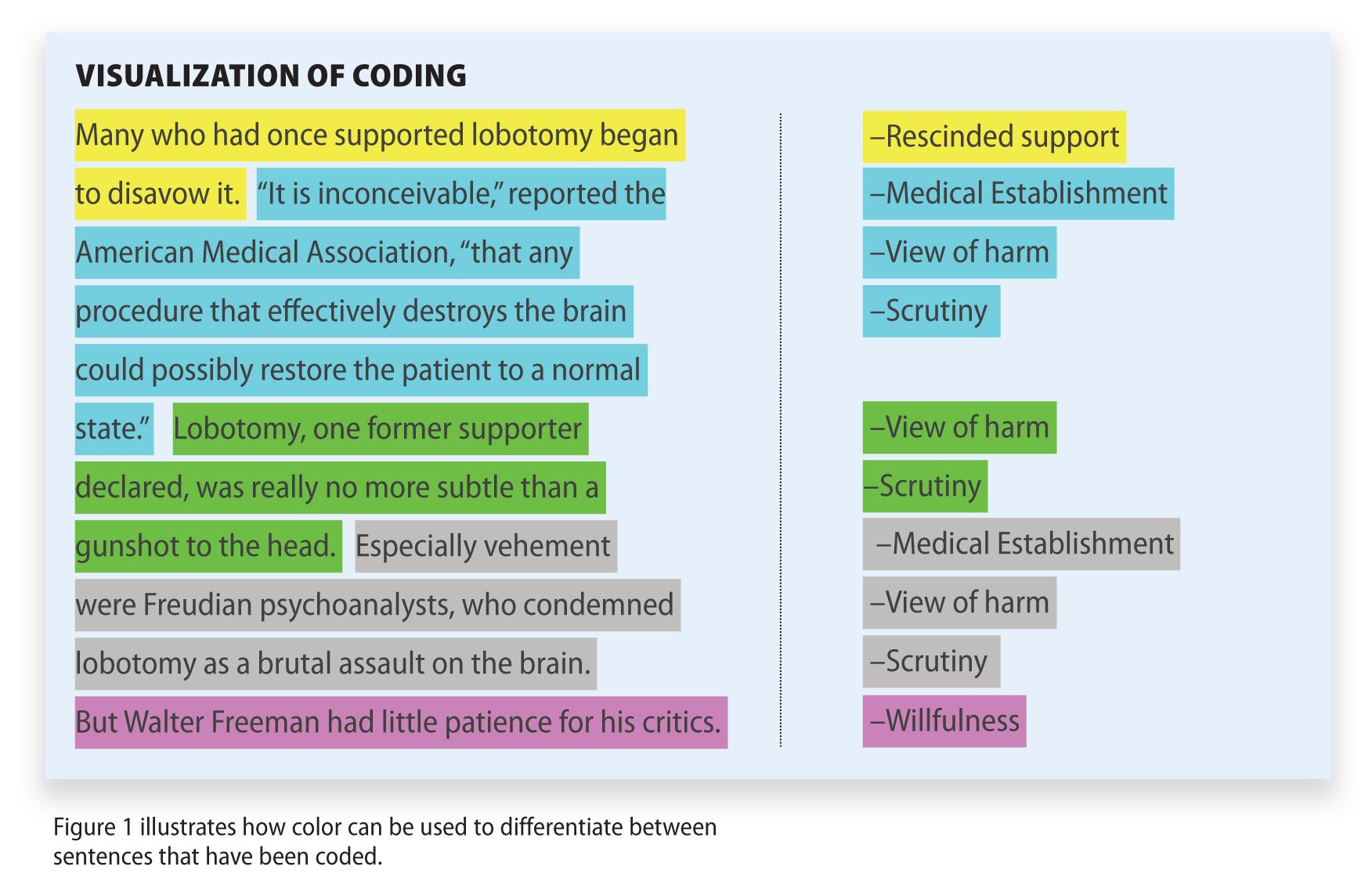
Research Methodology
Qualitative Data Coding

What Is a Focus Group?
Cross-Cultural Research Methodology In Psychology

What Is Internal Validity In Research?

Research Methodology , Statistics
What Is Face Validity In Research? Importance & How To Measure

Criterion Validity: Definition & Examples

IOE - Faculty of Education and Society
- Departments and centres
- Innovation and Enterprise
- Teacher Education College

Technicians Supporting Biology Practical Work for A Level
14 June 2024, 9:15 am–4:00 pm

This one-day practical course for science technicians focuses on building knowledge, key technical skills and techniques you will need to effectively support A level Biology teaching and student learning.
Event Information
Availability, course overview.
The course will cover:
- Understanding the current A level curriculum;
- Approaches to experiments known to be problematic and to help ensure successful results;
- Ways to incorporate information and communication technology (ICT) into lessons;
- Health and safety, risk assessments and keeping up to date.
You'll also look at three experiments in detail:
- Respiration;
- A circus of other practical activities with hints and tips including a heart rate investigation, making dilutions, colorimetry, photosynthesis and ecology tips.
Who the course is for
This course is suitable for all secondary school and college science technicians, especially those working with students at Key Stages 4 and 5.
Entry requirements
You'll need to be working as a science technician in a school or college.
Learning outcomes
This course will help you to:
- Improve your awareness of the practical demands of A level biology;
- Consider strategies for providing effective technical support;
- Gain confidence in the support you provide for the science department and students;
- Learn new skills and techniques to ensure good quality practical work and share good practice;
- Gain experience in the set-up and demonstration of key biology experiments, including relevant safety precautions;
- Understand hazard identification and risk assessment and the importance of keeping up to date.
Course structure
The course is delivered mainly as a practical workshop with some lectures/discussion.
Certificates
You'll get a certificate of attendance on completing the course.
Costs and booking
Price per participant is £180 and it includes course materials and a packed lunch.
Book your place
If you are not able to pay by credit/debit card, an invoice will be raised for upfront payment. If this is your preferred method of payment, email [email protected] to enquiry how to book this course if you wish to pay by invoice.
About the Speaker
Stephanie shaw.
Stephanie is a professional tutor and consultant for science technicians who work in an educational setting.
Related News
Related events, related case studies, related research projects, press and media enquiries.
UCL Media Relations +44 (0)7747 565 056

IMAGES
VIDEO
COMMENTS
The NCCSTS Case Collection, created and curated by the National Center for Case Study Teaching in Science, on behalf of the University at Buffalo, contains over a thousand peer-reviewed case studies on a variety of topics in all areas of science. Cases (only) are freely accessible; subscription is required for access to teaching notes and ...
These four case studies are interactivities based on actual scientific research projects carried out by leading teams in the field. Each case study takes the participant through a series of steps in a research project — just follow the step-by-step instructions to test a hypothesis or analyze data. The case studies provide an interactive ...
In any case, the book is delightful biology, delightfully presented. It is suited for the scientific novice, but even biologists are likely to find it rewarding. This book is listed on my Book suggestions page, and as further reading for Intro Chem Ch 15, re intermolecular forces, and for Organic/Biochem Ch 15, re spider silk.
Students in my anatomy class complete many case studies throughout the year focused on body system units. Case studies are a way to add a personal story to (sometimes) technical information about physiology. For my high school students, I try to find cases that are about younger people or even children, cases like " A Tiny Heart ," which ...
I always allow students to talk and help one another during clicker questions to enhance their interaction and give them a choice to go along with a group opinion or answer based on their individual thinking. 4. Flip the classroom. Another effective way to use BioInteractive resources in large classes is to use videos to flip a class session.
HHMI and Case Studies. HHMI Biointeractive has many interactive resources, case studies, and data analysis. Here is a list of my favorites: The National Center for Case Study Teaching in Science is collaborating with HHMI BioInteractive to pair case studies in our collection with their resources.
Case Studies in Biology: Climate and Health Exploration Course. This is an online course offered in the summer and open to current high school students. As scientists, we take a lot of STEM classes, including biology, chemistry, physics, and math. But we often don't have time to connect all of this information together.
Hardcover Book USD 84.99. Price excludes VAT (USA) Durable hardcover edition. Dispatched in 3 to 5 business days. Free shipping worldwide - see info. Buy Hardcover Book. Tax calculation will be finalised at checkout. This unique book features topics or case studies can be used independently giving instructors and students flexibility in the ...
In this study, we used a case study approach to obtain an in-depth understanding of the change process of two university instructors (Julie and Alex) who were involved with redesigning a biology course. The instructors sought to transform the course from a teacher-centered, lecture-style class to one that incorporated learner-centered teaching.
This page titled 8.1: Case Study: Genes and Inheritance is shared under a CK-12 license and was authored, remixed, and/or curated by Suzanne Wakim & Mandeep Grewal via source content that was edited to the style and standards of the LibreTexts platform; a detailed edit history is available upon request. People tend to look similar to their ...
How Gregor Mendel discovered the laws of inheritance for certain types of traits. The science of heredity, known as genetics, and the relationship between genes and traits. How gametes, such as eggs and sperm, are produced through meiosis. How sexual reproduction works on the cellular level and how it increases genetic variation.
Human biology is the scientific study of the human species, which includes the fascinating story of human evolution and a detailed account of our genetics, anatomy, physiology, and ecology. In short, the study focuses on how we got here, how we function, and the role we play in the natural world. This helps us to better understand human health ...
The case studies provided on this page exploit the molecular graphics program VMD for teaching molecular cell biology. The case studies start out like a conventional textbook chapter, but utilize VMD molecular graphics to offer a much more detailed view of the subjects than commonly possible in textbooks. The case studies induce the reader to ...
NSF DBI-1827011 Project Goal and Objectives. Goal: To assemble a new network (Molecular CaseNet); for developing case studies at the interface of biology and chemistry, discussed at a molecular level, and in atomic detail; and engaging educators in using them for biology, chemistry, and biochemistry education at the undergraduate level. 1.
Despite many years of research, scientists have yet to determine the basis of persistence of most pathogens, and have therefore struggled to develop reliable prevention and treatment strategies. Systems biology provides a new and integrative tool that will help to achieve these goals. In this article, we use Mycobacterium tuberculosis as an ...
Lukas J Musher, Glaucia Del-Rio, Rafael S Marcondes, Robb T Brumfield, Gustavo A Bravo, Gregory Thom, Geogenomic Predictors of Genetree Heterogeneity Explain Phylogeographic and Introgression History: A Case Study in an Amazonian Bird (Thamnophilus aethiops), Systematic Biology, Volume 73, Issue 1, January 2024, Pages 36-52, https://doi.org ...
This case study was revised in 2023, get the NEW VERSION! This case study focuses on a baby boy who was born with a problem with his heart. The story is based on a real scenario, though some of the names have been changed, and the parents gave permission to include photos of the infant. Students will read about symptoms that occur when a baby ...
Study biology online free by downloading OpenStax's Concepts of Biology book and using our accompanying online resources including a biology study guide.
This article presents four case studies where patients underwent the same procedures. Patient selection was based on the American Society of Anesthesiologists (ASA) classification, including ASA I and II and excluding ASA III to VI. Adults of both sexes, aged 18 to 60 years, with a surgical tibial fracture, were included.
CBSE Case Based Questions Class 10 Science Chemistry Chapter 6. CASE STUDY : 1. Carbon and energy requirements of the autotrophic organism are fulfilled by photosynthesis. It is the process by which autotrophs take in substances from the outside and convert them into stored forms of energy. This material is taken in the form of carbon dioxide ...
CBSE Case Study Questions Class 9 Science - The Fundamental Unit of Life. CASE 1. All living Organisms are made up of cells and these cells perform all the functions essential for the survival of the Organism eg. Respiration, digestion, excretion etc. In Unicellular organisms, a single cell carries out all these functions and in multicellular ...
Biology document from DeVry University, Phoenix, 4 pages, 1 Case study week 3: Thyroid Karmina Avila Chamberlain University BIOS 252: Anatomy and physiology II Dr. Bruce Oberstein April 15, 2024 2 1. What does the thyroid and its associated hormones do? The thyroid and the hormones of the thyroid glands are the
Increased evidence suggests that long non-coding RNA (lncRNA) holds a vital position in intricate human diseases. Nonetheless, the current pool of identified lncRNA linked to diseases remains restricted. Hence, the scientific community emphasizes the need for a reliable and cost-effective computational approach to predict the probable correlations between lncRNA and diseases. It would ...
Human biology is the scientific study of the human species, which includes the fascinating story of human evolution and a detailed account of our genetics, anatomy, physiology, and ecology. In short, the study focuses on how we got here, how we function, and the role we play in the natural world. This helps us to better understand human health ...
Case studies are in-depth investigations of a person, group, event, or community. Typically, data is gathered from various sources using several methods (e.g., observations & interviews). The case study research method originated in clinical medicine (the case history, i.e., the patient's personal history). In psychology, case studies are ...
Gain experience in the set-up and demonstration of key biology experiments, including relevant safety precautions; Understand hazard identification and risk assessment and the importance of keeping up to date. Course structure. The course is delivered mainly as a practical workshop with some lectures/discussion. Certificates