
Microbiology News
Top headlines, latest headlines.
- Exploring Mouthfeel of Food With a Microscope
- Chicken Feathers Deliver Chemotherapy Drugs
- New Antibiotic Kills Pathogenic Bacteria
- Novel Gene-Editing Tool Created
- Amazing Expertise of Scent Detection Dogs
- H5N1 Bird Flu Transmitted from Cows to Humans
- Intermittent Fasting for Better Gut Health
- Finnish Vole Fever Spreading Further South
- Selective MS Treatment Strategy
- Genetic Mosaicism More Common Than Thought
Earlier Headlines
Friday, may 24, 2024.
- High H5N1 Influenza Levels Found in Mice Given Raw Milk from Infected Dairy Cows
- Observing Mammalian Cells With Superfast Soft X-Rays
Thursday, May 23, 2024
- Developing Novel Methods to Detect Antibiotics in Vegetables and Earthworms
- Key Role of Plant-Bacteria Communication for the Assembly of a Healthy Plant Microbiome Supporting Sustainable Plant Nutrition
- New Approach to Epstein-Barr Virus and Resulting Diseases
- New 'atlas' Provides Unprecedented Insights on How Genes Function in Early Embryo Development
- Mistaken Identity Cleared Up of Foodborne Pathogen Causing Severe Symptoms in Children
- Novel Approach to Interrogate Tissue-Specific Protein-Protein Interactions
- Tracking Down the Genetic Causes of Lupus to Personalize Treatment
Wednesday, May 22, 2024
- Exploring Diversity in Cell Division
- Sweet Move: A Modified Sugar Enhances Antisense Oligonucleotide Safety and Efficacy
- How the 'home' Environment Influences Microbial Interactions
- Milk from Before Antibiotic Era Were Resistant to Antibiotic Tetracycline
- New Insights Into the Degradation Dynamics of Organic Material in the Seafloor
Tuesday, May 21, 2024
- Study Finds Widespread 'cell Cannibalism,' Related Phenomena Across Tree of Life
- Drug-Like Inhibitor Shows Promise in Preventing Flu
- How Plants 'mate' For Life and Repel Other Suitors
- Hope for a Cure for Visceral Leishmaniasis, an Often Fatal Infectious Disease
- Matcha Mouthwash Inhibits Bacteria That Causes Periodontitis
- Researchers Find Unique Adaptations of Fungus Associated With Bee Bread
- Studies Reveal Cell-by-Cell Changes Caused When Pig Hearts and Kidneys Are Transplanted Into Humans
- Clarifying the Cellular Mechanisms Underlying Periodontitis With an Improved Animal Model
Monday, May 20, 2024
- Digging Up Good News for Microbial Studies
- New Mechanisms Behind Antibiotic Resistance
- Evolutionary History of Extinct Duck Revealed
Friday, May 17, 2024
- Better Medical Record-Keeping Needed to Fight Antibiotic Overuse
- Chronic Wasting Disease Unlikely to Move from Animals to People
- Zombie Cells in the Sea: Viruses Keep the Most Common Marine Bacteria in Check
- Fruit Fly Wing Research Offers Window Into Birth Defects
Thursday, May 16, 2024
- A New 'rule of Biology' May Have Come to Light, Expanding Insight Into Evolution and Aging
- Natural Toxins in Food: Many People Are Not Aware of the Health Risks
- More Efficient Bioethanol Production Might Be Possible Using Persimmon Tannin to Help Yeast Thrive
- Bioengineered Enzyme Creates Natural Vanillin from Plants in One Step
Wednesday, May 15, 2024
- From Roots to Resilience: Investigating the Vital Role of Microbes in Coastal Plant Health
- H5N1 Virus from 2022 Mink Outbreak Capable of Inefficient Airborne Transmission
- Now We Know, What Gets Roots to Grow: Can Help in Future Droughts
- An Active Agent Against Hepatitis E
- Parasitic Worm Likely Playing Role in Decline of Moose Populations
- Drug Compounds to Combat Neurodegenerative Diseases
- Repurposed Beer Yeast May Offer a Cost-Effective Way to Remove Lead from Water
Tuesday, May 14, 2024
- Genetics Provide Key to Fight Crown-of-Thorns Starfish
- Virus That Causes COVID-19 Can Penetrate Blood-Retinal Barrier and Could Damage Vision
Monday, May 13, 2024
- Persistent Strain of Cholera Defends Itself Against Forces of Change, Scientists Find
- Fruit Fly Testes Offer Potential Tool Against Harmful Insects
- Commonly Used Antibiotic Brings More Complications, Death in the Sickest Patients
- New Viruses That Could Cause Epidemics on the Horizon
- How Do Genetically Identical Water Fleas Develop Into Male or Female?
- Plant Virus Treatment Shows Promise in Fighting Metastatic Cancers in Mice
Friday, May 10, 2024
- Cellular Activity Hints That Recycling Is in Our DNA
- Research Explores Ways to Mitigate the Environmental Toxicity of Ubiquitous Silver Nanoparticles
- New Light Shed on Carboxysomes in Key Discovery That Could Boost Photosynthesis
Thursday, May 9, 2024
- New Rhizobia-Diatom Symbiosis Solves Long-Standing Marine Mystery
- New Sex-Determining Mechanism in African Butterfly Discovered
- An Epigenome Editing Toolkit to Dissect the Mechanisms of Gene Regulation
Wednesday, May 8, 2024
- New Record Holder for Smallest Dispersers of Ingested Seeds: Woodlice
- An Adjuvant Made in Yeast Could Lower Vaccine Cost and Boost Availability
- Marine Bacteria Team Up to Produce a Vital Vitamin
- Limited Adaptability Makes Freshwater Bacteria Vulnerable to Climate Change
- Natural Compounds That Selectively Kill Parasites
Tuesday, May 7, 2024
- Why Is Breaking Down Plant Material for Biofuels So Slow?
- Bee Body Mass, Pathogens and Local Climate Influence Heat Tolerance
- Free-Forming Organelles Help Plants Adapt to Climate Change
- Fruit Fly Model Identifies Key Regulators Behind Organ Development
- Engineers Develop Innovative Microbiome Analysis Software Tools
Monday, May 6, 2024
- Using Advanced Genetic Techniques, Scientists Create Mice With Traits of Tourette Disorder
- Improved Nutrition, Sanitation Linked to Beneficial Changes in Child Stress and Epigenetic Programming
Friday, May 3, 2024
- Stony Coral Tissue Loss Disease Is Shifting the Ecological Balance of Caribbean Reefs
- Genomes of 'star Algae' Shed Light on Origin of Plants
- How E. Coli Get the Power to Cause Urinary Tract Infections
- Source of Pregnancy Complications from Infections Revealed by Placenta Map
- New Discovery of a Mechanism That Controls Cell Division
- Novel Chemical Tool for Understanding Membrane Remodeling in the Cell
Thursday, May 2, 2024
- For Microscopic Organisms, Ocean Currents Act as 'expressway' To Deeper Depths
- Scientists Track 'doubling' In Origin of Cancer Cells
- Deeper Understanding of Malaria Parasite Development Unlocks Opportunities to Block Disease Spread
- Promising New Treatment Strategy for Deadly Flu-Related Brain Disorders
- Activation of Innate Immunity: Important Piece of the Puzzle Identified
- When Good Bacteria Go Bad: New Links Between Bacteremia and Probiotic Use
- Medical School Scientist Creates Therapy to Kill Hypervirulent Bacteria
Wednesday, May 1, 2024
- Therapy to Kill Hypervirulent Bacteria Developed
- Archaea Can Be Picky Parasites
- Identifying Risks of Human Flea Infestations in Plague-Endemic Areas of Madagascar
- Key Functions of Therapeutically Promising Jumbo Viruses
- Marriage of Synthetic Biology and 3D Printing Produces Programmable Living Materials
- Calcium Can Protect Potato Plants from Bacterial Wilt
- Novel Genetic Plant Regeneration Approach Without the Application of Phytohormones
Tuesday, April 30, 2024
- Unlocking the Genetic Mysteries Behind Plant Adaptation: New Insights Into the Evolution of a Water-Saving Trait in the Pineapple Family (Bromeliaceae)
- New Technology Makes 3D Microscopes Easier to Use, Less Expensive to Manufacture
Monday, April 29, 2024
- Research on RNA Editing Illuminates Possible Lifesaving Treatments for Genetic Diseases
- Possible Alternative to Antibiotics Produced by Bacteria
Friday, April 26, 2024
- Study Details a Common Bacterial Defense Against Viral Infection
- Florida Dolphin Found With Highly Pathogenic Avian Flu
- Surprising Evolutionary Pattern in Yeast Study
Thursday, April 25, 2024
- Barley Plants Fine-Tune Their Root Microbial Communities Through Sugary Secretions
- These Jacks-of-All-Trades Are Masters, Too: Yeast Study Helps Answer Age-Old Biology Question
- Synthetic Droplets Cause a Stir in the Primordial Soup
- Advanced Cell Atlas Opens New Doors in Biomedical Research
- How Immune Cells Communicate to Fight Viruses
Wednesday, April 24, 2024
- Artificial Intelligence Can Develop Treatments to Prevent 'superbugs'
- Can Climate Change Accelerate Transmission of Malaria? Pioneering Research Sheds Light on Impacts of Temperature
- LATEST NEWS
- Top Science
- Top Physical/Tech
- Top Environment
- Top Society/Education
- Health & Medicine
- Mind & Brain
- Living Well
- Space & Time
- Matter & Energy
- Computers & Math
- Plants & Animals
- Agriculture & Food
- Beer and Wine
- Bird Flu Research
- Genetically Modified
- Pests and Parasites
- Cows, Sheep, Pigs
- Dolphins and Whales
- Frogs and Reptiles
- Insects (including Butterflies)
- New Species
- Spiders and Ticks
- Veterinary Medicine
- Business & Industry
- Biotechnology and Bioengineering
- CRISPR Gene Editing
- Food and Agriculture
- Endangered Animals
- Endangered Plants
- Extreme Survival
- Invasive Species
- Wild Animals
- Education & Learning
- Animal Learning and Intelligence
- Life Sciences
- Behavioral Science
- Biochemistry Research
- Biotechnology
- Cell Biology
- Developmental Biology
- Epigenetics Research
- Evolutionary Biology
- Marine Biology
- Mating and Breeding
- Molecular Biology
- Microbes and More
- Microbiology
- Zika Virus Research
- Earth & Climate
- Fossils & Ruins
- Science & Society
Strange & Offbeat
- Resting Brain: Neurons Rehearse for Future
- Observing Single Molecules
- A Greener, More Effective Way to Kill Termites
- One Bright Spot Among Melting Glaciers
- Martian Meteorites Inform Red Planet's Structure
- Volcanic Events On Jupiter's Moon Io: High Res
- What Negative Adjectives Mean to Your Brain
- 'Living Bioelectronics' Can Sense and Heal Skin
- Extinct Saber-Toothed Cat On Texas Coast
- Some Black Holes Survive in Globular Clusters
Trending Topics
Microbiology
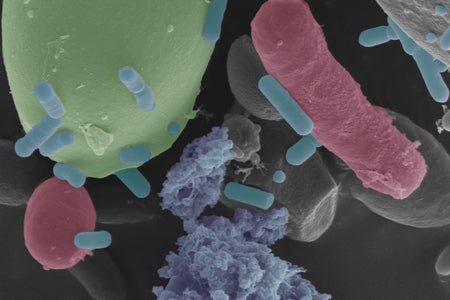
See What Gives Sourdough Its Distinctive Taste and Smell
You can thank yeast and bacteria cultivated over generations for the distinctive taste and smell of the oldest leavened bread in history
Daniel Veghte, The Conversation US
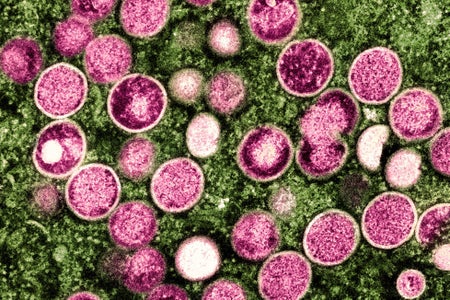
Viral Genetics Confirms What On-the-Ground Activists Knew Early in the Mpox Outbreak
Molecular biology could have changed the mpox epidemic—and could stop future outbreaks
Joseph Osmundson
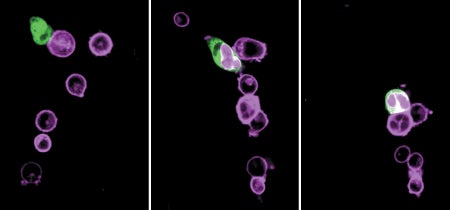
Cannibal Cells Inspire Cancer Treatment Improvement
Giving cells an appetite for cancer could enhance treatments
Kate Graham-Shaw

Is Raw-Milk Cheese Safe to Eat?
Recent bacterial outbreaks from consuming cheese made from unpasteurized milk, or “raw milk,” raise questions about the safety of eating these artisanal products
Riis Williams
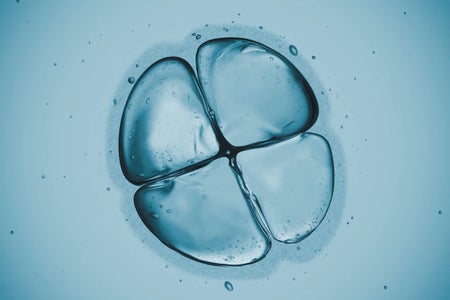
Many Pregnancy Losses Are Caused by Errors in Cell Division
Odd cell divisions could help explain why even young, healthy couples might struggle to get pregnant
Gina Jiménez

'Microbiome of Death' Uncovered on Decomposing Corpses Could Aid Forensics
Microbes that lurk in decomposing human corpses could help forensic detectives establish a person's time of death
Christoph Schwaiger, LiveScience
Weird ‘Obelisks’ Found in Human Gut May be Virus-Like Entities
Rod-shaped fragments of RNA called “obelisks” were discovered in gut and mouth bacteria for the first time
Joanna Thompson
Semen Has Its Own Microbiome—And It Might Influence Fertility
Recent research found a species of bacteria living in semen that’s associated with infertility and has links to the vaginal microbiome
Andrew Chapman

Bacteria Make Decisions Based on Generational Memories
Bacteria choose to swarm based on what happened to their great-grandparents
Allison Parshall

Your Body Has Its Own Built-In Ozempic
Popular weight-loss and diabetes drugs, such as Ozempic and Wegovy, target metabolic pathways that gut microbes and food molecules already play a key role in regulating
Christopher Damman, The Conversation US

See Your Body’s Cells in Size and Number
The larger a cell type is, the rarer it is in the body—and vice versa—a new study shows
Clara Moskowitz, Jen Christiansen, Ni-ka Ford

Subterranean ‘Microbial Dark Matter’ Reveals a Strange Dichotomy
The genes of microbes living as deep as 1.5 kilometers below the surface reveal a split between minimalist and maximalist lifestyles
Stephanie Pappas

Medical Microbiology and Immunology
- Covers major topics in microbial and viral pathogenesis and the immunological response to infections.
- Founded in 1886 by Robert Koch and Carl Flügge, it was renamed multiple times before adopting its current name in 1971.
- Publishes valuable information from related fields of microbiology, including mycology and parasitology.
- Encompasses basic, translational and clinical research in infectious diseases and infectious disease immunology.
Latest articles
Lipopolysaccharide with long o-antigen is crucial for salmonella enteritidis to evade complement activity and to facilitate bacterial survival in vivo in the galleria mellonella infection model.
- Eva Krzyżewska-Dudek
- Vinaya Dulipati
- Jacek Rybka

One-dose intradermal rabies booster enhances rabies antibody production and avidity maturation
- Chidchamai Kewcharoenwong
- Saranta Freeouf
- Ganjana Lertmemongkolchai

Quantitative assay to analyze neutralization and inhibition of authentic Middle East respiratory syndrome coronavirus
- Helena Müller-Kräuter
- Jolanda Mezzacapo
- Verena Krähling

Understanding host–pathogen interaction: paving the path for individualized anti-infective therapy
- Isabelle Bekeredjian-Ding
Topical application of ozonated sunflower oil accelerates the healing of lesions of cutaneous leishmaniasis in mice under meglumine antimoniate treatment
- Ana Paula Pivotto
- Lucas Bonatto de Souza Lima
- Rafael Andrade Menolli

Journal updates
New manuscript submission system.
Please note that as of 19th October 2023, manuscripts will need to be submitted through our new peer review system Snapp, the Springer Nature Article Platform.
New Editor-in-Chief Announcement
Springer is pleased to announce that Isabelle Bekeredjian-Ding is taking over as the Editor-in-Chief of Medical Microbiology and Immunology , effective as of January 1, 2024.
Medical Microbiology and Immunology – Young Scientist Award
The Awardees of Young Scientist Award funded by Springer Nature for the excellent scientific work were nominated! Congratulations go to Dr. Simone Herp (2020 Award), Dr. rer. nat Dominik Brokatzky (2021 Award), Dr. med. sci. Lorenz Kretschmer (2022 Award). The articles of our Awardees can be found in dedicated Collection.
Commissioned reviews
We would like to announce that starting from May 2022 we are accepting only commissioned reviews - review papers that are invited by our Editors.
Journal information
- Biological Abstracts
- CAB Abstracts
- Chemical Abstracts Service (CAS)
- Current Contents/Life Sciences
- Google Scholar
- Japanese Science and Technology Agency (JST)
- OCLC WorldCat Discovery Service
- Pathway Studio
- Science Citation Index Expanded (SCIE)
- TD Net Discovery Service
- UGC-CARE List (India)
Rights and permissions
Editorial policies
© Springer-Verlag GmbH Germany, part of Springer Nature
- Find a journal
- Publish with us
- Track your research
- Search Menu
- Sign in through your institution
- FEMS Microbiology Ecology
- FEMS Microbiology Letters
- FEMS Microbiology Reviews
- FEMS Yeast Research
- Pathogens and Disease
- FEMS Microbes
- Awards & Prizes
- Editor's Choice Articles
- Thematic Issues
- Virtual Special Issues
- Call for Papers
- Journal Policies
- Open Access Options
- Submit to the FEMS Journals
- Why Publish with the FEMS Journals
- About the Federation of European Microbiological Societies
- About the FEMS Journals
- Advertising and Corporate Services
- Conference Reports
- Editorial Boards
- Investing in Science
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic
Six Key Topics in Microbiology: 2020
Read an essential collection of papers showcasing high-quality content from across the five FEMS Journals, which together provide an overview of current research trends in microbiology. Follow the topic area links below for access to articles:
Antimicrobial Resistance
Environmental microbiology, pathogenicity and virulence, biotechnology and synthetic biology, microbiomes, food microbiology.

Affiliations
- Copyright © 2024
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
- Alzheimer's disease & dementia
- Arthritis & Rheumatism
- Attention deficit disorders
- Autism spectrum disorders
- Biomedical technology
- Diseases, Conditions, Syndromes
- Endocrinology & Metabolism
- Gastroenterology
- Gerontology & Geriatrics
- Health informatics
- Inflammatory disorders
- Medical economics
- Medical research
- Medications
- Neuroscience
- Obstetrics & gynaecology
- Oncology & Cancer
- Ophthalmology
- Overweight & Obesity
- Parkinson's & Movement disorders
- Psychology & Psychiatry
- Radiology & Imaging
- Sleep disorders
- Sports medicine & Kinesiology
- Vaccination
- Breast cancer
- Cardiovascular disease
- Chronic obstructive pulmonary disease
- Colon cancer
- Coronary artery disease
- Heart attack
- Heart disease
- High blood pressure
- Kidney disease
- Lung cancer
- Multiple sclerosis
- Myocardial infarction
- Ovarian cancer
- Post traumatic stress disorder
- Rheumatoid arthritis
- Schizophrenia
- Skin cancer
- Type 2 diabetes
- Full List »
share this!
June 1, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
Antibody discovery promises new hope in influenza B battle, paves way for universal vaccine
by Bill Snyder, Vanderbilt University Medical Center
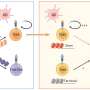
Researchers at Vanderbilt University Medical Center have isolated human monoclonal antibodies against influenza B, a significant public health threat that disproportionately affects children, the elderly and other immunocompromised individuals.
Seasonal flu vaccines cover influenza B and the more common influenza A but do not stimulate the broadest possible range of immune responses against both viruses. In addition, people whose immune systems have been weakened by age or illness may not respond effectively to the flu shot.
Small-molecule drugs that block neuraminidase, a major surface glycoprotein of the influenza virus, can help treat early infection, but they provide limited benefit when the infection is more severe, and they are generally less effective in treating influenza B infections. Thus, another way to combat this virus is needed.
Reporting in the journal Immunity , the VUMC researchers describe how, from the bone marrow of an individual previously vaccinated against influenza, they isolated two groups of monoclonal antibodies that bound to distinct parts of the neuraminidase glycoprotein on the surface of influenza B.
One of the antibodies, FluB-400, broadly inhibited virus replication in laboratory cultures of human respiratory epithelial cells. It also protected against influenza B in animal models when given by injection or through the nostrils.
Intranasal antibody administration may be more effective and have fewer systemic side effects than more typical routes— intravenous infusion or intramuscular injection —in part because intranasal antibodies may "trap" the virus in the nasal mucus, thereby preventing infection of the underlying epithelial surface, the researchers suggested.
These findings support the development of FluB-400 for the prevention and treatment of influenza B and will help guide efforts to develop a universal influenza vaccine, they said.
"Antibodies increasingly have become an interesting medical tool to prevent or treat viral infections," said the paper's corresponding author, James Crowe Jr., MD. "We set out to find antibodies for the type B influenza virus, which continues to be a medical problem, and we were happy to find such especially powerful molecules in our search."
Crowe, who holds the Ann Scott Carell Chair, is University Distinguished Professor of Pediatrics and director of the Vanderbilt Vaccine Center, which has isolated monoclonal antibodies against a host of viral infections , including COVID-19.
The paper's first author, Rachael Wolters, DVM, Ph.D., is a former graduate student in the Crowe lab. Other VUMC co-authors are Elaine Chen, Ph.D., Ty Sornberger, Luke Myers, Laura Handal, Taylor Engdahl, Nurgen Kose, Lauren Williamson, Ph.D., Buddy Creech, MD, and Katherine Gibson-Corley, DVM, Ph.D.
Explore further
Feedback to editors

Almost 1 in 3 Americans know someone who's died from a drug overdose
3 hours ago

Eye-tracking techniques could help primary care providers diagnose autism sooner, more accurately
21 hours ago
This self-powered sensor could make MRIs more efficient
22 hours ago

Not eating can hinder weight loss, study in fruit flies suggests
23 hours ago

Neuroscience research suggests ketones can enhance cognitive function and protect brain networks
May 31, 2024
New research finds antidepressants may help deliver other drugs into the brain

Mediterranean diet tied to one-fifth lower risk of death in women

Scientists find new method to enhance efficacy of bispecific antibodies for solid tumors

Cardiomyocytes study discovers new way to regenerate damaged heart cells
Study: The route into the cell influences the outcome of SARS-CoV-2 infection
Related stories.

New antibodies target 'dark side' of influenza virus protein
Mar 1, 2024

Antibody-based therapies may be effective in fight against influenza B
Aug 4, 2023

Designing antibodies to fight the flu
Feb 1, 2019

Measles-based vector vaccine protects mice against influenza A (H7N9) virus
Jul 7, 2023

Time-dependent viral interference between influenza virus and coronavirus in the infection of differ
Jun 2, 2021

New research provides clues to developing better intranasal vaccines for COVID-19 and flu
Aug 17, 2021
Recommended for you
AI model confirms vaccination is key to cutting COVID in prisons
Novel vaccine concept generates immune responses that could produce multiple types of HIV neutralizing antibodies
May 30, 2024
Scientists find 'Goldilocks' binding strength determines anti-cancer T-cell efficacy and fate

Existing drug shows promise as treatment for rare genetic disorder
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Medical Xpress in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Springer Nature - PMC COVID-19 Collection

Modern clinical microbiology: new challenges and solutions
Pierre-edouard fournier.
Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes, UM63, CNRS7278, IRD198, INSERMU1095, Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille Université, Faculté de Médecine, 27 Boulevard Jean Moulin, Marseille, 13385 France
Michel Drancourt
Philippe colson, jean-marc rolain, bernard la scola, didier raoult.
- Clinical microbiology laboratories (CMLs) now have new roles, notably in early patient and outbreak management.
- The use of rationalized diagnostic kits can simplify sampling and analysis.
- The development of new assays (notably, point-of-care tests) enables CMLs to obtain a diagnosis at the time of care.
- The use of new technologies, in particular MALDI–TOF mass spectrometry, phenotypic microarrays and real-time genome sequencing, can help to improve the workflow and accuracy of clinical-isolate characterization.
- CMLs can detect the emergence of unusual microorganisms, particularly new pathotypes or organisms with antibiotic resistance, and can have a crucial role in outbreak control by warning the medical authorities.
- CMLs also have a role in syndromic surveillance, which can lead to the discovery of unexplained illnesses.
- Large CMLs can also act as strain repositories.
Supplementary information
The online version of this article (doi:10.1038/nrmicro3068) contains supplementary material, which is available to authorized users.
Raoult and colleagues review recent developments in clinical microbiology, including the development of mass spectrometry-based diagnostics and point-of-care tests, which might change clinical practice.
In the twenty-first century, the clinical microbiology laboratory plays a central part in optimizing the management of infectious diseases and surveying local and global epidemiology. This pivotal role is made possible by the adoption of rational sampling, point-of-care tests, extended automation and new technologies, including mass spectrometry for colony identification, real-time genomics for isolate characterization, and versatile and permissive culture systems. When balanced with cost, these developments can improve the workflow and output of clinical microbiology laboratories and, by identifying and characterizing microbial pathogens, provide significant input to scientific discovery.
The roles of clinical microbiologists include the identification of bacterial, viral, fungal and parasitic agents that cause human disease, providing diagnostic and therapeutic support for the clinical management of patients, and preventing the transmission of infectious diseases in both the health care system and the community. The number of identified emerging infectious diseases has steadily climbed since the 1940s 1 , and in the early twenty-first century, tools such as PCR, high-throughput sequencing 2 and MALDI–TOF mass spectrometry (MS), along with new sampling and culture strategies, are changing clinical microbiology. This technological progress, which began in environmental microbiology 3 , 4 , has revealed a much larger microbial world than was believed to exist a few years ago. In 1980, only 1,800 validated bacterial species had been published, whereas more than 500 new species are now described annually 5 . Thus, over the past 4 years, more new bacterial species have been described than were described in the period up to 1980 ( Fig. 1 ). For example, important human pathogens such as Helicobacter pylori , which can cause gastric ulcers and cancer, and Tropheryma whipplei , the causative agent of Whipple's disease, were first isolated in 1985 and 2001, respectively. For viruses, the progress has been comparable; severe acute respiratory syndrome (SARS)-associated coronavirus 6 and the arenavirus Lujo virus, which causes haemorrhagic fever 7 , were identified in 2003 and 2009, respectively.

The development of new technologies has had a substantial impact on the number of microbial species that are identified each year.
PowerPoint slide
Moreover, our view of microbial pathogenesis, in which one given microorganism causes one given disease — a view that was inherited from Pasteur and Koch — began to change when new tools allowed us to observe the diversity of microorganisms and the impact that this diversity has on human diseases and their treatment 8 . For example, we now know that nine Mycobacterium species can cause tuberculosis, some of which, such as Mycobacterium bovis , require specific antibiotic treatments.
In this Review, we detail the main changes that have recently occurred in the technologies and techniques available to clinical microbiology laboratories (CMLs), including syndrome- and disease-based sampling kits, point-of-care (POC) testing, and isolate typing and characterization by MALDI–TOF MS or by next-generation sequencing (NGS). We also try to predict the most important developments for the future.
Developments in sampling
Syndrome- and disease-based kits. The conventional diagnosis of infectious diseases usually relies on a stepwise approach in which the physician examines the patient, diagnoses a clinical syndrome and then tests for pathogens that are potentially responsible for that syndrome, until a diagnosis is made. However, the growing number of emerging pathogens makes it difficult for physicians to memorize the actual list of pathogens for each infectious disease and thus prescribe all the appropriate diagnostic microbiology tests. To reduce the delays associated with resampling or retesting, sampling can be carried out using diagnostic kits that are standardized according to the syndrome or disease. Such a strategy enables optimization of the type and number of specimens collected from the patient, simplification of the laboratory test prescription for the physician, and the creation of a scheduled workflow for the nurse and the doctor, and it also allows the clinical laboratory to easily trace samples. To date, such diagnostic kits have been developed for endocarditis, pericarditis, diarrhoea, osteitis, meningitis, encephalitis, uveitis and keratitis 9 , 10 , 11 , and their design has been based on the repertoire of pathogens responsible for the syndrome or disease, and on the optimal methods to achieve the direct or indirect detection and identification of these pathogens. Further syndrome-based kits can be designed for specific groups of febrile patients, such as those presenting to the emergency room for POC testing, travellers, pilgrims to Mecca (Saudi Arabia), the homeless, or patients with cystic fibrosis and respiratory tract infections 12 . In addition to the broad syndrome- or disease-based kits, pathogen-specific kits can be used, as for pulmonary tuberculosis 13 .
Compared to a more traditional approach, in which the most common agents of an infectious syndrome are tested first, followed by testing for the rarer agents, a broad syndrome- or disease-based strategy might seem to be economically disadvantageous. But it is possible that the increased laboratory costs will be balanced by shorter hospitalization times owing to an earlier diagnosis and an earlier initiation of the appropriate antibiotic therapy, and by avoiding unnecessary treatment in the case of viral infections 14 .
In addition, specimens recovered from such kits can be preserved in biobanks for retrospective testing when new emerging pathogens are described, and data on kit usage can be used for epidemiological analyses such as seasonal changes in sampling or to evaluate the cost-effectiveness of the kits. To date, only a few CMLs have adopted syndrome- and disease-based kits; in France, for example, they are used in 25 university hospitals.
Strain collections. Currently, reference microorganisms are preserved in international collections such as the American Type Culture Collection, the Deutsche Sammlung von Mikroorganismen und Zellkulturen and the Japan Collection of Microorganisms. However, because of their number, most clinical isolates routinely cultivated in CMLs are neither conserved nor referred to these collections. The conservation of clinical isolates by CMLs might enable tracing of outbreaks and the characterization of emerging pathogens, and could thus be valuable for both public health (disease surveillance and prevention) and scientific purposes. Notably, bacterial strains cultivated from normally sterile organs and body fluids, such as blood and cerebrospinal fluid, might be conserved. However, because of the cost, few reference laboratories are currently equipped for long-term storage of samples in such collections.
Processing of clinical samples
Direct examination. Gram staining, used mostly for the direct examination of uncultured clinical samples, remains an essential step in detecting microorganisms and guiding empirical antibiotic therapy, although the adoption of MALDI–TOF MS to identify cultured colonies might restrict the use of this technique 15 . However, Gram staining can be automated — for example, by a robot such as the PREVI Colour Gram (BioMérieux), which can stain up to 300 slides per hour and reduces the chemical hazard for technicians.
Culturing clinical samples. Culturing remains the mainstay of clinical microbiology 16 . Several approaches, many of which were first implemented by environmental and then clinical microbiologists, have been used in recent years to improve the isolation of fastidious bacteria from human specimens 17 , 18 . The first approach, which remains historically the most efficient, is based on empiricism and uses existing media or media with enrichment components added. For instance, Borrelia recurrentis was reputed to be non-cultivable axenically until the blood of a patient with louse-borne relapsing borreliosis was inoculated on media dedicated to Borrelia burgdorferi 19 . In the case of Mycobacterium tuberculosis , the use of blood-enriched medium enabled the improved and accelerated recovery of colonies 20 . Recently, we developed a strategy named culturomics , which proved to be very successful in isolating previously unknown or uncultivated bacteria ( Box 1 ). This empirical approach has the advantage of using common media, but is poorly adapted to the detection of emerging fastidious bacteria.
The second, non-empirical approach is to use genome sequence data to develop media that are specifically adapted to a given fastidious pathogen 18 . For example, axenic culture of T. whipplei strains obtained by cell culture or directly from clinical samples was achieved using standard cell culture medium that had been supplemented with those amino acids for which there were no corresponding metabolism genes in the genomes of the cultured strains 21 . A comparable approach was recently initiated for the culture of two other strictly intracellular bacteria, Coxiella burnetii and Chlamydia trachomatis 22 , 23 , and might be implemented for other fastidious pathogens such as Mycobacterium leprae and Treponema pallidum 24 .
A third approach (often occurring by accident) is to extend the incubation time of fastidious extracellular pathogens, such as the extracellular forms of the facultative intracellular Bartonella spp. 25 , and of intracellular pathogens, such as Bartonella spp., T. whipplei and mycobacteria 25 , 26 , 27 . A fourth approach is the use of cell culture, especially the shell vial assay. Briefly, in this assay, ground clinical specimens are inoculated by centrifugation into a layer of eukaryotic cells grown on a 1 cm 2 cover slip at the bottom of a small tube 28 . This method has been shown to be highly efficient for the isolation of fastidious agents, even those that are not strictly intracellular 28 . The versatility of the technique is related to the nearly unlimited choice of cells that can be used for culture, thus widening the spectrum of culture conditions and isolated microorganisms. For example, Rickettsia felis 29 can be grown only in XTC-2 cells, which require a low temperature, and Legionella spp. 30 can be grown only in amoebae. However, these novel cell culture approaches are restricted to the few laboratories that are equipped with specific facilities and well-trained personnel.
Box 1: Culturomics: the diversification of culture methods for microbial isolation
A major challenge in the twenty-first century is the need for a comprehensive characterization of both the human microbiome and its relationships with health and disease 108 . Recent genomic and metagenomic studies have demonstrated that approximately 80% of bacterial species detected in the human gut are either uncultured so far or even unculturable 109 . Therefore, in the omics era, culturing became outdated for the study of complex microbial communities. However, several discrepancies have been observed among metagenomic studies, apparently reflecting biases of the techniques used and the fact that clinically relevant minor populations were not considered. For example, potentially pathogenic bacteria (such as Salmonella enterica subsp. enterica serovar Typhi, Tropheryma whipplei and Yersinia enterocolitica ) that can be present in faeces at concentrations of <10 5 colony-forming units per ml, were ignored. Thus, we designed a new culture strategy that we named culturomics, which uses diversified culture conditions (medium, temperature and atmosphere) to complement culture-independent approaches and to complete our knowledge of the human microbiota 110 .
In the first culturomics study, which used 212 distinct culture conditions, 341 bacterial species were cultured from Senegalese patients and classified as belonging to seven phyla and 117 genera 110 . In a second study of a patient receiving massive antibiotic drug therapy for multidrug-resistant tuberculosis, 70 distinct culture conditions enabled the isolation of 39 bacterial species, including one new species and three species that had not previously been observed in the human gut 111 . To date, culturomics has yielded 175 bacterial species never previously found in the human gut, and 32 new bacterial genera and/or species, including Microvirga massiliensis , which has the largest genome of the known human-associated bacteria. Metagenomic analysis of the same specimens produced 698 phylotypes, including 282 known species, 51 of which overlapped with the microbiome identified by culturomics. However, despite these promising results and although culturomics might complement metagenomics, notably by overcoming the bias inherent to metagenomic approaches of ignoring minor microbial populations, this new technique is a time-consuming, expensive strategy that, as yet, has not been adapted to routine microbiology.
Identification and resistance testing of pathogens
Identification of bacteria, fungi and viruses. Phenotypic identification of bacterial isolates has long relied on a combination of biochemical properties such as oxygen requirement, Gram staining, carbohydrate metabolism and the presence of specific enzymes. However, phenotypic identification systems, such as miniaturized strips, are costly and time-consuming, even when automated, and thus over the past decade have been superseded by MALDI–TOF MS. Worldwide, many large CMLs have adopted MALDI–TOF MS owing to the increased workflow efficiency and associated cost reduction when using this approach 31 ( Fig. 2 ). MALDI–TOF MS can identify bacterial isolates in a few minutes (6–10 minutes) and for low costs (€1.43 per strain, compared with €2.2–€8.23 per strain using API strips or Vitek automates, both of which are commonly used phenotypic assays, and €137 per strain for 16S rRNA sequencing) 31 . Moreover, the approach can also efficiently identify clinical isolates of fungi 32 , as well as bacteria in blood culture vials 33 .

MALDI–TOF mass spectrometry (MS) allows the identification and antibiotic-susceptibility testing of microbial pathogens in urine samples or cultivated on agar or in blood culture bottles. Alternatively, strain identification can be carried out by PCR–ESI–QTOF MS (PCR followed by electrospray ionization–quadrupole TOF MS) characterization of PCR amplicons from the microbial DNA.
The identification of microbial isolates using MALDI–TOF MS relies on a comparison between the mass spectrum of the isolate and mass spectra in available databases. In addition to the rapidity and low cost of MALDI–TOF MS, other advantages include the precise identification of the isolate at the species level and, in some species, at the subspecies and strain levels. Currently, bacterial species from many genera have been identified by MALDI–TOF MS, including fastidious microorganisms (reviewed in Ref. 34 ). However, the discriminatory power of the method varies depending on the species and the exhaustiveness of the database used. Notably, some bacterial taxa are under-represented in databases, and technical problems (such as variations in culture conditions, sample preparation and the spectrometer used) further affect the discriminatory power of this technique 35 . In addition, streptococci, non-fermenting Gram-negative bacteria and anaerobic bacteria are generally harder to discriminate than other bacteria, although this weakness can be partially corrected by enriching the available databases with spectra from more strains 31 . MALDI–TOF MS can be used for typing community-acquired pathogens, such as Listeria monocytogenes , Legionella pneumophila and Yersinia enterocolitica ; health care-associated pathogens, such as Corynebacterium striatum 36 and Candida parapsilosis 37 ; and both community- and health care-associated pathogens, such as Staphylococcus aureus . Finally, MALDI–TOF MS is now routinely used in several large CMLs for the direct identification of bacteria in blood cultures 33 . Although MALDI–TOF MS can identify various viral species 38 , commercially available MS databases do not contain viral spectra; therefore, MALDI–TOF MS does not yet enable the identification of viruses in routine microbiology.
The recent development of high-throughput phenotypic microarrays, such as the Biologue system (Biologue, Inc.), which combines up to 2,000 phenotypic tests, might challenge the leading position of MALDI–TOF MS. Microarrays can be used for several applications, such as the determination of bacterial metabolic and chemical properties, the formal description of new species, and the development and optimization of culture media. The Biologue system has been demonstrated to be highly efficient for phenotypic identification of organisms, although it is less discriminatory than16S rRNA sequencing 39 and slower than MALDI–TOF MS 31 .
In the past 10 years, Raman spectroscopy has been demonstrated to accurately and rapidly identify bacterial, fungal and yeast isolates at the species and subspecies levels 40 , 41 . In this method, light scattering from a laser-illuminated bacterial colony is analysed and transformed into a spectrum that is compared to a specific database. Although this method is inexpensive, reagent-free and as rapid as MS, Raman spectroscopy is not currently widely used in CMLs owing to technical hurdles such as low signal strength, and perhaps also owing to the initial cost of a new instrument. In addition, as for MALDI–TOF MS, the reliability of Raman spectroscopy depends on the quality and coverage of the reference databases.
Detection of antibiotic resistance. Classical methods, such as phenotypic antibiotic-susceptibility testing of a clinical isolate based on minimum inhibitory concentrations (MICs) and clinical breakpoints, remain the gold standard for in vitro prediction of antibiotic resistance, but many factors can influence the results obtained for a given isolate and can lead to a false positive or false negative result. MIC is defined as the lowest concentration of antibiotic able to inhibit the growth of a bacterial isolate in vitro , whereas breakpoints are used to predict the clinical outcome of an antibiotic treatment in vivo . Some new and/or emerging resistance mechanisms might be missed because they are not easily detected phenotypically in vitro . Instead, they require additional molecular testing. Moreover, results from the classical resistance tests might not be rapidly available, especially for fastidious bacteria such as M. tuberculosis 42 . The application of real-time PCR (RT-PCR) to the rapid screening and detection of common antibiotic resistance genes such as mecA or ndm1 (which provide resistance to methicillin in S. aureus 43 and carbapenem in bacteria of the family Enterobacteriaceae 44 , respectively) was revolutionary. In a similar manner, non-fluorescent DNA microarrays 45 and pyrosequencing 46 can detect gene mutations or SNPs that are known to be associated with M. tuberculosis resistance to antituberculous compounds. DNA microarrays are another new technology that can detect, in a single step, many resistance genes in Gram-negative and Gram-positive bacteria 47 . However, microarrays are expensive (∼300€ per Mb for microarrays versus 15€ per Mb for high-throughput genome sequencing) and can detect known genes only. Recently, MALDI–TOF MS has also been used clinically for the rapid phenotypic detection of antibiotic resistance, especially of β-lactamase activity 48 . For example, it is possible to detect carbapenemase activity in bacteria of the family Enterobacteriaceae, in Pseudomonas aeruginosa and in Acinetobacter baumannii through the identification of carbapenem compounds and their degradation products by MALDI–TOF MS in only 1–4 hours and with high sensitivity and specificity 49 , 50 , 51 ( Fig. 2 ). One of the main advantages of this technique is that any enzymatic activity associated with antibiotic resistance can be detected, even if the causative enzyme is unknown, and this could lead to the discovery of new antibiotic resistance determinants.
In addition, antibiotic resistance diagnostics can be further improved to detect and interpret resistance patterns using automated intra-laboratory surveys of antibiograms. This allows the detection of unusual phenotypes or emerging clusters of strains with a similar profile in clinical isolates. We recently developed a computer tool for real-time surveillance of resistance 52 , 53 . On a weekly basis, this program automatically and systematically compares the resistance patterns obtained from clinical samples in our routine microbiology laboratory to a Microsoft Excel database of resistance patterns based on results from 12,000 clinical isolates tested in our laboratory in 2012. The aim is to detect any atypical, abnormal or rare patterns of resistance or susceptibility. Such a tool might improve the diagnosis, monitoring and characterization of antibiotic resistance.
Sequencing of microbial genomes
Genomic sequence information from cultivated microorganisms is widely used for epidemiological studies ( Table 1 ). In clinical microbiology, applications of genome sequencing include the development of detection, identification and genotyping tools, the design of culture media and the assessment of antibiotic resistance or virulence repertoires 42 , 54 , 55 . Recently, thanks to NGS technologies such as the MiSeq (Illumina), Ion Torrent Personal Genome Machine (PGM) (Life Technologies) and 454 GS Junior (Roche) bench-top sequencers, bacterial genome sequencing has become fast (only a few hours) and cheap (only a few hundred US dollars) 56 , making whole-genome sequencing compatible with the routine clinical microbiology workflow 42 , 55 , 57 , 58 . This strategy might offer rapid and exhaustive access to the virulence determinants 59 , antibiotic resistance markers 60 or genotypes 61 of unusual or difficult-to-grow bacterial strains isolated from clinical specimens. Recent examples of the use of whole-genome sequencing in clinical microbiology include the investigations of hospital outbreaks of A. baumannii , S. aureus and Clostridium difficile infections 62 , 63 , and the identification of the virulence determinants of a Staphylococcus epidermidis strain that was the aetiological agent of native valve endocarditis 64 . In addition, as demonstrated recently, NGS might also enable complete genome sequencing of a pathogen directly from a clinical specimen 65 .
However, despite the undeniable advantages, NGS requires extensive bioinformatics for sequence analysis, from assembly to annotation, and this remains a significant problem for the routine application of genomics to clinical microbiology. The development of automated tools for the analysis of genomic sequences and the compilation of databases containing whole genome sequences together with specific genetic criteria, such as virulence factors and resistance markers, might solve this problem in the near future and enable NGS to be used in routine practice 55 .
In addition, NGS has been used to begin to decipher complex human microbial communities in metagenomic studies 66 . Furthermore, a recent analysis of the gut flora provided hints that might enable the prediction of infection susceptibility or antibiotic resistance, or the assessment of the risk of developing diseases such as obesity 67 .
Genotyping. Genotyping consists of tracing clones by identifying sequence-specific signatures. Various genotyping methods have been developed, including several methods based on DNA banding patterns, such as pulsed-field gel electrophoresis 68 , PCR–restriction fragment-length polymorphism (RFLP) 69 , multiple locus variable number of tandem repeats analysis (MLVA) 70 , random amplification using arbitrary primers (RAPD) 71 and amplified fragment-length polymorphism (AFLP) 72 . However, these methods often lack reproducibility within and between laboratories. DNA hybridization-based methods using nucleotide probes, such as DNA macroarrays 73 and DNA microarrays 74 , are expensive, can identify only the genetic markers present on the arrays and thus might underestimate the genetic diversity among bacterial populations. DNA sequence-based methods such as multilocus sequence typing (MLST) 54 , multispacer sequence typing 2 and genome sequencing 62 , 63 are discriminatory and reproducible, but are time-consuming and can be expensive. Although genotyping all the pathogenic isolates from all cultures would probably have a significant impact on infection control policies and their execution, most genotyping techniques are not carried out routinely, except for the detection of specific clones of certain pathogens. For example, M. tuberculosis str. Beijing can be detected within 2 hours using specific RT-PCR 75 , enabling patient isolation, contact tracing and extended antituberculous treatment.
Direct microbial identification in specimens
Mass spectrometry. In 2009, for the first time, MALDI–TOF MS was reported to efficiently identify bacteria directly from blood collected in culture bottles, with results obtained less than 2 hours after the blood culture vial was determined to be positive, and with a 97.5% success rate 76 ( Fig. 2 ). However, the accuracy of bacterial identification might be influenced by unstandardized sample preparation, differences in bacterial concentrations, pre-incubation, prolonged incubation and the blood culture system used 35 . Commercially available extraction kits 77 and the implementation of a minimum concentration of 10 6 –10 8 colony-forming units (CFU) per ml (Ref. 78 ) might resolve some discrepancies. In 2010, MALDI–TOF MS was also used for the direct identification of bacteria in urine samples, with an accuracy rate of 91.8% and with bacterial concentrations as low as 10 3 CFU per ml (Ref. 78 ). Finally, in addition to identifying bacteria, MALDI–TOF MS has been shown to be capable of identifying fungi in blood cultures and nail specimens 79 . However, MALDI–TOF MS itself has certain limitations, including the inability to identify species among mixed populations or directly in solid clinical specimens, and poor discriminatory power among corynebacteria and streptococci 31 .
Molecular detection. Molecular detection methods — notably, bacterial PCR combined with sequencing — have played a major part in clinical microbiology and enabled the discovery of several non-cultivable pathogens, such as Bartonella quintana and T. whipplei 80 , 81 . Among molecular diagnostic methods, RT-PCR has proved highly valuable, as it enables diagnoses to be obtained in less than 5 hours, whereas traditional microbiological methods, including conventional molecular diagnostics such as broad PCR followed by sequencing, take a minimum of 1 day. RT-PCR also enables the quantification of pathogens 82 and is routinely used to quantify the viral load in patients infected with HIV. RT-PCR assays can target either specific pathogens and resistance- or virulence-encoding genes, or universal targets, such as 16S rDNA for bacteria and rDNA internal transcribed spacers for fungi. In addition, RT-PCR multiplex assays can simultaneously target several DNA fragments, thus enabling the detection of a whole panel of pathogens that are implicated in particular syndromes, such as bloodstream infections 83 , community-acquired pneumonia, meningitis 84 , sexually transmitted diseases 85 or urinary tract infections 86 .
Although RT-PCR-mediated detection of specific agents avoids sequencing, conventional PCR using broad-range targets combined with sequencing remains an irreplaceable tool for the identification of new pathogens in clinical specimens. Over the past decade, PCR amplicon sequencing has been increasingly implemented by CMLs for the identification or genotyping of microbial pathogens 87 .
Alternatively, PCR–ESI–QTOF MS (PCR followed by electrospray ionization–quadrupole TOF MS), is a recent application of mass spectrometry that enables bacterial identification following PCR amplification of species-specific DNA fragments 88 . This commercially available technology (in the form of the PLEX-ID machinery from Abbott) can identify important human pathogenic bacteria, including Bacillus anthracis , Francisella tularensis , Yersinia pestis , Burkholderia mallei , Burkholderia pseudomallei , Brucella spp. and C. burnetii , along with influenza A virus and influenza B virus. However, further studies are needed to validate the usefulness of PCR–ESI–QTOF MS in routine microbiology.
Electronic nose. By identifying the specific combination of volatile organic compounds produced by microorganisms within a clinical specimen, an electronic nose is able to detect the presence and type of bacteria responsible for infectious diseases such as urinary tract and pulmonary infections before other methods, and this approach thus allows early initiation of appropriate antibiotic therapy 89 , 90 . A limit of the electronic nose is that it cannot quantify microorganisms within a specimen and therefore cannot discriminate between infection and colonization.
The clinical microbiology laboratory
Core laboratories. In several countries, the concentration of CMLs into large core laboratories aims to lower the management costs while increasing the number of routine and specialized tests available for various diseases 91 . However, this organization distances patients and doctors from the laboratory, and this might result in delayed reporting of test results and inappropriate management of patients. In 2011, an outbreak of Shiga toxin-producing Escherichia coli O104:H4 in Germany, a country in which core CMLs have been created, highlighted this risk, as there was delayed reporting of clinical cases 92 .
Point-of-care laboratories. The need for automation and workflow improvement made it necessary to centralize biomedical analyses, but the increased distance from the laboratory to the site of patient care, as well as the batch processing of clinical specimens, has resulted in delayed delivery of results. As a consequence, core CMLs have been unable to contribute to timely decision-making for most infectious diseases. By contrast, POC laboratories (POCLs) are on-site laboratories that carry out rapid diagnostic tests within 4 hours (compared with >12 hours for culture methods or serology assays) and operate around the clock, and these laboratories are perfectly adapted to provide results in the early stages of patient care and can help with making appropriate hospitalization, isolation and therapy decisions ( Fig. 3 ). Initially, most POC tests relied on immunochromatographic or agglutination assays; however, the latest automated DNA extraction and RT-PCR devices using simplified procedures have made molecular detection assays compatible with POC objectives.

Specimens referred to the point-of-care (POC) laboratory are tested according to the clinical picture and the physicians' queries. POC tests that will detect the most likely causative pathogen or a pathogen-associated antigen are carried out according to the clinical syndrome and the patient history. BSL3, biosafety level 3.
To date, several POC tests, such as the immunochromatographic detection of Streptococcus pyogenes antigen in the throat and Plasmodium falciparum in the blood, have demonstrated their usefulness in physicians' offices and on board ships visiting malaria-endemic countries (D.R., unpublished observations). POCLs should provide an accurate and rapid answer to a limited number of clinical microbiology questions and have a clear impact on patient management 14 . POC assays address the requirements to hospitalize patients, to isolate contagious individuals, and to initiate and focus anti-infective therapy. Assays routinely used in POCLs include those developed to detect bacteria (for example, Bordetella pertussis , C. difficile , Neisseria meningitidis , Streptococcus pneumoniae and S. pyogenes ), viruses (including adenoviruses, dengue virus, enteroviruses and influenza viruses) and parasites (such as P. falciparum ). Early studies have demonstrated that on-site POCLs met physicians' needs and have influenced the management of up to 8% of the patients who presented to emergency wards 14 . This strategy might represent a major evolution of decision-making regarding the management of infectious diseases and patient care. In particular, it has been demonstrated that the POCL strategy enables the rapid adaptation of antibiotic therapy for patients with bacterial meningitis and reduces the length of the hospital stay for patients with viral meningitis 14 . Typically, POCLs feature operator-independent tests, including RT-PCR and immunochromatographic assays, most of which are cleared by the US Food and Drug Administration and the European Commission. Compared with standard microbiological procedures, most POC tests have a high positive predictive value, but some of them lack sensitivity 93 , 94 .
Syndrome-based kits can also be used in POCLs for the rapid diagnosis of respiratory tract infections 95 , meningitis 96 and sexually transmitted diseases 94 . The results are immediately transmitted to physicians by SMS and to the hospital information system. POCLs can be easily implemented in any type of environment, including remote areas, as demonstrated recently with the development of a POCL in rural Senegal 97 . Furthermore, test panels can even be adapted to the local epidemiology.
Automation. Most CMLs face common constraints, including the need to process increasing numbers of samples and increasing numbers of assays per sample. They need to maintain a continuous workflow, sometimes even under the pressure to reduce human resources. As many of the assays are characterized by repetitive manual operations, partial or full automation reduces human intervention and increases the quality of results by reducing human error, thus improving the workflow and output. For years, CMLs have automated various tasks, such as blood culture monitoring, biochemical phenotypic identification of bacteria and yeasts, antibiotic-susceptibility testing, Gram staining, DNA extraction and PCR amplification. The automated early detection of positive blood cultures and the identification of the pathogens and their antimicrobial susceptibilities can significantly reduce mortality in patients with bacteraemia and are among the most important functions of CMLs 98 . However, it is likely that MALDI–TOF MS automation will progressively replace phenotypic microbial identification 15 .
One of the most recent advances in automation is the development of plate streakers that couple a bi-directional interface with a laboratory information system, and the possibility of inoculating different plates with a unique sample. To date, the three available automated systems — PREVI Isola (BioMérieux), WASP (Walk-Away Specimen Processor; Copan) and InoqulA (Kiestra) — all allow better CFU recovery than manual inoculation 99 . However, most specimens (except urine specimens) require time-consuming pretreatment, such as fluid sputum sample preparation or solid specimen grinding, or the use of specific and expensive swabs. Concomitantly, there is an increasing need to freeze and store samples in biobanks after inoculation or DNA extraction. Therefore, the future generation of automated plate streakers should incorporate functionalities such as aliquoting or DNA extraction or should connect to automated DNA extraction and/or amplification systems, such as the GeneXpert system (Cepheid). Finally, with the aim of complete automation, these systems should be coupled with automated urinary flow cytometry 100 and automated incubation in adapted atmospheres. Currently, only the fully automated Kiestra platform partially allows this.
In addition, the steps following incubation — that is, plate sorting according to positivity, colony picking for MALDI–TOF MS identification, and antimicrobial-susceptibility testing — should also be automated. Automatic incubators and plate sorters are currently under development, but are not yet available. A system that can discard (at the minimum) negative samples, similar to blood culture automated systems, would allow significant time savings. In our laboratory, 52% of the 161,240 samples inoculated annually are sterile. Automated colony picking that enables both MALDI–TOF MS identification and antimicrobial-susceptibility testing is already available using the Kiestra platform 101 .
Quality assurance and quality control. CMLs should implement a system of quality management that monitors all aspects of the pre- and post-analytical service provided by the CML, from the receipt of patient samples to the reporting of results 102 , ideally in accordance with the common International Standard Organization (ISO 9001) framework. In addition, all organizational and technical operating procedures should be standardized, made permanently available to the laboratory personnel and updated on a quarterly basis. Moreover, laboratories should participate in external and internal quality-assurance schemes.
Result reporting
Result interpretation and reporting is essential in the management of infectious diseases at both the patient and community levels and thus should be a constant priority of CMLs and health authorities.
Interpretation. For patient management, it is essential that the laboratory provide reliable results which the clinician can trust. Therefore, the correct interpretation of microbiology results is crucial. Interpretation can be improved by automated interpretation software that reports any unknown or impossible phenotype or genotype. Automated systems such as VITEK2 (BioMérieux), Phoenix (Becton, Dickinson and Co.) and EPIMIC 52 enable the detection of bacterial isolates that are either taxonomically unusual in a certain clinical syndrome or specimen type, or exhibit a new or rare antibiotic resistance profile. CMLs either use commercially available MALDI–TOF MS spectrum databases (such as those from Bruker Daltonik, Shimadzu or BioMérieux) for identifying bacterial isolates, or develop their own databases 15 . In addition, results from genotyping methods can be compared to online databases 69 . Databases from these three surveillance systems can be updated with the results from the laboratory. In our laboratory, a medical microbiologist validates each result with the help of these tools and a survey of the international scientific literature before the result is transmitted to clinicians.
Reporting. Early reporting of microbiology results should be a priority of all CMLs, in particular for critical results (a positive direct examination, culture, or PCR from blood, cerebrospinal fluid or tissue). Faster reporting of identification and antimicrobial-susceptibility results can significantly reduce the length of hospital stay and the overall costs 103 . Of course, faster reporting implies that CMLs should follow technological progress and be equipped with the fastest microbiological methods 104 . Reporting can be carried out using laboratory information systems, electronic dashboards or mobile phones (for oral or SMS communications), thereby enabling physicians to receive timely alerts. In our centre, following the development of POCLs, we implemented SMS-based transmission of results to inform physicians as quickly as possible. Physicians might also use personal information systems to access other laboratory or clinical-patient data, or local epidemiological data.
Another important aspect of reporting is warning the local, national and international medical communities (the CDC, the European Centre for Disease Prevention and Control (ECDC) and the WHO) in case of the emergence of unusual infectious diseases with epidemic potential. Alert websites, such as the Program for Monitoring Emerging Diseases ( ProMED-mail ) and the European Travel Medicine Network ( EuroTravNet ), or journals such as Eurosurveillance are suitable channels for such alerts.
Conclusions and perspectives
With the introduction of omics technologies (genomics, proteomics, culturomics, transcriptomics and metabolomics), CMLs face new challenges, such as obtaining a diagnosis at the time of care ( Fig. 4 ). For example, until recently, the usefulness of blood cultures in the emergency room was limited, as the results of identification and antibiotic-susceptibility testing were only available 72 hours after sampling 105 . Under these conditions, either the empirically prescribed antibiotic treatment was effective, or it had to be changed to another treatment, the worst-case scenario being the patient's death before the diagnosis was established. Clearly, CMLs can only have a major impact on early patient management when diagnostic speed enables the appropriate medical decisions to be made rapidly. In terms of treatment, rapid pathogen identification combined with knowledge about where the patient contracted the infection (for example, whether it was hospital acquired or community acquired) enables the presumptive deduction of antimicrobial susceptibility, and such antibiotic stewardship based on rapid diagnostics can reduce hospitalization costs 106 . Another crucial challenge for clinical microbiology, at a time when the need for cost reduction favours laboratory concentration, is to avoid a disconnection between core CMLs and clinical settings. Preserving permanent interactions between clinical microbiologists and primary care physicians ensures timely reporting, proper result interpretation and optimal therapy management.

The figure describes the processing of specimens from patients who are thought to have an infectious disease. Sampling is carried out using syndrome- and disease-based sampling kits, and the resulting specimens are tested using point-of-care assays, when available. Culturing, strain characterization (phenotypic or molecular identification, and antibiotic-susceptibility testing) and molecular detection in clinical samples are carried out in the core laboratory. Unusual pathogens, conserved by the laboratory, are reported to the physician in charge and possibly to the sanitary authorities, and are further characterized by genome sequencing. MS, mass spectrometry.
In the case of outbreaks, CMLs play an important part in warning the medical authorities, which then can corroborate the results across a particular region, country or continent. These laboratories can detect the emergence of unknown species, particular pathotypes and antibiotic resistance, and thus should detect the development of outbreaks rapidly. Early information sharing is mandatory for outbreak control, as demonstrated by a global outbreak of cotrimoxazole-resistant E. coli , the source of which remains mysterious, although links with poultry have been proposed 107 . CMLs also have a role in syndromic surveillance (that is, monitoring the type of specimens and the associated syndromes) and alerting, which can lead to the discovery of unexplained illnesses. As an example, an increase in the number of cerebrospinal fluid samples referred to the laboratory in the absence of an aetiologically labelled outbreak of meningitis should alert laboratory technicians and prompt the search for a new pathogen. Likewise, the emergence of an atypical organism must be reported through warning systems such as ProMED-mail or EuroTravNet, but probably also at the national and European levels. To date, Europe lacks such a centrally organized warning system.
In addition, large laboratories should act as strain repositories. Although European, American and Japanese reference strain collections exist, the collection of pathogenic microorganisms detected in CMLs should be extended to conserve clinical strains of interest. Moreover, large CMLs have an educational role and should offer training and up-to-date courses on currently used and evolving technologies. Finally, as CMLs identify and characterize microbial pathogens, sequence genomes and are involved in scientific discoveries in general, they can also have an important impact on the scientific literature.
Biographies
Pierre-Edouard Fournier is a medical microbiologist. He is the head of the Molecular Diagnosis Department at the Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes (URMITE) and the Méditerranée-Infection institute at the University Hospital in Marseille, France. His research topics include the study of bacterial genomes and the diagnosis of blood culture-negative endocarditis.
Michel Drancourt is a clinical microbiologist and co-director of Didier Raoult's laboratories. He is the author of 300 papers on fastidious organisms, including mycobacteria, archaea and planctomycetes, and on paleomicrobiology.
Philippe Colson is a clinical virologist at the Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes (URMITE) and at the Méditerranée-Infection institute at the University Hospital in Marseille, France. His main research interests are giant viruses that infect phagotrophic protists, and viral hepatitis.
Jean-Marc Rolain obtained his Pharm.D. and Ph.D. in 2000 and is currently a professor of clinical microbiology at the Méditerranée-Infection institute at the University Hospital in Marseille, France. His fields of interest include antibiotic resistance (notably, the detection and characterization of new and/or emerging mechanisms of resistance at the molecular level for fastidious bacteria) and multidrug-resistant bacteria (notably, those infecting patients with cystic fibrosis).
Bernard La Scola is a clinical microbiologist. He initially trained in routine clinical microbiology and hospital-acquired infection. He is now the head of a department of 'crisis in infectious diseases' and of a department of culture-identification of fastidious bacteria. He also runs a research group on emerging pneumonia-associated microorganisms, including giant viruses.
Didier Raoult is a clinical microbiologist. He initially trained in internal medicine and infectious diseases. He is now the head of a large, 3,000-bed clinical microbiology laboratory at the University Hospital in Marseille, France, and also runs a research laboratory that studies rickettsial diseases and emerging pathogens (the WHO Collaborative Center for Rickettsial Reference and Research). He has published more than 1,500 peer-reviewed articles.
Related links
Further information.
Eurosurveillance
EuroTravNet
ProMED-mail
PowerPoint slides
PowerPoint slide for Fig. 1 (128K, ppt) PowerPoint slide for Fig. 2 (145K, ppt) PowerPoint slide for Fig. 3 (196K, ppt) PowerPoint slide for Fig. 4 (231K, ppt)
PowerPoint slide for Table 1 (122K, ppt)
Competing interests
The authors declare no competing financial interests.
33 New Medical microbiology project topics
Medical microbiology plays a crucial role in understanding and combating infectious diseases. Conducting a well-planned and meaningful project in this field is essential for advancing our knowledge and finding innovative solutions.
Choosing the right project topic is a critical first step in ensuring the success and impact of your research. In this article, we will explore various medical microbiology project topic ideas, research methodologies, project planning, data analysis, and more.
Importance of Medical Microbiology Projects
Medical microbiology projects contribute significantly to our understanding of pathogens, antimicrobial resistance, host-microbe interactions, and disease mechanisms.
By conducting research in this field, we can unravel the mysteries of infectious diseases and develop strategies for prevention, diagnosis, and treatment.
Significance of Choosing the Right Topic
Selecting an appropriate project topic is paramount to the success of your research. It should align with your interests, address a relevant research gap, and have practical implications .
The right topic will not only engage you throughout the project but also contribute valuable insights to the field of medical microbiology.
33 intriguing project topic ideas to consider
Here’s a non-exhaustive list of medical microbiology project topics for undergraduates and MSc students.
Topic 1: Investigating the Role of Gut Microbiota in Autoimmune Diseases
Autoimmune diseases have been linked to alterations in the gut microbiota. This project aims to explore the relationship between gut microbiota composition and the development, progression, and management of autoimmune diseases.
By studying microbial diversity, immune responses, and metabolomic profiles, we can gain valuable insights into potential therapeutic interventions.
Topic 2: Antibiotic Resistance Patterns of Common Pathogens
Antibiotic resistance poses a significant global health threat. This project focuses on investigating the antibiotic resistance patterns of common pathogens, such as Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa, in a local hospital setting.
By identifying the prevalent resistance mechanisms and associated risk factors, we can optimize antimicrobial therapy and develop strategies for infection control.
Topic 3: Impact of Microbial Biofilms on Chronic Wound Infections
Biofilms play a crucial role in chronic wound infections, leading to treatment challenges.
Recommended articles:
- 43 Project Topics on Food Microbiology: Latest
- 33 Microbiology Project Topics: You haven’t thought of
- Medical Treatment in Canada for Foreigners
- 5 Best Medical Loans of 2024: Top Options
- Best Banks for Medical Practice Loans: Top Lenders…
This project aims to understand the formation, composition, and antimicrobial resistance mechanisms of biofilms associated with chronic wounds.
By elucidating the intricate interactions between biofilm communities and host factors, we can develop novel approaches to manage and prevent these infections.
4. Characterization of antibiotic resistance mechanisms in multidrug-resistant bacteria.
5. Investigation of the role of biofilms in chronic infections.
6. Exploring the impact of probiotics on gut microbiota composition and health.
7. Elucidating the molecular basis of viral-host interactions in viral infections.
8. Analysis of the microbiome in patients with autoimmune diseases.
9. Assessing the effectiveness of phage therapy against antibiotic-resistant bacteria.
10. Identification and characterization of novel antimicrobial compounds from natural sources.
11. Investigating the role of the microbiome in the development of allergies and asthma.
12. Understanding the mechanisms of horizontal gene transfer in antibiotic resistance dissemination.
13. Evaluation of the efficacy of novel disinfectants in healthcare settings.
14. Molecular epidemiology of nosocomial infections and surveillance of antibiotic-resistant pathogens.
15. Investigating the impact of climate change on the spread of vector-borne diseases.
16. Studying the role of microbiota in the development and progression of colorectal cancer.
17. Analysis of the microbial diversity in dental plaque and its association with oral health.
18. Exploring the potential of bacteriophages as alternatives to antibiotics in treating bacterial infections.
19. Investigating the impact of environmental factors on the microbiota of the skin.
20. Characterization of the role of gut microbiota in metabolic disorders, such as obesity and diabetes.
21. Understanding the mechanisms of antifungal drug resistance in Candida species.
22. Evaluation of the efficacy of different sterilization techniques in medical device manufacturing.
23. Investigation of the role of microbial communities in chronic wound infections.
24. Analysis of the impact of vaccination on the prevalence and diversity of infectious diseases.
25. Identification and characterization of novel drug targets in pathogenic bacteria.
26. Study of the interaction between the microbiome and the immune system in autoimmune disorders.
27. Exploring the microbiota composition and its association with mental health disorders.
28. Investigating the role of viral infections in the development of cancer.
29. Evaluation of the efficacy of antimicrobial coatings in preventing healthcare-associated infections.
30. Study of the genetic basis of virulence in bacterial pathogens.
31. Analysis of the impact of antimicrobial stewardship programs on antibiotic resistance patterns.
32. Investigation of the microbial diversity in respiratory tract infections.
33. Understanding the role of the microbiome in inflammatory bowel diseases.
Remember to choose a topic that aligns with your interests and research goals. These topics offer a range of exciting avenues for exploration in the field of medical microbiology.
Recommended articles
- 10 benefits of studying microbiology
- Can microbiologists work in hospitals?
- Can microbiologists make vaccines?
- Can a microbiologist work in NNPC?
- How to be a good microbiologist?
- Can microbiologists become doctors?
Project Planning and Execution
Once you have chosen a project topic , it is essential to plan and execute your research effectively.
This involves setting clear objectives and goals, designing appropriate experiments and protocols, and considering ethical aspects. Collaborating with experts in the field can enhance the quality and impact of your research.
Data Interpretation and Analysis
After collecting data, it is crucial to analyze and interpret the findings accurately. Statistical analysis and data visualization techniques can help identify trends, patterns, and statistical significance.
Ensuring the validity and reliability of your results is essential for drawing meaningful conclusions.
Discussion and Conclusion
In the discussion section, interpret the significance of your research findings in the context of existing literature.
Compare and contrast your results with previous studies, highlight limitations, and propose future directions.
Conclude your project by summarizing key findings, emphasizing their implications, and discussing potential applications in medical microbiology.
How do I choose the right medical microbiology project topic?
To choose the right project topic, consider your interests, relevance to the field, and potential impact. Research current trends and gaps in medical microbiology, consult with mentors and select a topic that aligns with your goals.
What are the ethical considerations in medical microbiology research?
Ethical considerations include obtaining informed consent, ensuring patient confidentiality, following ethical guidelines and regulations, and conducting research with integrity and transparency.
How can I ensure the validity of my research findings?
To ensure validity, employ rigorous experimental design, use appropriate controls, validate methods, replicate experiments, analyze data critically, and consider potential biases and confounding factors.
Can I collaborate with other researchers on my project?
Collaboration with other researchers is encouraged as it brings diverse expertise, resources, and perspectives to your project. Collaborative efforts can enhance the quality and impact of your research.
How can I present my project findings effectively?
To present your project findings effectively, prepare clear and concise visual aids, practice your presentation, engage with your audience, and highlight the significance and implications of your research.
Conducting a medical microbiology project provides an opportunity to contribute to scientific knowledge and make a positive impact in the field of healthcare.
By selecting a compelling topic, planning your research, analyzing data diligently, and engaging in meaningful discussions, you can successfully execute a project that advances our understanding of infectious diseases and leads to improved patient outcomes.
- Is Campylobacter Contagious?
- How to Use Bone Broth for Gut Health?
- Does Campylobacter stay in your system?
- Best greens supplement for bloating and gut health
- Is a plant-based diet good for gut health?
Last Updated on June 3, 2023 by Our Editorial Team
- See us on facebook
- See us on twitter
- See us on youtube
- See us on linkedin
- See us on instagram
Study reveals brain mechanisms behind speech impairment in Parkinson’s
Most Parkinson’s disease patients struggle with speech problems. New research by Stanford Medicine scientists uncovers the brain connections that could be essential to preserving speech.
May 28, 2024 - By Nina Bai

Research by Stanford Medicine scientists may explain why some treatments for Parkinson’s — developed mainly to target motor symptoms — can improve speech impairments while other treatments make them worse. Lightspring /Shutterstock.com
Parkinson’s disease is most well-known and well-studied for its motor impairments — tremors, stiffness and slowness of movement. But less visible symptoms such as trouble with memory, attention and language, which also can profoundly impact a person’s quality of life, are less understood. A new study by Stanford Medicine researchers reveals the brain mechanisms behind one of the most prevalent, yet often overlooked, symptoms of the disease — speech impairment.
Based on brain imaging from Parkinson’s patients, the researchers identified specific connections in the brain that may determine the extent of speech difficulties.
The findings , reported May 20 in the Proceedings of the National Academy of Sciences , could help explain why some treatments for Parkinson’s — developed mainly to target motor symptoms — can improve speech impairments while other treatments make them worse.
More than a motor disorder
“Parkinson’s disease is a very common neurological disorder, but it’s mostly considered a motor disorder,” said Weidong Cai , PhD, clinical associate professor of psychiatry and behavioral sciences and the lead author of the new study. “There’s been lots of research on how treatments such as medications and deep brain stimulation can help improve motor function in patients, but there was limited understanding about how these treatments affect cognitive function and speech.”
Over 90% of people with Parkinson’s experience difficulties with speech, an intricate neurological process that requires motor and cognitive control. Patients may struggle with a weak voice, slurring, mumbling and stuttering.
“Speech is a complex process that involves multiple cognitive functions, such as receiving auditory feedback, organizing thoughts and producing the final vocal output,” Cai said.
The senior author of the study is Vinod Menon , PhD, professor of psychiatry and behavioral sciences and director of the Stanford Cognitive and Systems Neuroscience Laboratory .
The researchers set out to study how levodopa, a common Parkinson’s drug that replaces the dopamine lost from the disease, affects overall cognitive function. They focused on the subthalamic nucleus, a small, pumpkin-seed-shaped region deep within the brain.

Weidong Cai
The subthalamic nucleus is known for its role in inhibiting motor activity, but there are clues to its involvement in other functions. For example, deep brain stimulation, which uses implanted electrodes to stimulate the subthalamic nucleus, has proven to be a powerful way to relieve motor symptoms for Parkinson’s patients — but a common side effect is worsened speech impairment.
Same test, different scores
In the new study, 27 participants with Parkinson’s disease and 43 healthy controls, all older than 60, took standard tests of motor and cognitive functioning. The participants with Parkinson’s took the tests while on and off their medication.
As expected, the medication improved motor functioning in the patients, with those having the most severe symptoms improving the most.
The test for cognitive functioning offered a surprise. The test, known as the Symbol Digit Modalities Test, is given in two forms — oral and written. Patients are provided with nine symbols, each matched with a number — a plus sign for the number 7, for example. They are then asked to translate a string of symbols into numbers, either speaking or writing down their answers, depending on the version of the test.
As a group, the patients’ performance on both versions of the cognitive test was little affected by medication. But taking a closer look, the researchers noticed that the subset of patients who performed particularly poorly on the spoken version of the test without medication improved their spoken performance on the medication. Their written test scores did not change significantly.
“It was quite interesting to find this dissociation between the written and oral version of the same test,” Cai said.
The dissociation suggested that the medication was not enhancing general cognitive functions such as attention and working memory, but it was selectively improving speech.
“Our research unveiled a previously unrecognized impact of dopaminergic drugs on the speech function of Parkinson’s patients,” Menon said.
Uncovering connections
Next, the researchers analyzed fMRI brain scans of the participants, looking at how the subthalamic nucleus interacted with brain networks dedicated to various functions, including hearing, vision, language and executive control.

Vinod Menon
They found that different parts of the subthalamic nucleus interacted with different networks.
In particular, they discovered that improvements on the oral version of the test correlated with better functional connectivity between the right side of the subthalamic nucleus and the brain’s language network.
Using a statistical model, they could even predict a patient’s improvement on the oral test based on changes in their brain’s functional connectivity.
“Here we’re not talking about an anatomical connection,” Cai explained. Rather, functional connectivity between brain regions means the activity in these regions is closely coordinated, as if they are talking to each other.
“We discovered that these medications influence speech by altering the functional connectivity between the subthalamic nucleus and crucial language networks,” Menon said. “This insight opens new avenues for therapeutic interventions tailored specifically to improve speech without deteriorating other cognitive abilities.”
This newly identified interaction between the subthalamic nucleus and the language network could serve as a biological indicator of speech behavior — in Parkinson’s as well as other speech disorders like stuttering.
Such a biomarker could be used to monitor treatment outcomes and inspire new therapies. “Of course, you can directly observe the outcome of a medication by observing behavior, but I think to have a biomarker in the brain will provide more useful information for the future development of drugs,” Cai said.
The findings also provide a detailed map of the subthalamic nucleus, which could guide neurosurgeons performing deep brain stimulation in avoiding damage to an area critical to speech function. “By identifying key neural maps and connections that predict speech improvement, we can craft more effective treatment plans that are both precise and personalized for Parkinson’s disease patients,” Menon said.
The study received funding from the National Institutes of Health (grants P50 AG047366, P30 AG066515, RF1 NS086085, R21 DC017950-S1, R01 NS115114, R01 MH121069 and K99 AG071837) and the Alzheimer’s Association.

About Stanford Medicine
Stanford Medicine is an integrated academic health system comprising the Stanford School of Medicine and adult and pediatric health care delivery systems. Together, they harness the full potential of biomedicine through collaborative research, education and clinical care for patients. For more information, please visit med.stanford.edu .
Hope amid crisis
Psychiatry’s new frontiers

Suggestions or feedback?
MIT News | Massachusetts Institute of Technology
- Machine learning
- Social justice
- Black holes
- Classes and programs
Departments
- Aeronautics and Astronautics
- Brain and Cognitive Sciences
- Architecture
- Political Science
- Mechanical Engineering
Centers, Labs, & Programs
- Abdul Latif Jameel Poverty Action Lab (J-PAL)
- Picower Institute for Learning and Memory
- Lincoln Laboratory
- School of Architecture + Planning
- School of Engineering
- School of Humanities, Arts, and Social Sciences
- Sloan School of Management
- School of Science
- MIT Schwarzman College of Computing
Understanding why autism symptoms sometimes improve amid fever
Press contact :.

Previous image Next image
Scientists are catching up to what parents and other caregivers have been reporting for many years: When some people with autism spectrum disorders experience an infection that sparks a fever, their autism-related symptoms seem to improve.
With a pair of new grants from The Marcus Foundation, scientists at MIT and Harvard Medical School hope to explain how this happens in an effort to eventually develop therapies that mimic the “fever effect” to similarly improve symptoms.
“Although it isn’t actually triggered by the fever, per se, the ‘fever effect’ is real, and it provides us with an opportunity to develop therapies to mitigate symptoms of autism spectrum disorders,” says neuroscientist Gloria Choi , associate professor in the MIT Department of Brain and Cognitive Sciences and affiliate of The Picower Institute for Learning and Memory.
Choi will collaborate on the project with Jun Huh, associate professor of immunology at Harvard Medical School. Together the grants to the two institutions provide $2.1 million over three years.
“To the best of my knowledge, the ‘fever effect’ is perhaps the only natural phenomenon in which developmentally determined autism symptoms improve significantly, albeit temporarily,” Huh says. “Our goal is to learn how and why this happens at the levels of cells and molecules, to identify immunological drivers, and produce persistent effects that benefit a broad group of individuals with autism.”
The Marcus Foundation has been involved in autism work for over 30 years, helping to develop the field and addressing everything from awareness to treatment to new diagnostic devices.
“I have long been interested in novel approaches to treating and lessening autism symptoms, and doctors Choi and Huh have honed in on a bold theory,” says Bernie Marcus, founder and chair of The Marcus Foundation. “It is my hope that this Marcus Foundation Medical Research Award helps their theory come to fruition and ultimately helps improve the lives of children with autism and their families.”
Brain-immune interplay
For a decade, Huh and Choi have been investigating the connection between infection and autism. Their studies suggest that the beneficial effects associated with fever may arise from molecular changes in the immune system during infection, rather than on the elevation of body temperature, per se.
Their work in mice has shown that maternal infection during pregnancy, modulated by the composition of the mother’s microbiome, can lead to neurodevelopmental abnormalities in the offspring that result in autism-like symptoms, such as impaired sociability. Huh’s and Choi’s labs have traced the effect to elevated maternal levels of a type of immune-signaling molecule called IL-17a, which acts on receptors in brain cells of the developing fetus, leading to hyperactivity in a region of the brain’s cortex called S1DZ. In another study , they’ve shown how maternal infection appears to prime offspring to produce more IL-17a during infection later in life.
Building on these studies, a 2020 paper clarified the fever effect in the setting of autism. This research showed that mice that developed autism symptoms as a result of maternal infection while in utero would exhibit improvements in their sociability when they had infections — a finding that mirrored observations in people. The scientists discovered that this effect depended on over-expression of IL-17a, which in this context appeared to calm affected brain circuits. When the scientists administered IL-17a directly to the brains of mice with autism-like symptoms whose mothers had not been infected during pregnancy, the treatment still produced improvements in symptoms.
New studies and samples
This work suggested that mimicking the “fever effect” by giving extra IL-17a could produce similar therapeutic effects for multiple autism-spectrum disorders, with different underlying causes. But the research also left wide-open questions that must be answered before any clinically viable therapy could be developed. How exactly does IL-17a lead to symptom relief and behavior change in the mice? Does the fever effect work in the same way in people?
In the new project, Choi and Huh hope to answer those questions in detail.
“By learning the science behind the fever effect and knowing the mechanism behind the improvement in symptoms, we can have enough knowledge to be able to mimic it, even in individuals who don’t naturally experience the fever effect,” Choi says.
Choi and Huh will continue their work in mice seeking to uncover the sequence of molecular, cellular and neural circuit effects triggered by IL-17a and similar molecules that lead to improved sociability and reduction in repetitive behaviors. They will also dig deeper into why immune cells in mice exposed to maternal infection become primed to produce IL-17a.
To study the fever effect in people, Choi and Huh plan to establish a “biobank” of samples from volunteers with autism who do or don’t experience symptoms associated with fever, as well as comparable volunteers without autism. The scientists will measure, catalog, and compare these immune system molecules and cellular responses in blood plasma and stool to determine the biological and clinical markers of the fever effect.
If the research reveals distinct cellular and molecular features of the immune response among people who experience improvements with fever, the researchers could be able to harness these insights into a therapy that mimics the benefits of fever without inducing actual fever. Detailing how the immune response acts in the brain would inform how the therapy should be crafted to produce similar effects.
"We are enormously grateful and excited to have this opportunity," Huh says. "We hope our work will ‘kick up some dust’ and make the first step toward discovering the underlying causes of fever responses. Perhaps, one day in the future, novel therapies inspired by our work will help transform the lives of many families and their children with ASD [autism spectrum disorder]."
Share this news article on:
Related links.
- Gloria Choi
- The Picower Institute for Learning and Memory
- Department of Brain and Cognitive Sciences
Related Topics
- Brain and cognitive sciences
- Picower Institute
- Neuroscience
- Psychiatric disorders
Related Articles

Building a bridge between neuroscience and immunology
Research finds potential mechanism linking autism, intestinal inflammation

Study may explain how infections reduce autism symptoms
Studies help explain link between autism, severe infection during pregnancy
Previous item Next item
More MIT News

MIT Corporation elects 10 term members, two life members
Read full story →

Diane Hoskins ’79: How going off-track can lead new SA+P graduates to become integrators of ideas

Chancellor Melissa Nobles’ address to MIT’s undergraduate Class of 2024

Noubar Afeyan PhD ’87 gives new MIT graduates a special assignment

Commencement address by Noubar Afeyan PhD ’87

President Sally Kornbluth’s charge to the Class of 2024
- More news on MIT News homepage →
Massachusetts Institute of Technology 77 Massachusetts Avenue, Cambridge, MA, USA
- Map (opens in new window)
- Events (opens in new window)
- People (opens in new window)
- Careers (opens in new window)
- Accessibility
- Social Media Hub
- MIT on Facebook
- MIT on YouTube
- MIT on Instagram


News and Events
News archives, faculty media experts, social media directory, publications, style guidelines, public relations team, requests for public relations services, tara jarboe, m.s. ‘19, ph.d. ‘24, shares her plans beyond commencement and her commitment to cancer research, jarboe addressed the gsbms class of 2024 and is ready for her career in cancer research.

Tara Jarboe, M.S. ‘19, Ph.D. ‘24, addressed her fellow classmates at the 165th Commencement Ceremony for the Graduate School of Biomedical Sciences on May 23. From pliés to publications, Dr. Jarboe’s dedication to cancer research exemplifies her commitment to advancing scientific knowledge.
What inspired you to pursue your degree? I always was really interested in science when I was a child. I enjoyed conducting experiments at home and knew I wanted to do that for a career. When I was a freshman in high school, my mom's cousin was diagnosed with pancreatic cancer. Watching her go through treatment and seeing how fast the disease progressed before her passing made me realize that I wanted to pursue cancer research. I also had even more family members afterwards who were diagnosed with different types of cancer and it reinforced the reasoning for my path in cancer research. What type of research have you been involved in during your time at NYMC?
Most of my research at NYMC has been my dissertation, which is looking at how the natural compound berberine can remodel fatal anaplastic thyroid cancer properties to make it more amenable to therapy. I’ve also explored different types of research within the topic of berberine and anaplastic thyroid cancer. My dissertation was a collaborative project with my principal investigator Raj K. Tiwari, Ph.D., professor of pathology, microbiology, and immunology and associate professor of otolaryngology; and Xiu-Min Li, M.D., M.S., professor of pathology, microbiology, and immunology, who brought me into the world of natural products. I was also involved in research with Jan Geliebter, Ph.D., professor ofpathology, microbiology, and immunology and associate professor of otolaryngology; and Mark Hurwitz, M.D., professor and chair of the Department of Radiation Medicine, exploring hyperthermia for melanoma treatment, and research with Dr. Li regarding allergy and asthma. What has helped motivate you along your educational journey? Have you encountered any challenges along the way?
Research can get a little frustrating when things don't work the first time around, which leads you to get creative when dealing with challenges. The members of my lab were supportive and brainstormed with me, which helped me through these past few years. I’m thankful for them. My family also was a huge support system for me as well. They checked in on me and brought positivity when I was stresssed. The COVID-19 pandemic was obviously a pretty big hurdle during the completion of my Ph.D., because we were unable to go into the lab for months. When all your work primarily relies on active research inside of a lab, that becomes really problematic, but Dr. Tiwari ensured we were still able to be productive during this time out of the lab through journal clubs, grant writing, and studying for qualifying exams. After you graduate, what is your dream career?
I certainly want to stay in academia. I really love teaching. I am an adjunct professor at Manhattanville College. I would also love to have my own active research lab as well one day. What made you choose NYMC?
I knew I wanted to work at a cancer lab. When I did research on labs in the field, I came across NYMC. I loved the work that the faculty were doing. The more I read, the more I felt like the College was the right fit – and it was. I pursued my master’s and Ph.D. in microbiology and immunology. The faculty and students have been warm and inviting ever since I stepped foot here seven years ago. What has been your favorite aspect of being an NYMC student?
I love how collaborative everything is. We work with so many people in our lab who are amazing and inspiring. My mentors are wonderful. Working Dr. Tiwari alone would have certainly been enough. All the people in the labs that I work with are so wonderful. We've developed a friendship during that time, which is cool. The people I’ve met here will certainly remain in my life as friends. What faculty member has had the greatest influence on you here?
Dr. Tiwari is an incredible mentor. He's everything that a mentor should be for a student. He gives such good scientific guidance, but he also gives you the tools you need, where you can be independent at the same time. I've had a lot of family illnesses that I've dealt with and he was understanding and really encouraging and positive. When my mom received a breast cancer diagnosis, he sent her a care package the day that he found out and made sure that if I needed to take her to an appointment that it was fine with him. She’s in the clear now, but I really appreciate that. Dr. Geliebter and Dr. Li have had a great influence me as well. They were both on my committee and advisors on my project. Dr. Geliebter is one of the funniest and passionate people I’ve met. Julie S. Di Martino, Ph.D., assistant professor of cell biology and anatomy, who was on my committee as well, and Tetyana Cheairs, M.D., M.S.P.H., assistant dean of the GSBMS and assistant professor of pathology, immunology and microbiology, gave me a lot of guidance. They’re great role models as women in STEM. What advice would you give applicants or incoming students? Make sure that you find the mentor that's the right fit for you. Mine was so perfect, and it really shaped my experience, but everyone's looking for something different in a mentor and all the different labs here run a little bit differently. We do a ton of research here and we have to be really passionate about it and really love what we're doing. Find the research field that really drives you to keep pushing through and tests your brain.
Outside of your studies, what are your hobbies or interests?
I love to dance ballet. I am a professional ballerina. I also really love reading. I'm an avid audiobook listener. I enjoy putting my headphones in, listening to a book, while pipetting something 7,000 times in a row. I also like going to the beach and hiking. Are you a part of any student organizations or interest groups?
I was one of the founders of the Ph.D. Representatives. The Graduate Student Association is amazing, but we wanted to have a smaller group of Ph.D. students who can connect, since we are in the minority in the GSBMS. What is a fun fact about you?
Being a ballerina is a well-known fact about me, but a less known fact is that I'm pregnant. I’m having a boy this September.

NOTICE OF NONDISCRIMINATORY POLICY AS TO STUDENTS The New York Medical College admits students of any race, color, national and ethnic origin to all the rights, privileges, programs, and activities generally accorded or made available to students at the college. It does not discriminate on the basis of race, color, national and ethnic origin in administration of its educational policies, admissions policies, scholarship and loan programs, and athletic and other school-administered programs. See full non-discrimination statement with contact info .

Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
Collection 10 March 2022
Top 100 in Microbiology
This collection highlights our most downloaded* microbiology papers published in 2021. Featuring authors from around the world, these papers showcase valuable research from an international community.
*Data obtained from SN Inights, which is based on Digital Science's Dimensions.

The in-vitro effect of famotidine on SARS-CoV-2 proteases and virus replication
- Madeline Loffredo
- Hector Lucero
- Ali H. Munawar
In vitro efficacy of artemisinin-based treatments against SARS-CoV-2
- Yuyong Zhou
- Kerry Gilmore
- Peter H. Seeberger
Identification of CRF89_BF, a new member of an HIV-1 circulating BF intersubtype recombinant form family widely spread in South America
- Elena Delgado
- Aurora Fernández-García
- Michael M. Thomson
The effectiveness of various gargle formulations and salt water against SARS-CoV-2
- Vunjia Tiong
- Pouya Hassandarvish
- Ilina Isahak
The serotonin reuptake inhibitor Fluoxetine inhibits SARS-CoV-2 in human lung tissue
- Melissa Zimniak
- Luisa Kirschner
- Jochen Bodem
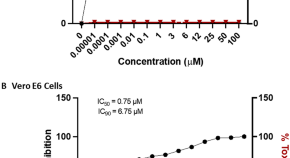
Probenecid inhibits SARS-CoV-2 replication in vivo and in vitro
- Jackelyn Murray
- Robert J. Hogan
- Ralph A. Tripp
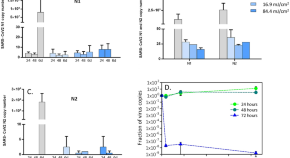
UV-C irradiation is highly effective in inactivating SARS-CoV-2 replication
- Mara Biasin
- Andrea Bianco
- Mario Clerici
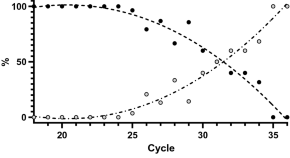
Diagnostic performance of rapid antigen tests (RATs) for SARS-CoV-2 and their efficacy in monitoring the infectiousness of COVID-19 patients
- John G. Routsias
- Maria Mavrouli
- Athanasios Tsakris
Epidemiology and outcomes of COVID-19 in HIV-infected individuals: a systematic review and meta-analysis
- Paddy Ssentongo
- Emily S. Heilbrunn
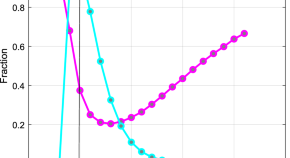
Temporal dynamics of viral load and false negative rate influence the levels of testing necessary to combat COVID-19 spread
- Katherine F. Jarvis
- Joshua B. Kelley
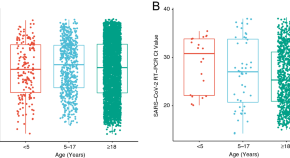
Nasopharyngeal SARS-CoV-2 viral loads in young children do not differ significantly from those in older children and adults
- Sharline Madera
- Emily Crawford
- Joseph L. DeRisi
Morphometry of SARS-CoV and SARS-CoV-2 particles in ultrathin plastic sections of infected Vero cell cultures
- Michael Laue
- Anne Kauter
- Andreas Nitsche
Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry
- Dhiraj Mannar
- Karoline Leopold
- Sriram Subramaniam
The Spanish gut microbiome reveals links between microorganisms and Mediterranean diet
- Adriel Latorre-Pérez
- Marta Hernández
- Luis Collado
In vitro inactivation of SARS-CoV-2 by commonly used disinfection products and methods

Methylene Blue has a potent antiviral activity against SARS-CoV-2 and H1N1 influenza virus in the absence of UV-activation in vitro
- Valeria Cagno
- Chiara Medaglia
- Caroline Tapparel
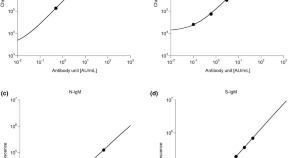
A novel highly quantitative and reproducible assay for the detection of anti-SARS-CoV-2 IgG and IgM antibodies
- Kouki Matsuda
- Akinobu Hamada

Colorimetric RT-LAMP SARS-CoV-2 diagnostic sensitivity relies on color interpretation and viral load
- Mateus Nóbrega Aoki
- Bruna de Oliveira Coelho
- Lucas Blanes
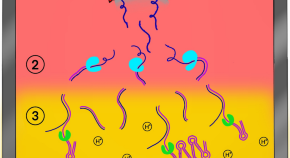
Field-deployable, rapid diagnostic testing of saliva for SARS-CoV-2
- Hemant Suryawanshi
- Zev Williams

No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor
- Jarrod Shilts
- Thomas W. M. Crozier
- Gavin J. Wright
Carvacrol exhibits rapid bactericidal activity against Streptococcus pyogenes through cell membrane damage
- Niluni M. Wijesundara
- Song F. Lee
- H. P. Vasantha Rupasinghe
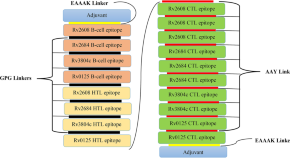
In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology
- Shaheen Bibi
- Inayat Ullah
- Shiquan Niu
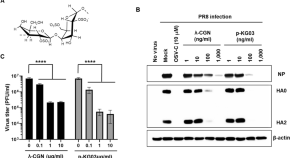
Antiviral activity of lambda-carrageenan against influenza viruses and severe acute respiratory syndrome coronavirus 2
- Heegwon Shin
- Meehyein Kim
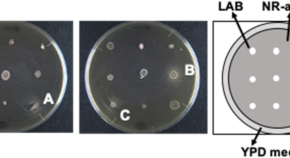
Nicotinamide mononucleotide production by fructophilic lactic acid bacteria
- Kazane Sugiyama
- Kana Iijima
- Nobuyuki Yoshida
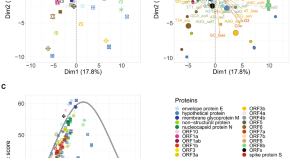
Molecular features similarities between SARS-CoV-2, SARS, MERS and key human genes could favour the viral infections and trigger collateral effects
- Lucas L. Maldonado
- Andrea Mendoza Bertelli
- Laura Kamenetzky
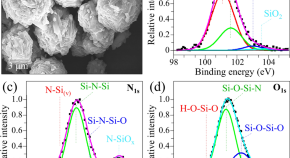
Silicon nitride: a potent solid-state bioceramic inactivator of ssRNA viruses
- Giuseppe Pezzotti
- Francesco Boschetto
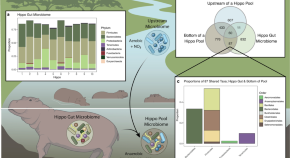
The meta-gut: community coalescence of animal gut and environmental microbiomes
- Christopher L. Dutton
- Amanda L. Subalusky
- David M. Post

Ingestion of probiotic ( Lactobacillus helveticus and Bifidobacterium longum ) alters intestinal microbial structure and behavioral expression following social defeat stress
- Katherine A. Partrick
- Anna M. Rosenhauer
- Kim L. Huhman
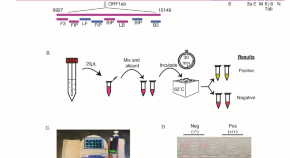
Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction
- Esther Kohl
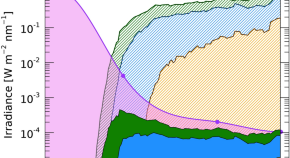
Solar UV-B/A radiation is highly effective in inactivating SARS-CoV-2
- Fabrizio Nicastro
- Giorgia Sironi
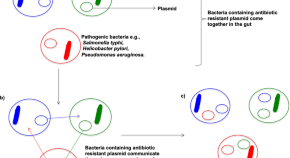
High prevalence of antibiotic resistance in commensal Escherichia coli from healthy human sources in community settings
- Emmanuel Nji
- Joseph Kazibwe
- La Thi Quynh Lien
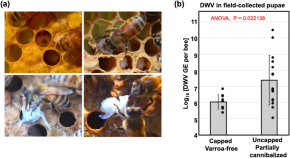
Pupal cannibalism by worker honey bees contributes to the spread of deformed wing virus
- Francisco Posada-Florez
- Zachary S. Lamas
- Eugene V. Ryabov

Network analysis of Down syndrome and SARS-CoV-2 identifies risk and protective factors for COVID-19
- Ilario De Toma
- Mara Dierssen
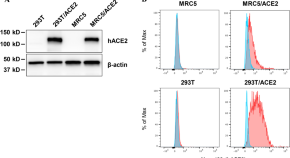
MRC5 cells engineered to express ACE2 serve as a model system for the discovery of antivirals targeting SARS-CoV-2
- Kentaro Uemura
- Michihito Sasaki
- Akihiko Sato

A descriptive analysis of antimicrobial resistance patterns of WHO priority pathogens isolated in children from a tertiary care hospital in India
- Vijayalaxmi V. Mogasale
- Prakash Saldanha
- Vittal Mogasale
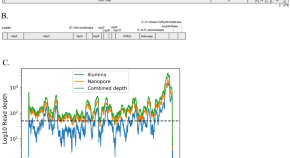
Metagenomic identification of a new sarbecovirus from horseshoe bats in Europe
- Jack M. Crook
- Ivana Murphy
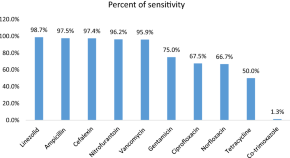
Comprehensive study of antimicrobial susceptibility pattern and extended spectrum beta-lactamase (ESBL) prevalence in bacteria isolated from urine samples
- Mohammad Javad Gharavi
- Javad Zarei
- Niloufar Rashidi
A PCR amplicon-based SARS-CoV-2 replicon for antiviral evaluation
- Tomohiro Kotaki
- Masanori Kameoka
An inter-laboratory study to investigate the impact of the bioinformatics component on microbiome analysis using mock communities
- Denise M. O’Sullivan
- Ronan M. Doyle
- Jim F. Huggett

Microplastics accumulate fungal pathogens in terrestrial ecosystems
- Gerasimos Gkoutselis
- Stephan Rohrbach
- Gerhard Rambold
The lower respiratory tract microbiome of critically ill patients with COVID-19
- Paolo Gaibani
- Elisa Viciani
- Simone Ambretti
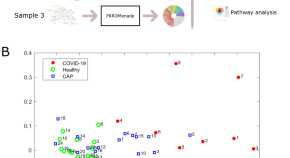
Functional profiling of COVID-19 respiratory tract microbiomes
- Niina Haiminen
- Filippo Utro
- Laxmi Parida
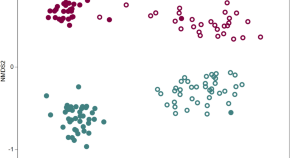
The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions
- Cristina Lazcano
- Kelly Ivors
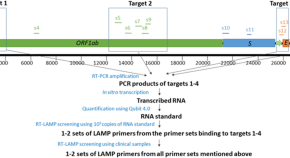
Comparative evaluation of 19 reverse transcription loop-mediated isothermal amplification assays for detection of SARS-CoV-2
- Yajuan Dong
- Chiyu Zhang
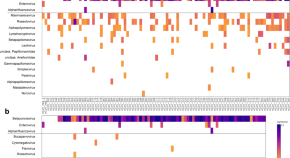
Respiratory viral co-infections among SARS-CoV-2 cases confirmed by virome capture sequencing
- Ki Wook Kim
- Ira W. Deveson
- William D. Rawlinson
UVC disinfects SARS-CoV-2 by induction of viral genome damage without apparent effects on viral morphology and proteins
- Chieh-Wen Lo
- Ryosuke Matsuura
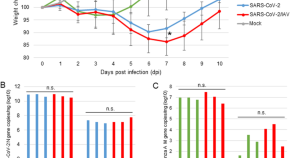
Co-infection of SARS-CoV-2 and influenza virus causes more severe and prolonged pneumonia in hamsters
- Takaaki Kinoshita
- Kenichi Watanabe
- Jiro Yasuda
Identification of ebselen and its analogues as potent covalent inhibitors of papain-like protease from SARS-CoV-2
- Ewelina Weglarz-Tomczak
- Jakub M. Tomczak
- Stanley Brul
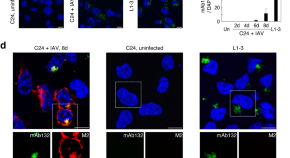
Neurotropic influenza A virus infection causes prion protein misfolding into infectious prions in neuroblastoma cells
- Hideyuki Hara
- Junji Chida
- Suehiro Sakaguchi
The Edible Plant Microbiome represents a diverse genetic reservoir with functional potential in the human host
- Maria J. Soto-Giron
- Gerardo Toledo
Blockade of SARS-CoV-2 spike protein-mediated cell–cell fusion using COVID-19 convalescent plasma
Dynamics of SARS-CoV-2 mutations reveals regional-specificity and similar trends of N501 and high-frequency mutation N501Y in different levels of control measures
- Santiago Justo Arevalo
- Daniela Zapata Sifuentes
- Roberto Pineda Chavarría
Lactobacillus reuteri AN417 cell-free culture supernatant as a novel antibacterial agent targeting oral pathogenic bacteria
- Kyung Mi Yang
- Kwang-Hak Bae
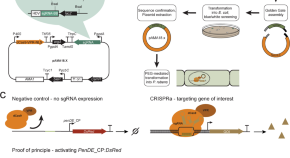
CRISPR-based transcriptional activation tool for silent genes in filamentous fungi
- László Mózsik
- Mirthe Hoekzema
- Arnold J. M. Driessen
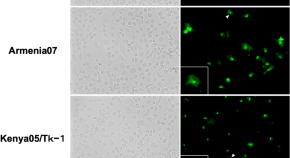
An immortalized porcine macrophage cell line competent for the isolation of African swine fever virus
- Kentaro Masujin
- Tomoya Kitamura
- Takehiro Kokuho
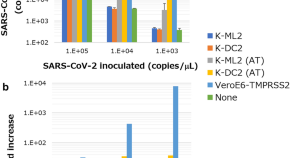
The potential of COVID-19 patients’ sera to cause antibody-dependent enhancement of infection and IL-6 production
- Jun Shimizu
- Tadahiro Sasaki
- Tatsuo Shioda
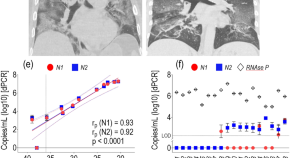
Digital PCR for high sensitivity viral detection in false-negative SARS-CoV-2 patients
- Paolo Poggio
- Paola Songia
- Maurizio Pesce
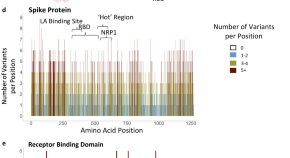
Natural variants in SARS-CoV-2 Spike protein pinpoint structural and functional hotspots with implications for prophylaxis and therapeutic strategies
- Suman Pokhrel
- Benjamin R. Kraemer
- Daria Mochly-Rosen
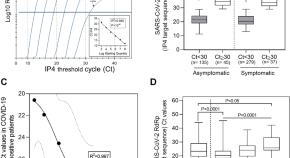
Asymptomatic COVID-19 Adult Outpatients identified as Significant Viable SARS-CoV-2 Shedders
- Marie Glenet
- Anne-Laure Lebreil
- Laurent Andreoletti
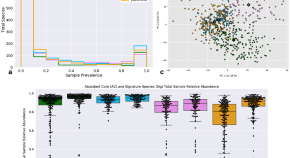
Bacterial associations in the healthy human gut microbiome across populations
- Mark Loftus
- Sayf Al-Deen Hassouneh
- Shibu Yooseph
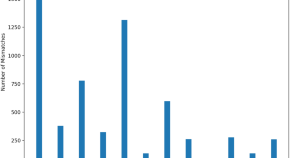
Analysis and forecasting of global real time RT-PCR primers and probes for SARS-CoV-2
- Gowri Nayar
- Edward E. Seabolt
- James H. Kaufman
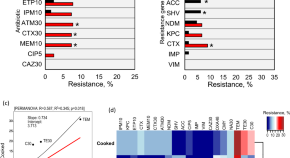
Antibiotic-resistant bacteria and gut microbiome communities associated with wild-caught shrimp from the United States versus imported farm-raised retail shrimp
- Laxmi Sharma
- Ravinder Nagpal
- Prashant Singh
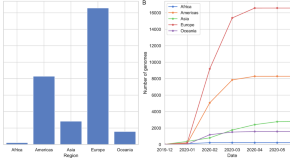
Emergence of novel SARS-CoV-2 variants in the Netherlands
- Aysun Urhan
- Thomas Abeel
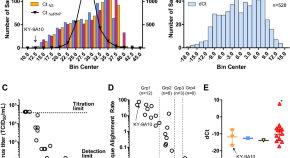
Induction of interferon response by high viral loads at early stage infection may protect against severe outcomes in COVID-19 patients
- Eric C. Rouchka
- Julia H. Chariker
- Donghoon Chung
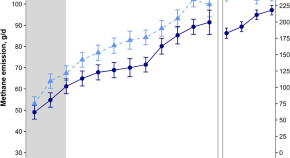
Early life dietary intervention in dairy calves results in a long-term reduction in methane emissions
- S. J. Meale
- D. P. Morgavi
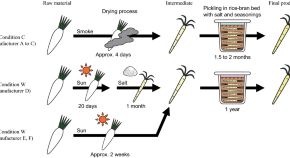
The relationships between microbiota and the amino acids and organic acids in commercial vegetable pickle fermented in rice-bran beds
- Kazunori Sawada
- Hitoshi Koyano
- Takuji Yamada
Response of bacterial and fungal communities to high petroleum pollution in different soils
- Polina Galitskaya
- Liliya Biktasheva
- Svetlana Selivanovskaya
A streamlined clinical metagenomic sequencing protocol for rapid pathogen identification
- Xiaofang Jia
- Xiaonan Zhang
Sensitive detection and quantification of SARS-CoV-2 in saliva
- Mustafa Fatih Abasiyanik
- Blake Flood
- Evgeny Izumchenko
Whole genome sequencing of clinical samples reveals extensively drug resistant tuberculosis (XDR TB) strains from the Beijing lineage in Nigeria, West Africa
- Idowu B. Olawoye
- Jessica N. Uwanibe
- Christian T. Happi

The SARS-CoV-2 viral load in COVID-19 patients is lower on face mask filters than on nasopharyngeal swabs
- Agnieszka Smolinska
- David S. Jessop
- Marc P. van der Schee
Metagenomics analysis of bacteriophages and antimicrobial resistance from global urban sewage
- Josephine E. S. Strange
- Pimlapas Leekitcharoenphon
- Frank M. Aarestrup
Limited intestinal inflammation despite diarrhea, fecal viral RNA and SARS-CoV-2-specific IgA in patients with acute COVID-19
- Graham J. Britton
- Alice Chen-Liaw
- Jeremiah J. Faith
Genomic evolution of antimicrobial resistance in Escherichia coli
- Markus Hans Kristofer Johansson

Outer membrane vesicles containing OmpA induce mitochondrial fragmentation to promote pathogenesis of Acinetobacter baumannii
- Varnesh Tiku
- Eric M. Kofoed
- Man-Wah Tan

Meta-analysis of host transcriptional responses to SARS-CoV-2 infection reveals their manifestation in human tumors
- Fengju Chen
- Yiqun Zhang
- Chad J. Creighton
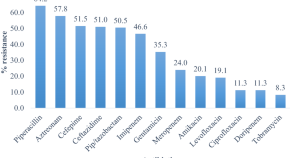
Detection of extended spectrum beta-lactamase genes in Pseudomonas aeruginosa isolated from patients in rural Eastern Cape Province, South Africa
- Mojisola C. Hosu
- Sandeep D. Vasaikar
- Teke Apalata
Production of bacterial cellulose using Gluconacetobacter kombuchae immobilized on Luffa aegyptiaca support
- Sameeha Syed Abdul Rahman
- T. Vaishnavi
- Sugumaran Karuppiah

An efficient direct screening system for microorganisms that activate plant immune responses based on plant–microbe interactions using cultured plant cells
- Mari Kurokawa
- Masataka Nakano
- Toshiki Furuya
Glucose confers protection to Escherichia coli against contact killing by Vibrio cholerae
- Cristian V. Crisan
- Holly L. Nichols
- Brian K. Hammer
Neutral evolution test of the spike protein of SARS-CoV-2 and its implications in the binding to ACE2
- Georgina I. López-Cortés
- Miryam Palacios-Pérez
- Marco V. José
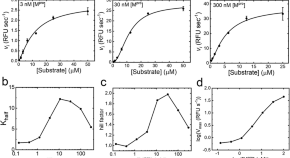
Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19
- Martin A. Redhead
- C. David Owen
- Martin A. Walsh
Assessing the efficacy of eDNA metabarcoding for measuring microbial biodiversity within forest ecosystems
- Zachary S. Ladin
- Barbra Ferrell
- K. Eric Wommack
Comparative analysis of the diagnostic performance of five commercial COVID-19 qRT PCR kits used in India
- A. K. Yadav
Dissecting industrial fermentations of fine flavour cocoa through metagenomic analysis
- Miguel Fernández-Niño
- María Juliana Rodríguez-Cubillos
- Andrés Fernando González Barrios
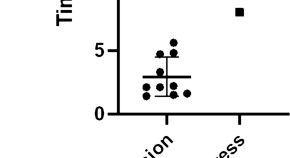
Isolation of viable Babesia bovis merozoites to study parasite invasion
- Hassan Hakimi
- Masahito Asada
- Shinichiro Kawazu
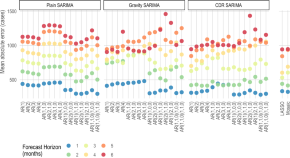
Incorporating human mobility data improves forecasts of Dengue fever in Thailand
- Mathew V. Kiang
- Mauricio Santillana
- Caroline O. Buckee

A rapid near-patient detection system for SARS-CoV-2 using saliva
- Noah B. Toppings
- Abu Naser Mohon
- Dylan R. Pillai
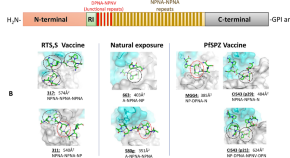
In vitro and in vivo inhibition of malaria parasite infection by monoclonal antibodies against Plasmodium falciparum circumsporozoite protein (CSP)
- Merricka C. Livingstone
- Alexis A. Bitzer
- Sheetij Dutta
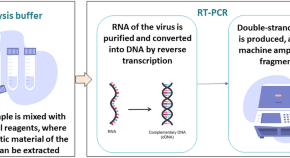
Covid-19 diagnosis by combining RT-PCR and pseudo-convolutional machines to characterize virus sequences
- Juliana Carneiro Gomes
- Aras Ismael Masood
- Wellington Pinheiro dos Santos
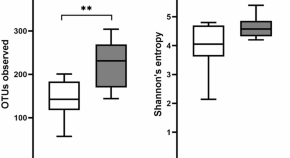
Dysbiosis of gut microbiota in Polish patients with ulcerative colitis: a pilot study
- Oliwia Zakerska-Banaszak
- Hanna Tomczak
- Marzena Skrzypczak-Zielinska
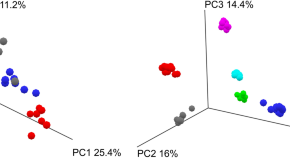
Fecal sample collection methods and time of day impact microbiome composition and short chain fatty acid concentrations
- Jacquelyn Jones
- Stacey N Reinke
- Claus T. Christophersen

An ultra-portable, self-contained point-of-care nucleic acid amplification test for diagnosis of active COVID-19 infection
- Asanka Jayawardena
- Patrick Kwan
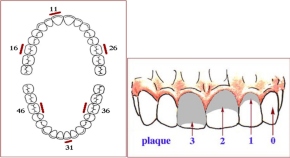
Oral hygiene and oral microbiota in children and young people with neurological impairment and oropharyngeal dysphagia
- Luiz Fernando Fregatto
- Isabela Bazzo Costa
- Paula Cristina Cola
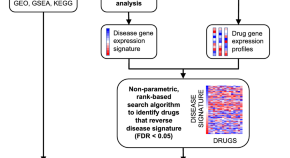
Transcriptomics-based drug repositioning pipeline identifies therapeutic candidates for COVID-19
- Brian L. Le
- Gaia Andreoletti
- Marina Sirota
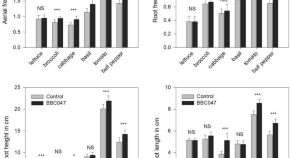
Importance of crop phenological stages for the efficient use of PGPR inoculants
- Alexandra Stoll
- Ricardo Salvatierra-Martínez
- Jaime Bravo
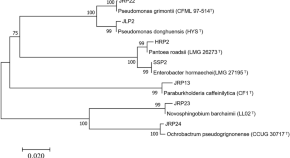
Isolation and screening of multifunctional phosphate solubilizing bacteria and its growth-promoting effect on Chinese fir seedlings
- Guangyu Zhao
- Ruzhen Jiao
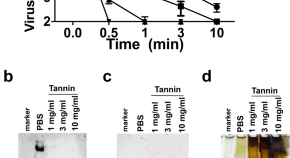
Persimmon-derived tannin has antiviral effects and reduces the severity of infection and transmission of SARS-CoV-2 in a Syrian hamster model
- Ryutaro Furukawa
- Masahiro Kitabatake
- Toshihiro Ito
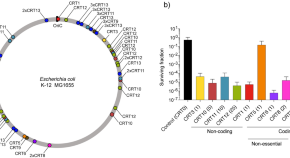
Bacterial resistance to CRISPR-Cas antimicrobials
- Ruben V. Uribe
- Christin Rathmer
- Morten Otto Alexander Sommer
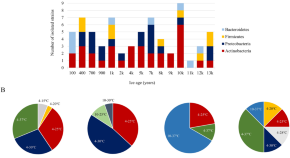
First report on antibiotic resistance and antimicrobial activity of bacterial isolates from 13,000-year old cave ice core
- Victoria I. Paun
- Paris Lavin
- Cristina Purcarea
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
share this!
May 28, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
New research shows soil microorganisms could produce additional greenhouse gas emissions from thawing permafrost
by Colorado State University

As the planet has warmed, scientists have long been concerned about the potential for harmful greenhouse gases to seep out of thawing Arctic permafrost. Recent estimates suggest that by 2100 the amount of carbon dioxide and methane released from these perpetually frozen lands could be on par with emissions from large industrial countries. However, new research led by a team of Colorado State University microbiome scientists suggests those estimates might be too low.
Microorganisms are responsible for the process that will generate greenhouse gases from thawing northern peatlands, which contain about 50% of the world's soil carbon . For now, many of the microbes in this environment are frozen and inactive.
But as the land thaws, the microbes will "wake up" and begin to churn through carbon in the ground. This natural process, known as microbial respiration, is what produces the carbon dioxide and methane emissions forecasted by climate modelers.
Currently, these models assume that this community of microorganisms—known as a microbiome—will break down some types of carbon but not others. But the CSU-led work published this week in the journal Nature Microbiology provides new insight into how these microbes will behave once activated. The research demonstrates that the soil microbes embedded in the permafrost will go after a class of compounds previously thought to be untouchable under certain conditions: polyphenols.
"There were these pools of carbon—say, donuts, pizza and chips—and we were comfortable with the idea that microbes were going to use this stuff," said Bridget McGivern, a CSU postdoctoral researcher and the paper's first author.
"But then there was this other stuff, spicy food; we didn't think the organisms liked spicy food. But what our work is showing is that actually there are organisms that are eating it, and so it's not going to just stay as carbon, it's going to be broken down."
More carbon being broken down by microbial respiration will produce additional greenhouse gas emissions. But this new finding has other implications, too. Some scientists had previously theorized that adding polyphenols to the thawing Arctic permafrost could potentially "turn off" these microorganisms altogether, effectively trapping a massive cache of potentially problematic carbon in the ground. The concept is known as the enzyme latch theory.
That no longer appears to be a viable option, said Kelly Wrighton, associate professor in the College of Agricultural Sciences' Department of Soil and Crop Sciences, whose lab led the work.
"Not only did we think these microbes didn't eat polyphenols," Wrighton said, "we thought that if the polyphenols were there it was like they were toxic and would lock the microbes into inactivity."
The soil microbiome has often been considered something of a black box due to its complexity. Wrighton hopes this new information about the role of polyphenols in permafrost helps shift that perception.
"I'd like to move past these black box assumptions," she said. "We can't engineer solutions if we don't understand the underlying wiring and plumbing of a system."
Probing the permafrost in Sweden
Unlocking the relationship between soil microbes and polyphenols has been years in the making for McGivern, who began examining this topic while working on her doctoral degree in Wrighton's lab in 2017.
McGivern started with a simple question. Scientists presumed that without oxygen, soil microbes could not break down polyphenols. Gut microbes, however, don't need oxygen to churn up the compound—that's how humans extract healthy antioxidant benefits from polyphenol -rich substances such as chocolate and red wine.
McGivern wondered why the process would be different in soils, a question that is particularly relevant to permafrost or waterlogged lands that contain little or no oxygen.
"The motivation for a lot of my Ph.D. was how could these two things exist?" McGivern said. "Organisms in our gut can breakdown polyphenols but organisms in the soil can't? The reality was that nobody in soils had really ever looked at it."
McGivern and Wrighton successfully tested the theory in a lab experiment and published a proof of concept study in 2021. The next step was testing it in the field. The team gained access to core samples from a research site in northern Sweden, a place that scientists have used for years to examine questions related to permafrost and the soil microbiome.
But before McGivern could look for evidence of polyphenol degradation in the core samples, she first had to create a database of gene sequences that corresponded to polyphenol metabolism. McGivern mined thousands of pages of existing scientific literature , cataloging the enzymes in cattle, the human gut, and some soils that were known to be responsible for the process.
Once she built the database, McGivern compared the results to the gene sequences expressed by the microbes in the core samples . Sure enough, she said, polyphenol metabolism was happening.
"What we found was that genes across 58 different polyphenol pathways were expressed," McGivern said. "So, we're saying not only can the microorganisms potentially do it, but they actually are, in the field, expressing the genes for this metabolism."
Still, more work is needed, McGivern said. They don't know what might constrain the process or the rates at which the metabolism is happening—both important factors for eventually quantifying the amount of additional greenhouse gas emissions that could be released from permafrost.
"The whole point of this is to build a better predictive understanding so that we have a framework we can actually manipulate," Wrighton said. "The climate crisis we're facing is so fast. But can we model it? Can we predict it? The only way we're going to get there is to actually understand how something works."
Journal information: Nature Microbiology
Provided by Colorado State University
Explore further
Feedback to editors

Saturday Citations: The sound of music, sneaky birds, better training for LLMs. Plus: Diversity improves research
3 hours ago

Study investigates a massive 'spider' pulsar
5 hours ago

Greener, more effective termite control: Natural compound attracts wood eaters
7 hours ago

Shear genius: Researchers find way to scale up wonder material, which could do wonders for the Earth

New vestiges of the first life on Earth discovered in Saudi Arabia
21 hours ago

Mussels downstream of wastewater treatment plant contain radium, study reports
22 hours ago

A new way to see viruses in action: Super-resolution microscopy provides a nano-scale look

Martian meteorites deliver a trove of information on red planet's structure
23 hours ago

New imager acquires amplitude and phase information without digital processing
May 31, 2024

AI helps scientists understand cosmic explosions
Relevant physicsforums posts, commercial scintillator solutions and clinical deployments.
34 minutes ago
A DNA Animation
May 29, 2024
Probability, genetic disorder related
Looking for today's dna knowledge.
May 27, 2024
Covid Vaccines Reducing Infections
Human sperm, egg cells mass-generated using ips.
More from Biology and Medical
Related Stories

Soil microbes metabolize the same polyphenols found in chocolate, wine
Jun 9, 2021

Researchers first to sample permafrost CO2 emissions during fall and winter
Mar 25, 2022

Plant roots increase carbon emission from permafrost soils
Jul 20, 2020

In peatland soil, a warmer climate and elevated carbon dioxide rapidly alter soil organic matter
Mar 6, 2024

Source or sink? A review of permafrost's role in the carbon cycle
Mar 4, 2024
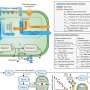
Improving climate predictions by unlocking the secrets of soil microbes
Feb 5, 2024
Recommended for you

Researchers discover 'Trojan Horse' virus hiding in human parasite

The world famous Roman Baths could help scientists counter the challenge of antibiotic resistance

Scientists map biodiversity changes in the world's forests
Stem cell study sheds new light on how the human embryo forms
Scientists discover virus-like nanoparticles control the multicellular organization and reproduction of host bacteria
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter

IMAGES
VIDEO
COMMENTS
Doxycycline post-exposure prophylaxis to prevent STIs is a novel promising intervention, which in a new study caused an ∼65% reduction in incident STIs. However, long-term effects on STI ...
Microbiology is the study of microscopic organisms, such as bacteria, viruses, archaea, fungi and protozoa. This discipline includes fundamental research on the biochemistry, physiology, cell ...
Six Key Topics in Microbiology: 2024. in Virtual Special Issues. This collection from the FEMS journals presents the latest high-quality research in six key topic areas of microbiology that have an impact across the world. All of the FEMS journals aim to serve the microbiology community with timely and authoritative research and reviews, and by ...
Researchers have developed a new antibiotic that reduced or eliminated drug-resistant bacterial infections in mouse models of acute pneumonia and sepsis while sparing healthy microbes in the mouse ...
Microbiology News. Articles and images on biochemistry research, micro-organisms, cell functions and related topics, updated daily.
Top 100 in Microbiology - 2022. This collection highlights our most downloaded* microbiology papers published in 2022. Featuring authors from around the world, these papers showcase valuable ...
Microbiology January 1, 2024 See Your Body's Cells in Size and Number The larger a cell type is, the rarer it is in the body—and vice versa—a new study shows
Section Information. This Section will focus on research in infectious diseases, pathogenic microorganisms-hosts interaction, bacteriology, mycology, virology and parasitology, including immunology and epidemiology as related to these fields and all microbial pathogens as well as the microbiota and its effect on health and disease in various hosts.
Reviews and Research in Medical Microbiology is a quarterly review journal which provides a balanced coverage of the whole field of medical microbiology. The Journal publishes state-of-the art reviews, mini-reviews, case presentations and original research from on-going research of the latest developments and techniques in medical microbiology, virology, mycology, parasitology, clinical ...
As microbiology collaborations increased among researchers from different department and labs, Neil Rasmussen, a longtime member of the MIT Corporation and a member of the visiting committees for a number of departments, realized there was still one more component needed to turn microbiome research into a force for human health.
First published: January 18, 2024. Antimicrobial resistance (AMR) is a major global health issue. Current measures for tackling it comprise mainly the prudent use of drugs, the development of new drugs, and rapid diagnostics. Relatively little attention has been given to forecasting the evolution of resistance.
Funding new ideas. Supported by Harvard Catalyst microbiome pilot funding over the past year through the Translational Innovator program, Gerber, Bry and Allegretti have been investigating whether a microbiome-focused clinical test can predict whose C. difficile infections will recur, which happens about 25 percent of the time.
Symbiotic Interactions in Microbial-facilitated Vegetation Restoration and Agricultural Management. Dan Xiao. Wei Zhang. Tong Li. Yongjun Tan. 325 views. The most cited microbiology journal which advances our understanding of the role microbes play in addressing global challenges such as healthcare, food security, and climate change.
ISSN: 0022-2615. E-ISSN: 1473-5644. Journal of Medical Microbiology is the go-to interdisciplinary journal for medical, dental and veterinary microbiology, at the bench and in the clinic. It provides comprehensive coverage of medical, dental and veterinary microbiology and infectious diseases, welcoming articles ranging from laboratory research ...
Reviews and Research in Medical Microbiology is a quarterly review journal which provides a balanced coverage of the whole field of medical microbiology. The Journal publishes state-of-the art reviews, mini-reviews, case presentations and original research from on-going research of the latest developments and techniques in medical microbiology, virology, mycology, parasitology, clinical ...
Medical Microbiology and Immunology is a comprehensive journal exploring the interrelationship between infectious agents and their hosts. Covers major topics in microbial and viral pathogenesis and the immunological response to infections. Founded in 1886 by Robert Koch and Carl Flügge, it was renamed multiple times before adopting its current ...
Journal of Medical Microbiology is the go-to interdisciplinary journal for medical, dental and veterinary microbiology, at the bench and in the clinic. It provides comprehensive coverage of medical, dental and veterinary microbiology and infectious diseases, welcoming articles ranging from laboratory research to clinical trials, including bacteriology, virology, mycology and parasitology ...
Six Key Topics in Microbiology: 2020. Read an essential collection of papers showcasing high-quality content from across the five FEMS Journals, which together provide an overview of current research trends in microbiology. Follow the topic area links below for access to articles: Antimicrobial Resistance. Environmental Microbiology.
This new result unearths a new class of biomarkers to forecast tumor progression and an entirely new way of understanding breast cancer origins." Curtis is the senior author of the study, which will be published May 31 in Science. Postdoctoral scholar Kathleen Houlahan, PhD, is the lead author of the research.
Researchers at Vanderbilt University Medical Center have isolated human monoclonal antibodies against influenza B, a significant public health threat that disproportionately affects children, the ...
Microbiology Top 100 of 2023. This collection highlights the most downloaded* microbiology research papers published by Scientific Reports in 2023. Featuring authors from around the world, these ...
Abstract. Medical microbiology involves the identification of microorganisms for the diagnosis of infectious diseases and the assessment of likely response to specific therapeutic interventions. Major categories of organisms include bacteria, mycobacteria, fungi, viruses, and parasites. Microbiological methods combined with clinical symptoms ...
He is now the head of a large, 3,000-bed clinical microbiology laboratory at the University Hospital in Marseille, France, and also runs a research laboratory that studies rickettsial diseases and emerging pathogens (the WHO Collaborative Center for Rickettsial Reference and Research). He has published more than 1,500 peer-reviewed articles.
Here's a non-exhaustive list of medical microbiology project topics for undergraduates and MSc students. Topic 1: Investigating the Role of Gut Microbiota in Autoimmune Diseases. Autoimmune diseases have been linked to alterations in the gut microbiota. This project aims to explore the relationship between gut microbiota composition and the ...
The subthalamic nucleus is known for its role in inhibiting motor activity, but there are clues to its involvement in other functions. For example, deep brain stimulation, which uses implanted electrodes to stimulate the subthalamic nucleus, has proven to be a powerful way to relieve motor symptoms for Parkinson's patients — but a common side effect is worsened speech impairment.
New studies and samples. This work suggested that mimicking the "fever effect" by giving extra IL-17a could produce similar therapeutic effects for multiple autism-spectrum disorders, with different underlying causes. But the research also left wide-open questions that must be answered before any clinically viable therapy could be developed.
I was also involved in research with Jan Geliebter, Ph.D., professor ofpathology, microbiology, and immunology and associate professor of otolaryngology; and Mark Hurwitz, M.D., professor and chair of the Department of Radiation Medicine, exploring hyperthermia for melanoma treatment, and research with Dr. Li regarding allergy and asthma.
Andhra Medical College hosted the 2024 Prof. Dr. E. Pedaveera Raju Gold Medal for Best Research Paper in Investigative Medicine for PGs in the departments of Pathology, Biochemistry, Microbiology ...
Top 100 in Microbiology. This collection highlights our most downloaded* microbiology papers published in 2021. Featuring authors from around the world, these papers showcase valuable research ...
The research demonstrates that the soil microbes embedded in the permafrost will go after a class of compounds previously thought to be untouchable under certain conditions: polyphenols.