

Literature Reviews: systematic searching at various levels
- for assignments
- for dissertations / theses
- Search strategy and searching
- Boolean Operators
- Search strategy template
- Screening & critiquing
- Citation Searching
- Google Scholar (with Lean Library)
- Resources for literature reviews
- Adding a referencing style to EndNote
- Exporting from different databases
PRISMA Flow Diagram
- Grey Literature
- What is the PRISMA Flow Diagram?
- How should I use it?
- When should I use it?
- PRISMA Links
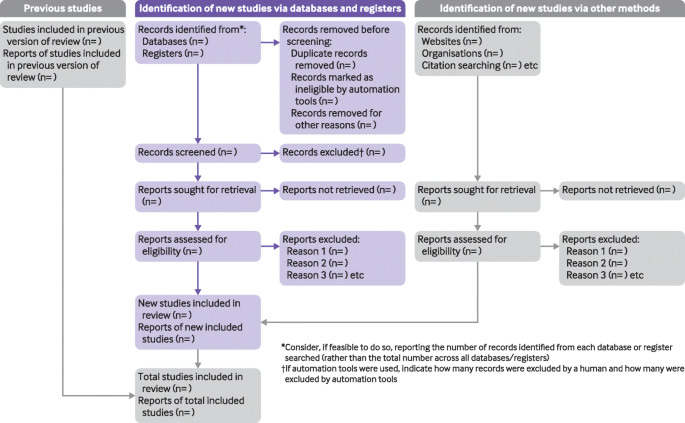
The PRISMA Flow Diagram is a tool that can be used to record different stages of the literature search process--across multiple resources--and clearly show how a researcher went from, 'These are the databases I searched for my terms', to, 'These are the papers I'm going to talk about'.
PRISMA is not inflexible; it can be modified to suit the research needs of different people and, indeed, if you did a Google images search for the flow diagram you would see many different versions of the diagram being used. It's a good idea to have a look at a couple of those examples, and also to have a look at a couple of the articles on the PRISMA website to see how it has--and can--be used.
The PRISMA 2020 Statement was published in 2021. It consists of a checklist and a flow diagram , and is intended to be accompanied by the PRISMA 2020 Explanation and Elaboration document.
In order to encourage dissemination of the PRISMA 2020 Statement, it has been published in several journals.
- How to use the PRISMA Flow Diagram for literature reviews A PDF [3.81MB] of the PowerPoint used to create the video. Each slide that has notes has a callout icon on the top right of the page which can be toggled on or off to make the notes visible.
There is also a PowerPoint version of the document but the file size is too large to upload here.
If you would like a copy, please email the Academic Librarians' mailbox from your university account to ask for it to be sent to you.
This is an example of how you could fill in the PRISMA flow diagram when conducting a new review. It is not a hard and fast rule but it should give you an idea of how you can use it.
For more detailed information, please have a look at this article:
Page, M.J., McKenzie, J.E., Bossuyt, P.M., Boutron, I., Hoffmann, T.C., Mulrow, C.D., Shamseer, L., Tetzlaff, J.M., Akl, E.A., Brennan, S.E., Chou, R., Glanville, J., Grimshaw, J.M., Hróbjartsson, A., Lalu, M.M., Li, T., Loder, E.W., Mayo-Wilson, E., McDonald, S., McGuinness, L.A., Stewart, L.A., Thomas, J., Tricco, A.C., Welch, V.A., Whiting,P. & Moher, D. (2021) 'The PRISMA 2020 statement: an updated guideline for reporting systematic reviews', BMJ 372:(71). doi: 10.1136/bmj.n71 .
- Example of PRISMA 2020 diagram This is an example of *one* of the PRISMA 2020 flow diagrams you can use when reporting on your research process. There is more than one form that you can use so for other forms and advice please look at the PRISMA website for full details.
Start using the flow diagram as you start searching the databases you've decided upon.
Make sure that you record the number of results that you found per database (before removing any duplicates) as per the filled in example. You can also do a Google images search for the PRISMA flow diagram to see the different ways in which people have used them to express their search processes.
- Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) PRISMA is an evidence-based minimum set of items for reporting in systematic reviews and meta-analyses. PRISMA focuses on the reporting of reviews evaluating randomized trials, but can also be used as a basis for reporting systematic reviews of other types of research, particularly evaluations of interventions.
- Prisma Flow Diagram This link will take you to downloadable Word and PDF copies of the flow diagram. These are modifiable and act as a starting point for you to record the process you engaged in from first search to the papers you ultimately discuss in your work. more... less... Do an image search on the internet for the flow diagram and you will be able to see all the different ways that people have modified the diagram to suit their personal research needs.
You can access the various checklists via the Equator website and the articles explaining PRISMA and its various extensions are available via PubMed.
Page, M.J., McKenzie, J.E., Bossuyt, P.M., Boutron, I., Hoffmann, T.C., Mulrow, C.D., Shamseer, L., Tetzlaff, J.M., Akl, E.A., Brennan, S.E., Chou, R., Glanville, J., Grimshaw, J.M., Hróbjartsson, A., Lalu, M.M., Li, T., Loder, E.W., Mayo-Wilson, E., McDonald, S., McGuinness, L.A., Stewart, L.A., Thomas, J., Tricco, A.C., Welch, V.A., Whiting, P., & Moher, D. (2021) ' The PRISMA 2020 statement: an updated guideline for reporting systematic reviews,' BMJ . Mar 29; 372:n71. doi: 10.1136/bmj.n71 .
Page, M.J., Moher, D., Bossuyt, P.M., Boutron, I., Hoffmann, T.C., Mulrow, C.D., Shamseer, L., Tetzlaff, J.M., Akl, E.A., Brennan, S.E., Chou, R., Glanville, J., Grimshaw, J.M., Hróbjartsson, A., Lalu, M.M., Li, T., Loder, E.W., Mayo-Wilson, E., McDonald, S., McGuinness, L.A., Stewart, L.A., Thomas, J., Tricco, A.C., Welch, V.A., Whiting, P., & McKenzie, J.E. (2021) 'PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews', BMJ, Mar 29; 372:n160. doi: 10.1136/bmj.n160 .
Page, M.J., McKenzie, J.E., Bossuyt, P.M., Boutron, I., Hoffmann, T.C., Mulrow, C.D., Shamseer, L., Tetzlaff, J.M., Akl, E.A., Brennan, S.E., Chou, R., Glanville, J., Grimshaw, J.M., Hróbjartsson, A., Lalu, M.M., Li, T., Loder, E.W., Mayo-Wilson, E., McDonald, S., McGuinness, L.A., Stewart, L.A., Thomas, J., Tricco, A.C., Welch, V.A., Whiting, P., & Moher, D. (2021) ' The PRISMA 2020 statement: An updated guideline for reporting systematic reviews,' Journal of Clinical Epidemiology, June; 134:178-189. doi: 10.1016/j.jclinepi.2021.03.001 .
- << Previous: Exporting from different databases
- Next: Grey Literature >>
- Last Updated: Apr 12, 2024 11:57 AM
- URL: https://libguides.derby.ac.uk/literature-reviews
- UNC Libraries
- HSL Academic Process
- Systematic Reviews
- Step 8: Write the Review
Systematic Reviews: Step 8: Write the Review
Created by health science librarians.

- Step 1: Complete Pre-Review Tasks
- Step 2: Develop a Protocol
- Step 3: Conduct Literature Searches
- Step 4: Manage Citations
- Step 5: Screen Citations
- Step 6: Assess Quality of Included Studies
- Step 7: Extract Data from Included Studies
About Step 8: Write the Review
Write your review, report your review with prisma, review sections, plain language summaries for systematic reviews, writing the review- webinars.
- Writing the Review FAQs
Check our FAQ's
Email us
Call (919) 962-0800
Make an appointment with a librarian
Request a systematic or scoping review consultation
Search the FAQs
In Step 8, you will write an article or a paper about your systematic review. It will likely have five sections: introduction, methods, results, discussion, and conclusion. You will:
- Review the reporting standards you will use, such as PRISMA.
- Gather your completed data tables and PRISMA chart.
- Write the Introduction to the topic and your study, Methods of your research, Results of your research, and Discussion of your results.
- Write an Abstract describing your study and a Conclusion summarizing your paper.
- Cite the studies included in your systematic review and any other articles you may have used in your paper.
- If you wish to publish your work, choose a target journal for your article.
The PRISMA Checklist will help you report the details of your systematic review. Your paper will also include a PRISMA chart that is an image of your research process.
Click an item below to see how it applies to Step 8: Write the Review.
Reporting your review with PRISMA
To write your review, you will need the data from your PRISMA flow diagram . Review the PRISMA checklist to see which items you should report in your methods section.
Managing your review with Covidence
When you screen in Covidence, it will record the numbers you need for your PRISMA flow diagram from duplicate removal through inclusion of studies. You may need to add additional information, such as the number of references from each database, citations you find through grey literature or other searching methods, or the number of studies found in your previous work if you are updating a systematic review.
How a librarian can help with Step 8
A librarian can advise you on the process of organizing and writing up your systematic review, including:
- Applying the PRISMA reporting templates and the level of detail to include for each element
- How to report a systematic review search strategy and your review methodology in the completed review
- How to use prior published reviews to guide you in organizing your manuscript
Reporting standards & guidelines
Be sure to reference reporting standards when writing your review. This helps ensure that you communicate essential components of your methods, results, and conclusions. There are a number of tools that can be used to ensure compliance with reporting guidelines. A few review-writing resources are listed below.
- Cochrane Handbook - Chapter 15: Interpreting results and drawing conclusions
- JBI Manual for Evidence Synthesis - Chapter 12.3 The systematic review
- PRISMA 2020 (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) The aim of the PRISMA Statement is to help authors improve the reporting of systematic reviews and meta-analyses.
Tools for writing your review
- RevMan (Cochrane Training)
- Methods Wizard (Systematic Review Accelerator) The Methods Wizard is part of the Systematic Review Accelerator created by Bond University and the Institute for Evidence-Based Healthcare.
- UNC HSL Systematic Review Manuscript Template Systematic review manuscript template(.doc) adapted from the PRISMA 2020 checklist. This document provides authors with template for writing about their systematic review. Each table contains a PRISMA checklist item that should be written about in that section, the matching PRISMA Item number, and a box where authors can indicate if an item has been completed. Once text has been added, delete any remaining instructions and the PRISMA checklist tables from the end of each section.
- The PRISMA 2020 statement: an updated guideline for reporting systematic reviews The PRISMA 2020 statement replaces the 2009 statement and includes new reporting guidance that reflects advances in methods to identify, select, appraise, and synthesise studies.
- PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews This document is intended to enhance the use, understanding and dissemination of the PRISMA 2020 Statement. Through examples and explanations, the meaning and rationale for each checklist item are presented.
The PRISMA checklist
The Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) is a 27-item checklist used to improve transparency in systematic reviews. These items cover all aspects of the manuscript, including title, abstract, introduction, methods, results, discussion, and funding. The PRISMA checklist can be downloaded in PDF or Word files.
- PRISMA 2020 Checklists Download the 2020 PRISMA Checklists in Word or PDF formats or download the expanded checklist (PDF).
The PRISMA flow diagram
The PRISMA Flow Diagram visually depicts the flow of studies through each phase of the review process. The PRISMA Flow Diagram can be downloaded in Word files.
- PRISMA 2020 Flow Diagrams The flow diagram depicts the flow of information through the different phases of a systematic review. It maps out the number of records identified, included and excluded, and the reasons for exclusions. Different templates are available depending on the type of review (new or updated) and sources used to identify studies.
Documenting grey literature and/or hand searches
If you have also searched additional sources, such as professional organization websites, cited or citing references, etc., document your grey literature search using the flow diagram template version 1 PRISMA 2020 flow diagram for new systematic reviews which included searches of databases, registers and other sources or the version 2 PRISMA 2020 flow diagram for updated systematic reviews which included searches of databases, registers and other sources .
Complete the boxes documenting your database searches, Identification of studies via databases and registers, according to the PRISMA flow diagram instructions. Complete the boxes documenting your grey literature and/or hand searches on the right side of the template, Identification of studies via other methods, using the steps below.
Need help completing the PRISMA flow diagram?
There are different PRISMA flow diagram templates for new and updated reviews, as well as different templates for reviews with and without grey literature searches. Be sure you download the correct template to match your review methods, then follow the steps below for each portion of the diagram you have available.
View the step-by-step explanation of the PRISMA flow diagram
Step 1: Preparation Download the flow diagram template version 1 PRISMA 2020 flow diagram for new systematic reviews which included searches of databases and registers only or the version 2 PRISMA 2020 flow diagram for updated systematic reviews which included searches of databases and registers only .
View the step-by-step explanation of the grey literature & hand searching portion of the PRISMA flow diagram
Step 1: Preparation Download the flow diagram template version 1 PRISMA 2020 flow diagram for new systematic reviews which included searches of databases, registers and other sources or the version 2 PRISMA 2020 flow diagram for updated systematic reviews which included searches of databases, registers and other sources .
View the step-by-step explanation of review update portion of the PRISMA flow diagram
Step 1: Preparation Download the flow diagram template version 2 PRISMA 2020 flow diagram for updated systematic reviews which included searches of databases and registers only or the version 2 PRISMA 2020 flow diagram for updated systematic reviews which included searches of databases, registers and other sources .
For more information about updating your systematic review, see the box Updating Your Review? on the Step 3: Conduct Literature Searches page of the guide.
Sections of a Scientific Manuscript
Scientific articles often follow the IMRaD format: Introduction, Methods, Results, and Discussion. You will also need a title and an abstract to summarize your research.
You can read more about scientific writing through the library guides below.
- Structure of Scholarly Articles & Peer Review • Explains the standard parts of a medical research article • Compares scholarly journals, professional trade journals, and magazines • Explains peer review and how to find peer reviewed articles and journals
- Writing in the Health Sciences (For Students and Instructors)
- Citing & Writing Tools & Guides Includes links to guides for popular citation managers such as EndNote, Sciwheel, Zotero; copyright basics; APA & AMA Style guides; Plagiarism & Citing Sources; Citing & Writing: How to Write Scientific Papers
Sections of a Systematic Review Manuscript
Systematic reviews follow the same structure as original research articles, but you will need to report on your search instead of on details like the participants or sampling. Sections of your manuscript are shown as bold headings in the PRISMA checklist.
Refer to the PRISMA checklist for more information.
Consider including a Plain Language Summary (PLS) when you publish your systematic review. Like an abstract, a PLS gives an overview of your study, but is specifically written and formatted to be easy for non-experts to understand.
Tips for writing a PLS:
- Use clear headings e.g. "why did we do this study?"; "what did we do?"; "what did we find?"
- Use active voice e.g. "we searched for articles in 5 databases instead of "5 databases were searched"
- Consider need-to-know vs. nice-to-know: what is most important for readers to understand about your study? Be sure to provide the most important points without misrepresenting your study or misleading the reader.
- Keep it short: Many journals recommend keeping your plain language summary less than 250 words.
- Check journal guidelines: Your journal may have specific guidelines about the format of your plain language summary and when you can publish it. Look at journal guidelines before submitting your article.
Learn more about Plain Language Summaries:
- Rosenberg, A., Baróniková, S., & Feighery, L. (2021). Open Pharma recommendations for plain language summaries of peer-reviewed medical journal publications. Current Medical Research and Opinion, 37(11), 2015–2016. https://doi.org/10.1080/03007995.2021.1971185
- Lobban, D., Gardner, J., & Matheis, R. (2021). Plain language summaries of publications of company-sponsored medical research: what key questions do we need to address? Current Medical Research and Opinion, 1–12. https://doi.org/10.1080/03007995.2021.1997221
- Cochrane Community. (2022, March 21). Updated template and guidance for writing Plain Language Summaries in Cochrane Reviews now available. https://community.cochrane.org/news/updated-template-and-guidance-writing-plain-language-summaries-cochrane-reviews-now-available
- You can also look at our Health Literacy LibGuide: https://guides.lib.unc.edu/healthliteracy
How to Approach Writing a Background Section
What Makes a Good Discussion Section
Writing Up Risk of Bias
Developing Your Implications for Research Section
- << Previous: Step 7: Extract Data from Included Studies
- Next: FAQs >>
- Last Updated: May 16, 2024 3:24 PM
- URL: https://guides.lib.unc.edu/systematic-reviews
- Open access
- Published: 29 March 2021
The PRISMA 2020 statement: an updated guideline for reporting systematic reviews
- Matthew J. Page ORCID: orcid.org/0000-0002-4242-7526 1 ,
- Joanne E. McKenzie 1 ,
- Patrick M. Bossuyt 2 ,
- Isabelle Boutron 3 ,
- Tammy C. Hoffmann 4 ,
- Cynthia D. Mulrow 5 ,
- Larissa Shamseer 6 ,
- Jennifer M. Tetzlaff 7 ,
- Elie A. Akl 8 ,
- Sue E. Brennan 1 ,
- Roger Chou 9 ,
- Julie Glanville 10 ,
- Jeremy M. Grimshaw 11 ,
- Asbjørn Hróbjartsson 12 ,
- Manoj M. Lalu 13 ,
- Tianjing Li 14 ,
- Elizabeth W. Loder 15 ,
- Evan Mayo-Wilson 16 ,
- Steve McDonald 1 ,
- Luke A. McGuinness 17 ,
- Lesley A. Stewart 18 ,
- James Thomas 19 ,
- Andrea C. Tricco 20 ,
- Vivian A. Welch 21 ,
- Penny Whiting 17 &
- David Moher 22
Systematic Reviews volume 10 , Article number: 89 ( 2021 ) Cite this article
310k Accesses
78k Citations
102 Altmetric
Metrics details
An Editorial to this article was published on 19 April 2021
The Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement, published in 2009, was designed to help systematic reviewers transparently report why the review was done, what the authors did, and what they found. Over the past decade, advances in systematic review methodology and terminology have necessitated an update to the guideline. The PRISMA 2020 statement replaces the 2009 statement and includes new reporting guidance that reflects advances in methods to identify, select, appraise, and synthesise studies. The structure and presentation of the items have been modified to facilitate implementation. In this article, we present the PRISMA 2020 27-item checklist, an expanded checklist that details reporting recommendations for each item, the PRISMA 2020 abstract checklist, and the revised flow diagrams for original and updated reviews. In order to encourage its wide dissemination this article is freely accessible on BMJ, PLOS Medicine, Journal of Clinical Epidemiology and International Journal of Surgery journal websites.
Systematic reviews serve many critical roles. They can provide syntheses of the state of knowledge in a field, from which future research priorities can be identified; they can address questions that otherwise could not be answered by individual studies; they can identify problems in primary research that should be rectified in future studies; and they can generate or evaluate theories about how or why phenomena occur. Systematic reviews therefore generate various types of knowledge for different users of reviews (such as patients, healthcare providers, researchers, and policy makers) [ 1 , 2 ]. To ensure a systematic review is valuable to users, authors should prepare a transparent, complete, and accurate account of why the review was done, what they did (such as how studies were identified and selected) and what they found (such as characteristics of contributing studies and results of meta-analyses). Up-to-date reporting guidance facilitates authors achieving this [ 3 ].
The Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement published in 2009 (hereafter referred to as PRISMA 2009) [ 4 , 5 , 6 , 7 , 8 , 9 , 10 ] is a reporting guideline designed to address poor reporting of systematic reviews [ 11 ]. The PRISMA 2009 statement comprised a checklist of 27 items recommended for reporting in systematic reviews and an “explanation and elaboration” paper [ 12 , 13 , 14 , 15 , 16 ] providing additional reporting guidance for each item, along with exemplars of reporting. The recommendations have been widely endorsed and adopted, as evidenced by its co-publication in multiple journals, citation in over 60,000 reports (Scopus, August 2020), endorsement from almost 200 journals and systematic review organisations, and adoption in various disciplines. Evidence from observational studies suggests that use of the PRISMA 2009 statement is associated with more complete reporting of systematic reviews [ 17 , 18 , 19 , 20 ], although more could be done to improve adherence to the guideline [ 21 ].
Many innovations in the conduct of systematic reviews have occurred since publication of the PRISMA 2009 statement. For example, technological advances have enabled the use of natural language processing and machine learning to identify relevant evidence [ 22 , 23 , 24 ], methods have been proposed to synthesise and present findings when meta-analysis is not possible or appropriate [ 25 , 26 , 27 ], and new methods have been developed to assess the risk of bias in results of included studies [ 28 , 29 ]. Evidence on sources of bias in systematic reviews has accrued, culminating in the development of new tools to appraise the conduct of systematic reviews [ 30 , 31 ]. Terminology used to describe particular review processes has also evolved, as in the shift from assessing “quality” to assessing “certainty” in the body of evidence [ 32 ]. In addition, the publishing landscape has transformed, with multiple avenues now available for registering and disseminating systematic review protocols [ 33 , 34 ], disseminating reports of systematic reviews, and sharing data and materials, such as preprint servers and publicly accessible repositories. To capture these advances in the reporting of systematic reviews necessitated an update to the PRISMA 2009 statement.
Development of PRISMA 2020
A complete description of the methods used to develop PRISMA 2020 is available elsewhere [ 35 ]. We identified PRISMA 2009 items that were often reported incompletely by examining the results of studies investigating the transparency of reporting of published reviews [ 17 , 21 , 36 , 37 ]. We identified possible modifications to the PRISMA 2009 statement by reviewing 60 documents providing reporting guidance for systematic reviews (including reporting guidelines, handbooks, tools, and meta-research studies) [ 38 ]. These reviews of the literature were used to inform the content of a survey with suggested possible modifications to the 27 items in PRISMA 2009 and possible additional items. Respondents were asked whether they believed we should keep each PRISMA 2009 item as is, modify it, or remove it, and whether we should add each additional item. Systematic review methodologists and journal editors were invited to complete the online survey (110 of 220 invited responded). We discussed proposed content and wording of the PRISMA 2020 statement, as informed by the review and survey results, at a 21-member, two-day, in-person meeting in September 2018 in Edinburgh, Scotland. Throughout 2019 and 2020, we circulated an initial draft and five revisions of the checklist and explanation and elaboration paper to co-authors for feedback. In April 2020, we invited 22 systematic reviewers who had expressed interest in providing feedback on the PRISMA 2020 checklist to share their views (via an online survey) on the layout and terminology used in a preliminary version of the checklist. Feedback was received from 15 individuals and considered by the first author, and any revisions deemed necessary were incorporated before the final version was approved and endorsed by all co-authors.
The PRISMA 2020 statement
Scope of the guideline.
The PRISMA 2020 statement has been designed primarily for systematic reviews of studies that evaluate the effects of health interventions, irrespective of the design of the included studies. However, the checklist items are applicable to reports of systematic reviews evaluating other interventions (such as social or educational interventions), and many items are applicable to systematic reviews with objectives other than evaluating interventions (such as evaluating aetiology, prevalence, or prognosis). PRISMA 2020 is intended for use in systematic reviews that include synthesis (such as pairwise meta-analysis or other statistical synthesis methods) or do not include synthesis (for example, because only one eligible study is identified). The PRISMA 2020 items are relevant for mixed-methods systematic reviews (which include quantitative and qualitative studies), but reporting guidelines addressing the presentation and synthesis of qualitative data should also be consulted [ 39 , 40 ]. PRISMA 2020 can be used for original systematic reviews, updated systematic reviews, or continually updated (“living”) systematic reviews. However, for updated and living systematic reviews, there may be some additional considerations that need to be addressed. Where there is relevant content from other reporting guidelines, we reference these guidelines within the items in the explanation and elaboration paper [ 41 ] (such as PRISMA-Search [ 42 ] in items 6 and 7, Synthesis without meta-analysis (SWiM) reporting guideline [ 27 ] in item 13d). Box 1 includes a glossary of terms used throughout the PRISMA 2020 statement.
PRISMA 2020 is not intended to guide systematic review conduct, for which comprehensive resources are available [ 43 , 44 , 45 , 46 ]. However, familiarity with PRISMA 2020 is useful when planning and conducting systematic reviews to ensure that all recommended information is captured. PRISMA 2020 should not be used to assess the conduct or methodological quality of systematic reviews; other tools exist for this purpose [ 30 , 31 ]. Furthermore, PRISMA 2020 is not intended to inform the reporting of systematic review protocols, for which a separate statement is available (PRISMA for Protocols (PRISMA-P) 2015 statement [ 47 , 48 ]). Finally, extensions to the PRISMA 2009 statement have been developed to guide reporting of network meta-analyses [ 49 ], meta-analyses of individual participant data [ 50 ], systematic reviews of harms [ 51 ], systematic reviews of diagnostic test accuracy studies [ 52 ], and scoping reviews [ 53 ]; for these types of reviews we recommend authors report their review in accordance with the recommendations in PRISMA 2020 along with the guidance specific to the extension.
How to use PRISMA 2020
The PRISMA 2020 statement (including the checklists, explanation and elaboration, and flow diagram) replaces the PRISMA 2009 statement, which should no longer be used. Box 2 summarises noteworthy changes from the PRISMA 2009 statement. The PRISMA 2020 checklist includes seven sections with 27 items, some of which include sub-items (Table 1 ). A checklist for journal and conference abstracts for systematic reviews is included in PRISMA 2020. This abstract checklist is an update of the 2013 PRISMA for Abstracts statement [ 54 ], reflecting new and modified content in PRISMA 2020 (Table 2 ). A template PRISMA flow diagram is provided, which can be modified depending on whether the systematic review is original or updated (Fig. 1 ).
PRISMA 2020 flow diagram template for systematic reviews. The new design is adapted from flow diagrams proposed by Boers [ 55 ], Mayo-Wilson et al. [ 56 ] and Stovold et al. [ 57 ] The boxes in grey should only be completed if applicable; otherwise they should be removed from the flow diagram. Note that a “report” could be a journal article, preprint, conference abstract, study register entry, clinical study report, dissertation, unpublished manuscript, government report or any other document providing relevant information
We recommend authors refer to PRISMA 2020 early in the writing process, because prospective consideration of the items may help to ensure that all the items are addressed. To help keep track of which items have been reported, the PRISMA statement website ( http://www.prisma-statement.org/ ) includes fillable templates of the checklists to download and complete (also available in Additional file 1 ). We have also created a web application that allows users to complete the checklist via a user-friendly interface [ 58 ] (available at https://prisma.shinyapps.io/checklist/ and adapted from the Transparency Checklist app [ 59 ]). The completed checklist can be exported to Word or PDF. Editable templates of the flow diagram can also be downloaded from the PRISMA statement website.
We have prepared an updated explanation and elaboration paper, in which we explain why reporting of each item is recommended and present bullet points that detail the reporting recommendations (which we refer to as elements) [ 41 ]. The bullet-point structure is new to PRISMA 2020 and has been adopted to facilitate implementation of the guidance [ 60 , 61 ]. An expanded checklist, which comprises an abridged version of the elements presented in the explanation and elaboration paper, with references and some examples removed, is available in Additional file 2 . Consulting the explanation and elaboration paper is recommended if further clarity or information is required.
Journals and publishers might impose word and section limits, and limits on the number of tables and figures allowed in the main report. In such cases, if the relevant information for some items already appears in a publicly accessible review protocol, referring to the protocol may suffice. Alternatively, placing detailed descriptions of the methods used or additional results (such as for less critical outcomes) in supplementary files is recommended. Ideally, supplementary files should be deposited to a general-purpose or institutional open-access repository that provides free and permanent access to the material (such as Open Science Framework, Dryad, figshare). A reference or link to the additional information should be included in the main report. Finally, although PRISMA 2020 provides a template for where information might be located, the suggested location should not be seen as prescriptive; the guiding principle is to ensure the information is reported.
Use of PRISMA 2020 has the potential to benefit many stakeholders. Complete reporting allows readers to assess the appropriateness of the methods, and therefore the trustworthiness of the findings. Presenting and summarising characteristics of studies contributing to a synthesis allows healthcare providers and policy makers to evaluate the applicability of the findings to their setting. Describing the certainty in the body of evidence for an outcome and the implications of findings should help policy makers, managers, and other decision makers formulate appropriate recommendations for practice or policy. Complete reporting of all PRISMA 2020 items also facilitates replication and review updates, as well as inclusion of systematic reviews in overviews (of systematic reviews) and guidelines, so teams can leverage work that is already done and decrease research waste [ 36 , 62 , 63 ].
We updated the PRISMA 2009 statement by adapting the EQUATOR Network’s guidance for developing health research reporting guidelines [ 64 ]. We evaluated the reporting completeness of published systematic reviews [ 17 , 21 , 36 , 37 ], reviewed the items included in other documents providing guidance for systematic reviews [ 38 ], surveyed systematic review methodologists and journal editors for their views on how to revise the original PRISMA statement [ 35 ], discussed the findings at an in-person meeting, and prepared this document through an iterative process. Our recommendations are informed by the reviews and survey conducted before the in-person meeting, theoretical considerations about which items facilitate replication and help users assess the risk of bias and applicability of systematic reviews, and co-authors’ experience with authoring and using systematic reviews.
Various strategies to increase the use of reporting guidelines and improve reporting have been proposed. They include educators introducing reporting guidelines into graduate curricula to promote good reporting habits of early career scientists [ 65 ]; journal editors and regulators endorsing use of reporting guidelines [ 18 ]; peer reviewers evaluating adherence to reporting guidelines [ 61 , 66 ]; journals requiring authors to indicate where in their manuscript they have adhered to each reporting item [ 67 ]; and authors using online writing tools that prompt complete reporting at the writing stage [ 60 ]. Multi-pronged interventions, where more than one of these strategies are combined, may be more effective (such as completion of checklists coupled with editorial checks) [ 68 ]. However, of 31 interventions proposed to increase adherence to reporting guidelines, the effects of only 11 have been evaluated, mostly in observational studies at high risk of bias due to confounding [ 69 ]. It is therefore unclear which strategies should be used. Future research might explore barriers and facilitators to the use of PRISMA 2020 by authors, editors, and peer reviewers, designing interventions that address the identified barriers, and evaluating those interventions using randomised trials. To inform possible revisions to the guideline, it would also be valuable to conduct think-aloud studies [ 70 ] to understand how systematic reviewers interpret the items, and reliability studies to identify items where there is varied interpretation of the items.
We encourage readers to submit evidence that informs any of the recommendations in PRISMA 2020 (via the PRISMA statement website: http://www.prisma-statement.org/ ). To enhance accessibility of PRISMA 2020, several translations of the guideline are under way (see available translations at the PRISMA statement website). We encourage journal editors and publishers to raise awareness of PRISMA 2020 (for example, by referring to it in journal “Instructions to authors”), endorsing its use, advising editors and peer reviewers to evaluate submitted systematic reviews against the PRISMA 2020 checklists, and making changes to journal policies to accommodate the new reporting recommendations. We recommend existing PRISMA extensions [ 47 , 49 , 50 , 51 , 52 , 53 , 71 , 72 ] be updated to reflect PRISMA 2020 and advise developers of new PRISMA extensions to use PRISMA 2020 as the foundation document.
We anticipate that the PRISMA 2020 statement will benefit authors, editors, and peer reviewers of systematic reviews, and different users of reviews, including guideline developers, policy makers, healthcare providers, patients, and other stakeholders. Ultimately, we hope that uptake of the guideline will lead to more transparent, complete, and accurate reporting of systematic reviews, thus facilitating evidence based decision making.
Box 1 Glossary of terms
Systematic review —A review that uses explicit, systematic methods to collate and synthesise findings of studies that address a clearly formulated question [ 43 ]
Statistical synthesis —The combination of quantitative results of two or more studies. This encompasses meta-analysis of effect estimates (described below) and other methods, such as combining P values, calculating the range and distribution of observed effects, and vote counting based on the direction of effect (see McKenzie and Brennan [ 25 ] for a description of each method)
Meta-analysis of effect estimates —A statistical technique used to synthesise results when study effect estimates and their variances are available, yielding a quantitative summary of results [ 25 ]
Outcome —An event or measurement collected for participants in a study (such as quality of life, mortality)
Result —The combination of a point estimate (such as a mean difference, risk ratio, or proportion) and a measure of its precision (such as a confidence/credible interval) for a particular outcome
Report —A document (paper or electronic) supplying information about a particular study. It could be a journal article, preprint, conference abstract, study register entry, clinical study report, dissertation, unpublished manuscript, government report, or any other document providing relevant information
Record —The title or abstract (or both) of a report indexed in a database or website (such as a title or abstract for an article indexed in Medline). Records that refer to the same report (such as the same journal article) are “duplicates”; however, records that refer to reports that are merely similar (such as a similar abstract submitted to two different conferences) should be considered unique.
Study —An investigation, such as a clinical trial, that includes a defined group of participants and one or more interventions and outcomes. A “study” might have multiple reports. For example, reports could include the protocol, statistical analysis plan, baseline characteristics, results for the primary outcome, results for harms, results for secondary outcomes, and results for additional mediator and moderator analyses
Box 2 Noteworthy changes to the PRISMA 2009 statement
• Inclusion of the abstract reporting checklist within PRISMA 2020 (see item #2 and Box 2 ).
• Movement of the ‘Protocol and registration’ item from the start of the Methods section of the checklist to a new Other section, with addition of a sub-item recommending authors describe amendments to information provided at registration or in the protocol (see item #24a-24c).
• Modification of the ‘Search’ item to recommend authors present full search strategies for all databases, registers and websites searched, not just at least one database (see item #7).
• Modification of the ‘Study selection’ item in the Methods section to emphasise the reporting of how many reviewers screened each record and each report retrieved, whether they worked independently, and if applicable, details of automation tools used in the process (see item #8).
• Addition of a sub-item to the ‘Data items’ item recommending authors report how outcomes were defined, which results were sought, and methods for selecting a subset of results from included studies (see item #10a).
• Splitting of the ‘Synthesis of results’ item in the Methods section into six sub-items recommending authors describe: the processes used to decide which studies were eligible for each synthesis; any methods required to prepare the data for synthesis; any methods used to tabulate or visually display results of individual studies and syntheses; any methods used to synthesise results; any methods used to explore possible causes of heterogeneity among study results (such as subgroup analysis, meta-regression); and any sensitivity analyses used to assess robustness of the synthesised results (see item #13a-13f).
• Addition of a sub-item to the ‘Study selection’ item in the Results section recommending authors cite studies that might appear to meet the inclusion criteria, but which were excluded, and explain why they were excluded (see item #16b).
• Splitting of the ‘Synthesis of results’ item in the Results section into four sub-items recommending authors: briefly summarise the characteristics and risk of bias among studies contributing to the synthesis; present results of all statistical syntheses conducted; present results of any investigations of possible causes of heterogeneity among study results; and present results of any sensitivity analyses (see item #20a-20d).
• Addition of new items recommending authors report methods for and results of an assessment of certainty (or confidence) in the body of evidence for an outcome (see items #15 and #22).
• Addition of a new item recommending authors declare any competing interests (see item #26).
• Addition of a new item recommending authors indicate whether data, analytic code and other materials used in the review are publicly available and if so, where they can be found (see item #27).
Gurevitch J, Koricheva J, Nakagawa S, Stewart G. Meta-analysis and the science of research synthesis. Nature. 2018;555:175–82. https://doi.org/10.1038/nature25753 .
Article CAS PubMed Google Scholar
Gough D, Thomas J, Oliver S. Clarifying differences between reviews within evidence ecosystems. Syst Rev. 2019;8:170. https://doi.org/10.1186/s13643-019-1089-2 .
Article PubMed PubMed Central Google Scholar
Moher D. Reporting guidelines: doing better for readers. BMC Med. 2018;16:233. https://doi.org/10.1186/s12916-018-1226-0 .
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–9, W64. https://doi.org/10.7326/0003-4819-151-4-200908180-00135 .
Article PubMed Google Scholar
Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. https://doi.org/10.1136/bmj.b2535 .
Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. https://doi.org/10.1371/journal.pmed.1000097 .
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62:1006–12. https://doi.org/10.1016/j.jclinepi.2009.06.005 .
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8:336–41. https://doi.org/10.1016/j.ijsu.2010.02.007 .
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Open Med. 2009;3:e123–30.
PubMed PubMed Central Google Scholar
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Reprint--preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Phys Ther. 2009;89:873–80. https://doi.org/10.1093/ptj/89.9.873 .
Moher D, Tetzlaff J, Tricco AC, Sampson M, Altman DG. Epidemiology and reporting characteristics of systematic reviews. PLoS Med. 2007;4:e78. https://doi.org/10.1371/journal.pmed.0040078 .
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62:e1–34. https://doi.org/10.1016/j.jclinepi.2009.06.006 .
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. https://doi.org/10.1136/bmj.b2700 .
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann Intern Med. 2009;151:W65–94. https://doi.org/10.7326/0003-4819-151-4-200908180-00136 .
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. https://doi.org/10.1371/journal.pmed.1000100 .
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting. systematic reviews and meta-analyses of studies that evaluate health care. interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. https://doi.org/10.1371/journal.pmed.1000100 .
Page MJ, Shamseer L, Altman DG, et al. Epidemiology and reporting characteristics of systematic reviews of biomedical research: a cross-sectional study. PLoS Med. 2016;13:e1002028. https://doi.org/10.1371/journal.pmed.1002028 .
Panic N, Leoncini E, de Belvis G, Ricciardi W, Boccia S. Evaluation of the endorsement of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement on the quality of published systematic review and meta-analyses. PLoS One. 2013;8:e83138. https://doi.org/10.1371/journal.pone.0083138 .
Article CAS PubMed PubMed Central Google Scholar
Agha RA, Fowler AJ, Limb C, et al. Impact of the mandatory implementation of reporting guidelines on reporting quality in a surgical journal: a before and after study. Int J Surg. 2016;30:169–72. https://doi.org/10.1016/j.ijsu.2016.04.032 .
Leclercq V, Beaudart C, Ajamieh S, Rabenda V, Tirelli E, Bruyère O. Meta-analyses indexed in PsycINFO had a better completeness of reporting when they mention PRISMA. J Clin Epidemiol. 2019;115:46–54. https://doi.org/10.1016/j.jclinepi.2019.06.014 .
Page MJ, Moher D. Evaluations of the uptake and impact of the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and extensions: a scoping review. Syst Rev. 2017;6:263. https://doi.org/10.1186/s13643-017-0663-8 .
O’Mara-Eves A, Thomas J, McNaught J, Miwa M, Ananiadou S. Using text mining for study identification in systematic reviews: a systematic review of current approaches. Syst Rev. 2015;4:5. https://doi.org/10.1186/2046-4053-4-5 .
Marshall IJ, Noel-Storr A, Kuiper J, Thomas J, Wallace BC. Machine learning for identifying randomized controlled trials: an evaluation and practitioner’s guide. Res Synth Methods. 2018;9:602–14. https://doi.org/10.1002/jrsm.1287 .
Marshall IJ, Wallace BC. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Syst Rev. 2019;8:163. https://doi.org/10.1186/s13643-019-1074-9 .
McKenzie JE, Brennan SE. Synthesizing and presenting findings using other methods. In: Higgins JPT, Thomas J, Chandler J, et al., editors. Cochrane handbook for systematic reviews of interventions. London: Cochrane; 2019. https://doi.org/10.1002/9781119536604.ch12 .
Chapter Google Scholar
Higgins JPT, López-López JA, Becker BJ, et al. Synthesising quantitative evidence in systematic reviews of complex health interventions. BMJ Glob Health. 2019;4(Suppl 1):e000858. https://doi.org/10.1136/bmjgh-2018-000858 .
Campbell M, McKenzie JE, Sowden A, et al. Synthesis without meta-analysis (SWiM) in systematic reviews: reporting guideline. BMJ. 2020;368:l6890. https://doi.org/10.1136/bmj.l6890 .
Sterne JAC, Savović J, Page MJ, et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366:l4898. https://doi.org/10.1136/bmj.l4898 .
Sterne JA, Hernán MA, Reeves BC, et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ. 2016;355:i4919. https://doi.org/10.1136/bmj.i4919 .
Whiting P, Savović J, Higgins JP, ROBIS group, et al. ROBIS: a new tool to assess risk of bias in systematic reviews was developed. J Clin Epidemiol. 2016;69:225–34. https://doi.org/10.1016/j.jclinepi.2015.06.005 .
Shea BJ, Reeves BC, Wells G, et al. AMSTAR 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ. 2017;358:j4008. https://doi.org/10.1136/bmj.j4008 .
Hultcrantz M, Rind D, Akl EA, et al. The GRADE working group clarifies the construct of certainty of evidence. J Clin Epidemiol. 2017;87:4–13. https://doi.org/10.1016/j.jclinepi.2017.05.006 .
Booth A, Clarke M, Dooley G, et al. The nuts and bolts of PROSPERO: an international prospective register of systematic reviews. Syst Rev. 2012;1:2. https://doi.org/10.1186/2046-4053-1-2 .
Moher D, Stewart L, Shekelle P. Establishing a new journal for systematic review products. Syst Rev. 2012;1:1. https://doi.org/10.1186/2046-4053-1-1 .
Page MJ, McKenzie JE, Bossuyt PM, et al. Updating guidance for reporting systematic reviews: development of the PRISMA 2020 statement. J Clin Epidemiol 2021;134:103–112. https://doi.org/10.1016/j.jclinepi.2021.02.003 .
Page MJ, Altman DG, Shamseer L, et al. Reproducible research practices are underused in systematic reviews of biomedical interventions. J Clin Epidemiol. 2018;94:8–18. https://doi.org/10.1016/j.jclinepi.2017.10.017 .
Page MJ, Altman DG, McKenzie JE, et al. Flaws in the application and interpretation of statistical analyses in systematic reviews of therapeutic interventions were common: a cross-sectional analysis. J Clin Epidemiol. 2018;95:7–18. https://doi.org/10.1016/j.jclinepi.2017.11.022 .
Page MJ, McKenzie JE, Bossuyt PM, et al. Mapping of reporting guidance for systematic reviews and meta-analyses generated a comprehensive item bank for future reporting guidelines. J Clin Epidemiol. 2020;118:60–8. https://doi.org/10.1016/j.jclinepi.2019.11.010 .
Tong A, Flemming K, McInnes E, Oliver S, Craig J. Enhancing transparency in reporting the synthesis of qualitative research: ENTREQ. BMC Med Res Methodol. 2012;12:181. https://doi.org/10.1186/1471-2288-12-181 .
France EF, Cunningham M, Ring N, et al. Improving reporting of meta-ethnography: the eMERGe reporting guidance. BMC Med Res Methodol. 2019;19:25. https://doi.org/10.1186/s12874-018-0600-0 .
Page MJ, Moher D, Bossuyt PM, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ. 2021;372:n160. https://doi.org/10.1136/bmj.n160 .
Rethlefsen ML, Kirtley S, Waffenschmidt S, et al. PRISMA-S Group PRISMA-S: an extension to the PRISMA statement for reporting literature searches in systematic reviews. Syst Rev. 2021;10:39. https://doi.org/10.1186/s13643-020-01542-z .
Higgins JPT, Thomas J, Chandler J, et al. Cochrane handbook for systematic reviews of interventions: version 6.0. London: Cochrane; 2019. Available from https://training.cochrane.org/handbook
Book Google Scholar
Dekkers OM, Vandenbroucke JP, Cevallos M, Renehan AG, Altman DG, Egger M. COSMOS-E: guidance on conducting systematic reviews and meta-analyses of observational studies of etiology. PLoS Med. 2019;16:e1002742. https://doi.org/10.1371/journal.pmed.1002742 .
Cooper H, Hedges LV, Valentine JV. The handbook of research synthesis and meta-analysis. New York: Russell Sage Foundation; 2019.
IOM (Institute of Medicine). Finding what works in health care: standards for systematic reviews. Washington, D.C.: The National Academies Press; 2011.
Google Scholar
Moher D, Shamseer L, Clarke M, PRISMA-P Group, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4:1. https://doi.org/10.1186/2046-4053-4-1 .
Shamseer L, Moher D, Clarke M, PRISMA-P Group, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ. 2015;350:g7647. https://doi.org/10.1136/bmj.g7647 .
Hutton B, Salanti G, Caldwell DM, et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med. 2015;162:777–84. https://doi.org/10.7326/M14-2385 .
Stewart LA, Clarke M, Rovers M, PRISMA-IPD Development Group, et al. Preferred reporting items for systematic review and meta-analyses of individual participant data: the PRISMA-IPD statement. JAMA. 2015;313:1657–65. https://doi.org/10.1001/jama.2015.3656 .
Zorzela L, Loke YK, Ioannidis JP, et al. PRISMAHarms Group PRISMA harms checklist: improving harms reporting in systematic reviews. BMJ. 2016;352:i157. https://doi.org/10.1136/bmj.i157 .
McInnes MDF, Moher D, Thombs BD, the PRISMA-DTA Group, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA. 2018;319:388–96. https://doi.org/10.1001/jama.2017.19163 .
Tricco AC, Lillie E, Zarin W, et al. PRISMA extension for scoping reviews (PRISMA-SCR): checklist and explanation. Ann Intern Med. 2018;169:467–73. https://doi.org/10.7326/M18-0850 .
Beller EM, Glasziou PP, Altman DG, et al. PRISMA for Abstracts Group PRISMA for Abstracts: reporting systematic reviews in journal and conference abstracts. PLoS Med. 2013;10:e1001419. https://doi.org/10.1371/journal.pmed.1001419 .
Boers M. Graphics and statistics for cardiology: designing effective tables for presentation and publication. Heart. 2018;104:192–200. https://doi.org/10.1136/heartjnl-2017-311581 .
Mayo-Wilson E, Li T, Fusco N, Dickersin K, MUDS investigators. Practical guidance for using multiple data sources in systematic reviews and meta-analyses (with examples from the MUDS study). Res Synth Methods. 2018;9:2–12. https://doi.org/10.1002/jrsm.1277 .
Stovold E, Beecher D, Foxlee R, Noel-Storr A. Study flow diagrams in Cochrane systematic review updates: an adapted PRISMA flow diagram. Syst Rev. 2014;3:54. https://doi.org/10.1186/2046-4053-3-54 .
McGuinness LA. mcguinlu/PRISMA-Checklist: Initial release for manuscript submission (Version v1.0.0). Geneva: Zenodo; 2020. https://doi.org/10.5281/zenodo.3994319 .
Aczel B, Szaszi B, Sarafoglou A, et al. A consensus-based transparency checklist. Nat Hum Behav. 2020;4:4–6. https://doi.org/10.1038/s41562-019-0772-6 .
Barnes C, Boutron I, Giraudeau B, Porcher R, Altman DG, Ravaud P. Impact of an online writing aid tool for writing a randomized trial report: the COBWEB (Consort-based WEB tool) randomized controlled trial. BMC Med. 2015;13:221. https://doi.org/10.1186/s12916-015-0460-y .
Chauvin A, Ravaud P, Moher D, et al. Accuracy in detecting inadequate research reporting by early career peer reviewers using an online CONSORT-based peer-review tool (COBPeer) versus the usual peer-review process: a cross-sectional diagnostic study. BMC Med. 2019;17:205. https://doi.org/10.1186/s12916-019-1436-0 .
Wayant C, Page MJ, Vassar M. Evaluation of reproducible research practices in oncology systematic reviews with meta-analyses referenced by national comprehensive cancer network guidelines. JAMA Oncol. 2019;5:1550–5. https://doi.org/10.1001/jamaoncol.2019.2564 .
Article PubMed Central PubMed Google Scholar
McKenzie JE, Brennan SE. Overviews of systematic reviews: great promise, greater challenge. Syst Rev. 2017;6:185. https://doi.org/10.1186/s13643-017-0582-8 .
Moher D, Schulz KF, Simera I, Altman DG. Guidance for developers of health research reporting guidelines. PLoS Med. 2010;7:e1000217. https://doi.org/10.1371/journal.pmed.1000217 .
Simera I, Moher D, Hirst A, Hoey J, Schulz KF, Altman DG. Transparent and accurate reporting increases reliability, utility, and impact of your research: reporting guidelines and the EQUATOR Network. BMC Med. 2010;8:24. https://doi.org/10.1186/1741-7015-8-24 .
Speich B, Schroter S, Briel M, et al. Impact of a short version of the CONSORT checklist for peer reviewers to improve the reporting of randomised controlled trials published in biomedical journals: study protocol for a randomised controlled trial. BMJ Open. 2020;10:e035114. https://doi.org/10.1136/bmjopen-2019-035114 .
Stevens A, Shamseer L, Weinstein E, et al. Relation of completeness of reporting of health research to journals’ endorsement of reporting guidelines: systematic review. BMJ. 2014;348:g3804. https://doi.org/10.1136/bmj.g3804 .
Hair K, Macleod MR, Sena ES, IICARus Collaboration. A randomised controlled trial of an Intervention to Improve Compliance with the ARRIVE guidelines (IICARus). Res Integr Peer Rev. 2019;4:12. https://doi.org/10.1186/s41073-019-0069-3 .
Blanco D, Altman D, Moher D, Boutron I, Kirkham JJ, Cobo E. Scoping review on interventions to improve adherence to reporting guidelines in health research. BMJ Open. 2019;9:e026589. https://doi.org/10.1136/bmjopen-2018-026589 .
Charters E. The use of think-aloud methods in qualitative research: an introduction to think-aloud methods. Brock Educ J. 2003;12:68–82. https://doi.org/10.26522/brocked.v12i2.38 .
Article Google Scholar
Welch V, Petticrew M, Tugwell P, PRISMA-Equity Bellagio group, et al. PRISMA-equity 2012 extension: reporting guidelines for systematic reviews with a focus on health equity. PLoS Med. 2012;9:e1001333. https://doi.org/10.1371/journal.pmed.1001333 .
Wang X, Chen Y, Liu Y, et al. Reporting items for systematic reviews and meta-analyses of acupuncture: the PRISMA for acupuncture checklist. BMC Complement Altern Med. 2019;19:208. https://doi.org/10.1186/s12906-019-2624-3 .
Download references
Acknowledgements
We dedicate this paper to the late Douglas G Altman and Alessandro Liberati, whose contributions were fundamental to the development and implementation of the original PRISMA statement.
We thank the following contributors who completed the survey to inform discussions at the development meeting: Xavier Armoiry, Edoardo Aromataris, Ana Patricia Ayala, Ethan M Balk, Virginia Barbour, Elaine Beller, Jesse A Berlin, Lisa Bero, Zhao-Xiang Bian, Jean Joel Bigna, Ferrán Catalá-López, Anna Chaimani, Mike Clarke, Tammy Clifford, Ioana A Cristea, Miranda Cumpston, Sofia Dias, Corinna Dressler, Ivan D Florez, Joel J Gagnier, Chantelle Garritty, Long Ge, Davina Ghersi, Sean Grant, Gordon Guyatt, Neal R Haddaway, Julian PT Higgins, Sally Hopewell, Brian Hutton, Jamie J Kirkham, Jos Kleijnen, Julia Koricheva, Joey SW Kwong, Toby J Lasserson, Julia H Littell, Yoon K Loke, Malcolm R Macleod, Chris G Maher, Ana Marušic, Dimitris Mavridis, Jessie McGowan, Matthew DF McInnes, Philippa Middleton, Karel G Moons, Zachary Munn, Jane Noyes, Barbara Nußbaumer-Streit, Donald L Patrick, Tatiana Pereira-Cenci, Ba′ Pham, Bob Phillips, Dawid Pieper, Michelle Pollock, Daniel S Quintana, Drummond Rennie, Melissa L Rethlefsen, Hannah R Rothstein, Maroeska M Rovers, Rebecca Ryan, Georgia Salanti, Ian J Saldanha, Margaret Sampson, Nancy Santesso, Rafael Sarkis-Onofre, Jelena Savović, Christopher H Schmid, Kenneth F Schulz, Guido Schwarzer, Beverley J Shea, Paul G Shekelle, Farhad Shokraneh, Mark Simmonds, Nicole Skoetz, Sharon E Straus, Anneliese Synnot, Emily E Tanner-Smith, Brett D Thombs, Hilary Thomson, Alexander Tsertsvadze, Peter Tugwell, Tari Turner, Lesley Uttley, Jeffrey C Valentine, Matt Vassar, Areti Angeliki Veroniki, Meera Viswanathan, Cole Wayant, Paul Whaley, and Kehu Yang. We thank the following contributors who provided feedback on a preliminary version of the PRISMA 2020 checklist: Jo Abbott, Fionn Büttner, Patricia Correia-Santos, Victoria Freeman, Emily A Hennessy, Rakibul Islam, Amalia (Emily) Karahalios, Kasper Krommes, Andreas Lundh, Dafne Port Nascimento, Davina Robson, Catherine Schenck-Yglesias, Mary M Scott, Sarah Tanveer and Pavel Zhelnov. We thank Abigail H Goben, Melissa L Rethlefsen, Tanja Rombey, Anna Scott, and Farhad Shokraneh for their helpful comments on the preprints of the PRISMA 2020 papers. We thank Edoardo Aromataris, Stephanie Chang, Toby Lasserson and David Schriger for their helpful peer review comments on the PRISMA 2020 papers.
Provenance and peer review
Not commissioned; externally peer reviewed.
Patient and public involvement
Patients and the public were not involved in this methodological research. We plan to disseminate the research widely, including to community participants in evidence synthesis organisations.
There was no direct funding for this research. MJP is supported by an Australian Research Council Discovery Early Career Researcher Award (DE200101618) and was previously supported by an Australian National Health and Medical Research Council (NHMRC) Early Career Fellowship (1088535) during the conduct of this research. JEM is supported by an Australian NHMRC Career Development Fellowship (1143429). TCH is supported by an Australian NHMRC Senior Research Fellowship (1154607). JMT is supported by Evidence Partners Inc. JMG is supported by a Tier 1 Canada Research Chair in Health Knowledge Transfer and Uptake. MML is supported by The Ottawa Hospital Anaesthesia Alternate Funds Association and a Faculty of Medicine Junior Research Chair. TL is supported by funding from the National Eye Institute (UG1EY020522), National Institutes of Health, United States. LAM is supported by a National Institute for Health Research Doctoral Research Fellowship (DRF-2018-11-ST2–048). ACT is supported by a Tier 2 Canada Research Chair in Knowledge Synthesis. DM is supported in part by a University Research Chair, University of Ottawa. The funders had no role in considering the study design or in the collection, analysis, interpretation of data, writing of the report, or decision to submit the article for publication.
Author information
Authors and affiliations.
School of Public Health and Preventive Medicine, Monash University, Melbourne, Australia
Matthew J. Page, Joanne E. McKenzie, Sue E. Brennan & Steve McDonald
Department of Clinical Epidemiology, Biostatistics and Bioinformatics, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam, Netherlands
Patrick M. Bossuyt
Université de Paris, Centre of Epidemiology and Statistics (CRESS), Inserm, F 75004, Paris, France
Isabelle Boutron
Institute for Evidence-Based Healthcare, Faculty of Health Sciences and Medicine, Bond University, Gold Coast, Australia
Tammy C. Hoffmann
Annals of Internal Medicine, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA
Cynthia D. Mulrow
Knowledge Translation Program, Li Ka Shing Knowledge Institute, Toronto, Canada; School of Epidemiology and Public Health, Faculty of Medicine, University of Ottawa, Ottawa, Canada
Larissa Shamseer
Evidence Partners, Ottawa, Canada
Jennifer M. Tetzlaff
Clinical Research Institute, American University of Beirut, Beirut, Lebanon; Department of Health Research Methods, Evidence, and Impact, McMaster University, Hamilton, Ontario, Canada
Elie A. Akl
Department of Medical Informatics and Clinical Epidemiology, Oregon Health & Science University, Portland, OR, USA
York Health Economics Consortium (YHEC Ltd), University of York, York, UK
Julie Glanville
Clinical Epidemiology Program, Ottawa Hospital Research Institute, Ottawa, Canada; School of Epidemiology and Public Health, University of Ottawa, Ottawa, Canada; Department of Medicine, University of Ottawa, Ottawa, Canada
Jeremy M. Grimshaw
Centre for Evidence-Based Medicine Odense (CEBMO) and Cochrane Denmark, Department of Clinical Research, University of Southern Denmark, JB Winsløwsvej 9b, 3rd Floor, 5000 Odense, Denmark; Open Patient data Exploratory Network (OPEN), Odense University Hospital, Odense, Denmark
Asbjørn Hróbjartsson
Department of Anesthesiology and Pain Medicine, The Ottawa Hospital, Ottawa, Canada; Clinical Epidemiology Program, Blueprint Translational Research Group, Ottawa Hospital Research Institute, Ottawa, Canada; Regenerative Medicine Program, Ottawa Hospital Research Institute, Ottawa, Canada
Manoj M. Lalu
Department of Ophthalmology, School of Medicine, University of Colorado Denver, Denver, Colorado, United States; Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA
Tianjing Li
Division of Headache, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA; Head of Research, The BMJ, London, UK
Elizabeth W. Loder
Department of Epidemiology and Biostatistics, Indiana University School of Public Health-Bloomington, Bloomington, Indiana, USA
Evan Mayo-Wilson
Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK
Luke A. McGuinness & Penny Whiting
Centre for Reviews and Dissemination, University of York, York, UK
Lesley A. Stewart
EPPI-Centre, UCL Social Research Institute, University College London, London, UK
James Thomas
Li Ka Shing Knowledge Institute of St. Michael’s Hospital, Unity Health Toronto, Toronto, Canada; Epidemiology Division of the Dalla Lana School of Public Health and the Institute of Health Management, Policy, and Evaluation, University of Toronto, Toronto, Canada; Queen’s Collaboration for Health Care Quality Joanna Briggs Institute Centre of Excellence, Queen’s University, Kingston, Canada
Andrea C. Tricco
Methods Centre, Bruyère Research Institute, Ottawa, Ontario, Canada; School of Epidemiology and Public Health, Faculty of Medicine, University of Ottawa, Ottawa, Canada
Vivian A. Welch
Centre for Journalology, Clinical Epidemiology Program, Ottawa Hospital Research Institute, Ottawa, Canada; School of Epidemiology and Public Health, Faculty of Medicine, University of Ottawa, Ottawa, Canada
David Moher
You can also search for this author in PubMed Google Scholar
Contributions
JEM and DM are joint senior authors. MJP, JEM, PMB, IB, TCH, CDM, LS, and DM conceived this paper and designed the literature review and survey conducted to inform the guideline content. MJP conducted the literature review, administered the survey and analysed the data for both. MJP prepared all materials for the development meeting. MJP and JEM presented proposals at the development meeting. All authors except for TCH, JMT, EAA, SEB, and LAM attended the development meeting. MJP and JEM took and consolidated notes from the development meeting. MJP and JEM led the drafting and editing of the article. JEM, PMB, IB, TCH, LS, JMT, EAA, SEB, RC, JG, AH, TL, EMW, SM, LAM, LAS, JT, ACT, PW, and DM drafted particular sections of the article. All authors were involved in revising the article critically for important intellectual content. All authors approved the final version of the article. MJP is the guarantor of this work. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Corresponding author
Correspondence to Matthew J. Page .
Ethics declarations
Competing interests.
All authors have completed the ICMJE uniform disclosure form at http://www.icmje.org/conflicts-of-interest/ and declare: EL is head of research for the BMJ ; MJP is an editorial board member for PLOS Medicine ; ACT is an associate editor and MJP, TL, EMW, and DM are editorial board members for the Journal of Clinical Epidemiology ; DM and LAS were editors in chief, LS, JMT, and ACT are associate editors, and JG is an editorial board member for Systematic Reviews . None of these authors were involved in the peer review process or decision to publish. TCH has received personal fees from Elsevier outside the submitted work. EMW has received personal fees from the American Journal for Public Health , for which he is the editor for systematic reviews. VW is editor in chief of the Campbell Collaboration, which produces systematic reviews, and co-convenor of the Campbell and Cochrane equity methods group. DM is chair of the EQUATOR Network, IB is adjunct director of the French EQUATOR Centre and TCH is co-director of the Australasian EQUATOR Centre, which advocates for the use of reporting guidelines to improve the quality of reporting in research articles. JMT received salary from Evidence Partners, creator of DistillerSR software for systematic reviews; Evidence Partners was not involved in the design or outcomes of the statement, and the views expressed solely represent those of the author.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1..
PRISMA 2020 checklist.
Additional file 2.
PRISMA 2020 expanded checklist.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Page, M.J., McKenzie, J.E., Bossuyt, P.M. et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Syst Rev 10 , 89 (2021). https://doi.org/10.1186/s13643-021-01626-4
Download citation
Accepted : 04 January 2021
Published : 29 March 2021
DOI : https://doi.org/10.1186/s13643-021-01626-4
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Systematic Reviews
ISSN: 2046-4053
- Submission enquiries: Access here and click Contact Us
- General enquiries: [email protected]
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

PRISMA-S: an extension to the PRISMA Statement for Reporting Literature Searches in Systematic Reviews
Melissa l. rethlefsen.
1 Health Science Center Libraries, George A. Smathers Libraries, University of Florida, Gainesville, USA
Shona Kirtley
2 UK EQUATOR Centre, Centre for Statistics in Medicine (CSM), Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences (NDORMS), Botnar Research Centre, University of Oxford, Windmill Road, Oxford, OX3 7LD UK
Siw Waffenschmidt
3 Institute for Quality and Efficiency in Health Care, Cologne, Germany
Ana Patricia Ayala
4 Gerstein Science Information Centre, University of Toronto, Toronto, Canada
David Moher
5 Centre for Journalology, Clinical Epidemiology Program, Ottawa Hospital Research Institute, The Ottawa Hospital, General Campus, Centre for Practice Changing Research Building, 501 Smyth Road, PO BOX 201B, Ottawa, Ontario K1H 8L6 Canada
Matthew J. Page
6 School of Public Health and Preventive Medicine, Monash University, Melbourne, Australia
Jonathan B. Koffel
7 University of Minnesota, Minneapolis, USA
Associated Data
All data is available via the PRISMA-S PRISMA Search Reporting Extension OSF site (10.17605/OSF.IO/YGN9W) [ 32 ]. This includes all data relating to item development, survey instruments, data from the Delphi surveys, and consent documents.
Literature searches underlie the foundations of systematic reviews and related review types. Yet, the literature searching component of systematic reviews and related review types is often poorly reported. Guidance for literature search reporting has been diverse, and, in many cases, does not offer enough detail to authors who need more specific information about reporting search methods and information sources in a clear, reproducible way. This document presents the PRISMA-S (Preferred Reporting Items for Systematic reviews and Meta-Analyses literature search extension) checklist, and explanation and elaboration.
The checklist was developed using a 3-stage Delphi survey process, followed by a consensus conference and public review process.
The final checklist includes 16 reporting items, each of which is detailed with exemplar reporting and rationale.
Conclusions
The intent of PRISMA-S is to complement the PRISMA Statement and its extensions by providing a checklist that could be used by interdisciplinary authors, editors, and peer reviewers to verify that each component of a search is completely reported and therefore reproducible.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13643-020-01542-z.
Introduction
One crucial component of a systematic review is the literature search. The literature search, or information retrieval process, not only informs the results of a systematic review; it is the underlying process that establishes the data available for analysis. Additional components of the systematic review process such as screening, data extraction, and qualitative or quantitative synthesis procedures are dependent on the identification of eligible studies. As such, the literature search must be designed to be both robust and reproducible to ensure the minimization of bias.
Guidelines exist for both the conduct of literature searches (Table (Table2) 2 ) for systematic reviews and their reporting [ 2 – 7 ]. Problematically, however, the many guidelines for reporting systematic review searches share few common reporting elements. In fact, Sampson et al. discovered that of the eleven instruments designed to help authors report literature searches well, only one item appeared in all eleven instruments [ 8 ]. Though Sampson et al.’s study was conducted in 2007, the problem has only been compounded as new checklists and tools have continued to be developed. The most commonly used reporting guidance for systematic reviews, which covers the literature search component, is the Preferred Reporting Items for Systematic reviews and Meta-Analyses Statement, or PRISMA Statement [ 9 ]. The 2009 PRISMA Statement checklist included three items related to literature search reporting, items 7, 8, and 17:
Item 7: Describe all information sources (e.g., databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched.
Item 8: Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated.
Item 17: Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram.
Despite wide usage of the PRISMA Statement [ 10 ], compliance with its items regarding literature search reporting is low [ 11 – 14 ]. Even for those studies which explicitly reference PRISMA, there is only slight, statistically non-significant evidence of improved reporting, as found by Page et al. [ 15 ]. Part of the challenge may be the multifactorial nature of each of the PRISMA items relating to searches; authors may feel if they completed one of the components of the item that they can check off that item altogether. Another part of the challenge may be that many systematic reviews do not include librarians or information specialists as members of the systematic review team or as authors on the final manuscript [ 11 , 16 – 18 ]. Preliminary research suggests that librarian or information specialist involvement is correlated with reproducibility of searches [ 16 – 18 ], likely due to their expertise surrounding search development and documentation. However, reviews where librarians are authors still include reproducible searches only 64% of the time [ 17 ].
A larger issue may be that, even amongst librarians and information specialists, debate exists as to what constitutes a reproducible search and how best to report the details of the search. Researchers assessing the reproducibility of the search have used varying methods to determine what constitutes a reproducible search [ 11 , 17 , 19 , 20 ]. Post-publication peer review of search methods, even amongst Cochrane reviews, which generally have superior reporting compared to non-Cochrane reviews [ 15 ], has shown that reporting that appears complete may still pose challenges for those wishing to reproduce searches [ 20 – 24 ]. Furthermore, little guidance on how to report searches using information sources or methods other than literature databases, such as searching web sites or study registries, exists [ 25 , 26 ].
Incomplete reporting of the literature search methods can introduce doubt and diminish trust in the final systematic review conclusions. If researchers are unable to understand or reproduce how information was gathered for a systematic review, they may suspect the authors of having introduced bias into their review by not conducting a thorough or pre-specified literature search. After observing the high number of systematic reviews with poorly reported literature searches, we sought to create an extension to the PRISMA statement. Our aims were four-fold:
- To provide extensive guidance on reporting the literature search components of a systematic review.
- To create a checklist that could be used by authors, editors, and peer reviewers to verify that each component of a search was completely reported and therefore reproducible.
- To develop an interdisciplinary checklist applicable to all method-driven literature searches for evidence synthesis.
- To complement the PRISMA Statement and its extensions.
Because we intend the checklist to be used in all fields and disciplines, we use “systematic reviews” throughout this document as a representative name for the entire family of evidence syntheses [ 27 ]. This includes, but is not limited to, scoping reviews, rapid reviews, realist reviews, metanarrative reviews, mixed methods reviews, umbrella reviews, and evidence maps [ 28 ]. We use the term “literature search” or “search” throughout to encompass the full range of possible search methods and information sources.
Part 1: Developing the Checklist
After consultation with members of the PRISMA Statement steering group (D.M. and D.G.A.), we formed an executive committee (M.L.R, J.K., S.K.) and developed a protocol [ 29 ] according to the steps outlined in the “Guidance for Developers of Health Research Reporting Guidelines [ 30 ].” The protocol was registered on the EQUATOR Network [ 29 ]. We identified 405 potential items relevant to reporting searches in systematic reviews from 61 sources (see Additional file 1 ) located through a search of MEDLINE via Ovid, Embase via Embase.com , and LISTA via EBSCOhost, in addition to reviewing all of the sources identified by the EQUATOR Network relating to systematic reviews. We also searched our personal files and examined references of included documents for additional sources. Details of the search are available in Additional file 1 . Sources included both explicit reporting guidelines and studies assessing reproducibility of search strategies. The 405 items were reviewed for overlap and consolidated into 123 remaining items for potential inclusion in a checklist.
To narrow the list into a usable checklist, we then used a three-step Delphi survey process [ 31 ]. The first survey included the initially identified 123 items and asked respondents to rate each item on a 4-point Likert-type scale. Items that 70% of experts rated as 3 or 4 (4 being “essential” and 1 “not important”) and that received a mean score of at least 3.25 were retained for rating in the second round of the Delphi process. Respondents to the first survey were invited to participate in the second and third rounds. The second round asked respondents to pick the 25 most essential items out of the remaining 53 potential items; the third round was identical, except respondents also selected the most appropriate location for reporting their selected items (e.g., in the main text, or a supplementary file). The items were ranked and categorized by general theme for discussion at an in-person consensus conference.
We created a list of one hundred and sixty-three international experts, including librarian and information specialists with expertise in systematic reviews, researchers who had written about systematic review reporting, journal editors, and systematic review methodologists, to whom we sent our initial Delphi survey. The list of experts was created using a combination of publications, mailing lists, conference proceedings, and knowledge of the authors to represent research groups and experts in 23 countries. We received 52 responses (32% response rate) to the first survey, and of these, 35 (67% response rate) completed both surveys two and three. This study was declared exempt by the University of Utah Institutional Review Board (IRB_00088425).
The results of the Delphi process were reported at a consensus conference meeting that took place in May 2016 concurrently with Mosaic ‘16, the joint meeting of the Medical Library Association, Canadian Health Libraries Association/Association des bibliothèques de la santé du Canada, and the International Clinical Librarian Conference (ICLC). 38 individuals attended the consensus conference, 14 (37%) of whom had participated in the Delphi surveys. At the consensus conference, the grouped and ranked remaining items were distributed to small groups who were asked to discuss, consolidate, remove, or add missing critical items under the guidance of a group leader. After two rounds of discussion, the group leaders presented the discussion and proposed list items from their small groups for consideration by the whole group of experts.
Upon completion of the consensus conference, 30 items remained from those identified during the Delphi process, with an additional three items that had been excluded during the Delphi process added back to the draft checklist because meeting attendees considered them critical to the guideline. The list was then consolidated and reviewed by executive committee members, including two new information specialist members (S.W. and A.P.A). The draft checklist and explanation and elaboration document was released to the public on March 20, 2019, along with all data and study materials [ 32 ]. All participants in the Delphi process and/or consensus conference were contacted via email with instructions on how to provide feedback on the draft checklist items and/or elaboration and explanation document by commenting directly on the explanation and elaboration draft using a private commenting system, Hypothesis [ 33 ], or if preferred, via email. Comments from other interested individuals were solicited via Twitter, conference presentations, and personal contacts. Comments were collected from the private Hypothesis group, the public Hypothesis comments, and via email. All comments were combined into a single document. Executive committee members reviewed each comment in duplicate to indicate what type of feedback was received (i.e., linguistic, major substantive, minor substantive, or unclear) and, for substantive comments, whether change was recommended or required further discussion.
During the draft and revision process (March 20–June 15, 2019), 358 separate comments were received from 22 individuals and organizations. Based upon the extensive feedback received, the executive team revised the checklist and developed the next iteration, which was released on December 6, 2019, to coincide with the 2019 Virtual Cochrane Colloquium Santiago. Additional feedback from this release was incorporated into the final checklist. Throughout the draft and revision process, several teleconferences were held with the lead of the PRISMA 2020 statement (M.J.P), as an update of the 2009 PRISMA statement was in development, to ensure that the content on search methods was consistent between the PRISMA 2020 and PRISMA-S guidelines [ 34 , 35 ].
Part 2: Checklist
PRISMA-S is a 16-item checklist that covers multiple aspects of the search process for systematic reviews. It is intended to guide reporting, not conduct, of the search. The checklist should be read in conjunction with the Explanation and Elaboration (Part 3), which provides more detail about each item. We also include two boxes, one a glossary of terms (see Table Table2) 2 ) and the other, guidance on depositing search data and method descriptions in online repositories (see Table Table3 3 ).
Supplementary Materials
The Explanation and Elaboration also includes examples of good reporting for each item. Each exemplar is drawn from published systematic reviews. For clarity, some exemplars are edited to match the style of this document, including any original citations, and abbreviations are spelled out to aid comprehension. Any other edits to the text are noted with square brackets. A description of the rationale behind the item is explained, followed by additional suggestions for clear reporting and a suggested location(s) for reporting the item.
Not every systematic review will make use of all of the items in the Information Sources and Methods section of the checklist, depending on the research question and the methods chosen by the authors. The checklist provides a framework for the current most common and recommended types of information sources and methods for systematic reviews, but authors should use and report those items relevant and appropriate to their review. The checklist may also be used for systematic review protocols to fully document the planned search, in conjunction with the PRISMA-P reporting guideline [ 36 ] (Table (Table1 1 ).
PRISMA-S checklist. A downloadable version of the checklist is available on the PRISMA website [ 37 ]
Part 3: Explanation and Elaboration
Item 1. database name.
Name each individual database searched, stating the platform for each.
“The following electronic databases were searched: MEDLINE (Ovid), CINAHL (EBSCOhost), PsycINFO (Ovid), Cochrane Central Register of Controlled Trials (Ovid), SPORTDiscus (EBSCOhost), EMBASE (Ovid) and ProQuest Dissertations and Theses Global (ProQuest).” [ 38 ]
Explanation
Databases are the most commonly used tool to locate studies to include in systematic reviews and meta-analyses [ 6 , 39 ]. There is no single database that is able to provide a complete and accurate list of all studies that meet systematic review criteria due to the differences in the articles included and the indexing methods used between databases (Table (Table2). 2 ). These differences have led to recommendations that systematic review teams search multiple databases to maximize the likelihood of finding relevant studies [ 6 , 39 , 40 ]. This may include using broad disciplinary databases (e.g., MEDLINE [ 41 ], Embase [ 42 ], Scopus [ 43 ]), specialized databases (e.g., PsycINFO [ 44 ] or EconLit [ 45 ]), or regional databases (e.g., LILACS [ 46 ] or African Index Medicus [ 47 ]).
Many of these literature databases are available through multiple different search platforms (Table (Table2). 2 ). For example, the MEDLINE database is available through at least 10 different platforms, including Ovid, EBSCOhost, Web of Science, and PubMed. Each platform offers different ways of searching the databases, such as platform-specific field codes (Table (Table2), 2 ), phrase searching, truncation, or searching full-text versus abstract and keyword only [ 48 ]. Different platforms may contain additional data that are not available in the original database, such as times cited, social media impact, or additional keywords. These differences between the platforms can have a meaningful impact on the results provided [ 48 – 50 ].
Authors should identify which specific literature databases were searched to locate studies included in the systematic review. It is important that authors indicate not only the database, but the platform through which the database was searched. This helps readers to evaluate the quality and comprehensiveness of the search and supports reproducibility and updating (Table (Table2) 2 ) in the future by allowing the strategy to be copied and pasted as recommended in Item 8, below.
The distinctions between database and platform may not always be clear to authors, especially when the database is the only one available through a platform (e.g., Scopus [ 43 ]). In these cases, authors may choose to include the web address of the database in the text or the bibliography to provide clarity for their readers.
Suggested location for reporting
Report each database name and platform in the methods section and any supplementary materials (Table (Table2). 2 ). If space permits, report key database names in the abstract.
Item 2. Multi-database searching
If databases were searched simultaneously on a single platform, state the name of the platform, listing all of the databases searched.
“The MEDLINE and Embase strategies were run simultaneously as a multi-file search in Ovid and the results de-duplicated using the Ovid de-duplication tool.” [ 51 ]
“A systematic literature search was performed in Web of Knowledge™ (including KCI Korean Journal Database, MEDLINE, Russian Science Citation Index, and SciELO Citation Index)….” [ 52 ]
Authors may choose to search multiple databases at once through a single search platform to increase efficiency. Along with the name of the platform, it is necessary to list the names of each of the individual databases included as part of the search. Including information about using this approach in the text of the manuscript helps readers immediately understand how the search was constructed and executed. This helps readers determine how effective the search strategy (Table (Table2) 2 ) will be for each database [ 1 ].
Report any multi-database search (Table (Table2) 2 ) in the methods section and any supplementary materials. If space permits, report key individual database names in the abstract, even if run through a multi-database search.
Item 3. Study registries
List any study registries searched.
“[We] searched several clinical trial registries ( ClinicalTrials.gov , Current Controlled Trials ( www.controlled-trials.com ), Australian New Zealand Clinical Trials Registry ( www.actr.org.au ), and University Hospital Medical Information Network Clinical Trials Registry ( www.umin.ac.jp/ctr )) to identify ongoing trials.” [ 53 ]
Study registries are a key source of information for systematic reviews and meta-analyses in the health sciences and increasingly in other disciplines. In the health sciences, study registries (Table (Table2) 2 ) allow researchers to locate ongoing clinical trials and studies that may have gone unpublished [ 54 – 56 ]. Some funders, including the National Institutes of Health, require principal investigators to share their data on study registries within a certain time frame after grant completion [ 57 ]. This data may not have been published in any other location, making study registries a critical component of an information strategy, though timely reporting remains a challenge [ 58 , 59 ]. Different countries have their own study registries, as do many pharmaceutical companies.
Outside the health sciences, study registries are becoming increasingly important as many disciplines adopt study pre-registration as a tactic for improving the rigor of research. Though not yet as established as in the health sciences, these study registries are continually expanding and will serve as key sources for finding unpublished studies in fields in the social sciences and beyond.
To fully describe the study registries searched, list the name of each study registry searched, and include a citation or link to the study registry.
Report any study registries searched in the methods section and any supplementary materials.
Item 4. Online resources and browsing
Describe any online or print source purposefully searched or browsed (e.g., tables of contents, print conference proceedings, web sites), and how this was done.
“ We also searched the grey literature using the search string: “public attitudes” AND “sharing” AND “health data” on Google (in June 2017). The first 20 results were selected and screened.” [ 60 ]
“The grey literature search was conducted in October 2015 and included targeted, iterative hand searching of 22 government and/or research organization websites that were suggested during the expert consultation and are listed in S1 Protocol. Twenty two additional citations were added to the review from the grey literature search.” [ 61 ]
“To locate unpublished studies, we searched Embase [via Embase.com ] for conference proceedings since 2000 and hand-searched meeting abstracts of the Canadian Conference on Physician Health and the International Conference on Physician Health (2012 to 2016).” [ 62 ]
Systematic reviews were developed to remove as much bias as possible from the literature review process. One of the most important ways they achieve this reduction in bias is by searching beyond literature databases, which are skewed towards English-language publications with positive results [ 63 , 64 ]. To achieve a fuller picture of what the research on a specific topic looks like, systematic reviewers could seek out research that may be in progress and research that was never published [ 6 ]. Using other methods of finding research also helps identify research that may have been indexed in literature databases, but went undiscovered when searching those sources [ 40 ]. Seeking out this research often involves a complex strategy, drawing on a wealth of online and print resources as well as personal contacts.
Web search engines and specific web sites
Searching general internet search engines and searching the contents of specific websites is a key component of many systematic reviews [ 26 , 65 ]. Government, non-profit organization, and pharmaceutical company websites, for example, contain a wealth of information not published elsewhere [ 6 , 66 ]. Though searching a general search engine like Google or using a general search engine to search a specific website may introduce some bias into the search methodology through the personalization algorithms inherent in many of these tools [ 67 , 68 ], it is still important to fully document how web searches were conducted [ 65 ].
Authors should list all websites searched, along with their corresponding web address. Readers should be able to clearly understand if researchers used a website’s native search interface or advanced search techniques within a general search engine. If authors used a general search engine, authors should declare whether steps were taken to reduce personalization bias (e.g., using “incognito” mode in a browser). Authors may choose whether to detail the websites searched within the text (i.e., Google ( http://www.google.com )), by citing the websites in the bibliography, or by listing the website with corresponding web address in supplementary material, as shown in the examples above.
Review teams may occasionally set an artificial limit to the number of items they will screen from a given search or source [ 65 ]. This is because searching web search engines and individual websites will often lead to an unmanageable number of results, the search engine itself may only display a restricted number of results (e.g., Google will only display 1000 results), or the team has a finite budget or timeline to complete the review. Thus, many systematic review teams utilizing web search engines will often pre-designate a limit to the number of results they review. If review teams choose to review a limited set of results, it should be noted in the text, along with the rationale.
Conference proceedings
Studies show that large percentages of research presented as papers and posters at conferences never make their way into the published literature, particularly if the study’s results were statistically negative [ 63 , 69 ]. Conference proceedings are often the only way to locate these studies. Including conference proceedings in a systematic review search helps minimize bias [ 70 ]. The introduction of online conference proceedings has been a boon to researchers and reduced the need to review printed abstract books. Additionally, some databases either include conference proceedings along with journal articles (i.e., Embase [ 42 ]) or contain only conference proceedings (i.e., ProceedingsFirst [ 71 ] or Directory of Published Papers [ 72 ]). Some conferences have made their abstracts available in a single database (i.e., International AIDS Society’s Abstract Archive [ 73 ]). When using these types of databases to search conference proceedings, authors can treat them as above in Item 1.
Individual conferences’ online proceedings may be password-protected for association members or conference attendees [ 74 ]. When reporting on conference proceedings searched or browsed (Table (Table2) 2 ) via a conference or association’s online or print proceedings, authors must specify the conference names, the dates of conferences included, and the method used to search the proceedings (i.e., browsing print abstract books or using an online source). If the conference proceedings are searched online, authors should specify the web address(es) for the conference proceedings and the date(s) of the conferences. If the conference proceedings are published in a journal, the authors should cite the journal. If the proceedings are a standalone publication, authors may choose to cite them using the same methods used to cite a book or by providing the full information about the conference (name, location, dates, etc.) in a supplementary file.

General browsing
Authors also commonly browse print or online tables of contents, full contents of journals, or other sources that are the most likely to contain research on the topic sought. When purposefully browsing, describe any method used, the name of the journal or other source, and the time frame covered by the search, if applicable.
Report online information sources (Table (Table2) 2 ) searched or browsed in the methods section and in any supplementary materials. Systematic reviews using several of these methods, or using multiple information sources for each method, may need to report their methods briefly in the methods section, but should fully report all necessary information to describe their approaches in supplementary materials.
Item 5. Citation searching
Indicate whether cited references or citing references were examined, and describe any methods used for locating cited/citing references (e.g., browsing reference lists, using a citation index, setting up email alerts for references citing included studies).
“Reference lists of included articles were manually screened to identify additional studies.” [ 75 ]
“[W]e used all shared decision making measurement instruments that were identified in Gärtner et al’s recent systematic review (Appendix A). We then performed a systematic citation search, collecting all articles that cited the original papers reporting on the development, validation, or translation of any the observational and/or self-reported shared decision making measurement instruments identified in that review. An experienced librarian (P.J.E.) searched Web of Science [Science Citation Index] and Scopus for articles published between January 2012 and February 2018.” [ 76 ]
“We [conducted] citation tracking of included studies in Web of Science Core Collection on an ongoing basis, using citation alerts in Web of Science Core Collection.” [ 77 ]
One of the most common search methods is reviewing the references or bibliographies of included studies [ 11 , 17 ]. This type of citation searching (looking for cited references) can be additive to other cited reference searching methods, such as examining bibliographies of relevant systematic reviews. In addition, researchers may choose to look for articles that cite specified studies [ 78 ]. This may include looking beyond one level forwards and backwards (e.g., examining the bibliographies of articles cited by specified articles) [ 78 ]. Looking at bibliographies of included articles or other specified articles is often conducted by examining full-text articles, but it can also be accomplished using online tools called citation indexes (Table (Table2 2 ).
The use of these methods can be complicated to describe, but the explanation should clearly state the database used, if applicable (i.e., Scopus, Google Scholar, Science Citation Index) and describe any other methods used. Authors also must cite the “base” article(s) that citation searching was performed upon, either for examining cited or citing articles (Table (Table2). 2 ). If the same database is used for both a topical search as well as citation searching, describe each use separately. For manually checking the reference lists for included articles, a simple statement as in the first example is sufficient.
Report citation searching details in the methods section and in any supplementary materials.
Item 6. Contacts
Indicate whether additional studies or data were sought by contacting authors, experts, manufacturers, or others.
“We contacted representatives from the manufacturers of erythropoietin-receptor agonists (Amgen, Ortho-Biotech, Roche), corresponding or first authors of all included trials and subject-area experts for information about ongoing studies.” [ 79 ]
“We also sought data via expert requests. We requested data on the epidemiology of injecting drug use and blood-borne viruses in October, 2016, via an email distribution process and social media. This process consisted of initial emails sent to more than 2000 key experts and organisations, including contacts in the global, regional, and country offices of WHO, UNAIDS, Global Fund, and UNODC (appendix p 61). Staff in those agencies also forwarded the request to their colleagues and other relevant contacts. One member of the research team (SL) posted a request for data on Twitter, which was delivered to 5525 individual feeds (appendix p 62).” [ 80 ]
Contacting manufacturers (e.g., pharmaceutical companies), or reaching out to authors or experts directly or through organizations, is a key method to locate unpublished and ongoing studies [ 6 ]. Contacting authors or manufacturers may also be necessary when publications, conference proceedings, or clinical trials registry records do not provide the complete information needed [ 63 , 81 ]. Contacting manufacturers or regulating agencies might be required to acquire complete trial data from the clinical study reports [ 82 , 83 ]. More broad calls for evidence may also be conducted when no specific groups or individuals are targeted.
Contact methods may vary widely, depending on the context, and may include personal contact, web forms, email mailing lists, mailed letters, social media contacts, or other methods. As these strategies are inherently difficult to reproduce, researchers should attempt to give as much detail as possible on what data or information was sought, who or what group(s) provided data or information, and how the individuals or groups were identified.
Report information about contacts to solicit additional information in the methods section and in any supplementary materials. Systematic reviews using elaborate calls for evidence or making extensive use of contacts as an information source may need to report their methods briefly in the methods section, but should fully report all necessary information to describe their approaches in supplementary materials.
Item 7. Other methods
Describe any additional information sources or search methods used.
“We also searched… our personal files.” [ 84 ]
“PubMed’s related articles search was performed on all included articles.” [ 85 ]
A thorough systematic review may utilize many additional methods of locating studies beyond database searching, many of which may not be reproducible methods. A key example is searching personal files. Another is using databases’ built in tools, such as PubMed’s Related Articles feature [ 86 ] or Clarivate Analytics’ Web of Science’s Related Records feature [ 87 ], to locate relevant articles based on commonalities with a starting article. Because these tools are often proprietary and their algorithms opaque, researchers may not be able to replicate the exact results at a later date. To attempt to be as transparent as possible, researchers should both note the tool that was used and cite any articles these operations were run upon. For all “other” methods, it is still important to declare that the method was used, even if it may not be fully replicable.
Report information about any other additional information sources or search methods used in the methods section and in any supplementary materials.
Item 8. Full search strategies
Include the search strategies for each database and information source, copied and pasted exactly as run.
Database search. Methods section description . “The reproducible searches for all databases are available at DOI:10.7302/Z2VH5M1H.” [ 88 ]
Database search. One of the full search strategies from supplemental materials in online repository . “ Embase.com (692 on Jan 19, 2017) 'social media'/exp OR (social NEAR/2 (media* OR medium* OR network*)):ti OR twitter:ti OR youtube:ti OR facebook:ti OR linkedin:ti OR pinterest:ti OR microblog*:ti OR blog:ti OR blogging:ti OR tweeting:ti OR 'web 2.0':ti 'professionalism'/exp OR 'ethics'/exp OR 'professional standard'/de OR 'professional misconduct'/de OR ethic*:ab,ti OR unprofessional*:ab,ti OR professionalism:ab,ti OR (professional* NEAR/3 (standard* OR misconduct)):ab,ti OR ((professional OR responsib*) NEAR/3 (behavi* OR act OR conduct*)):ab,ti #1 AND #2 AND [english]/lim NOT ('conference abstract':it OR 'conference paper':it) [ 88 ]
Online resources and browsing. Methods section description . “The approach to study identification from this systematic review is transparently reported in the Electronic Supplementary Material Appendix S1.” [ 89 ]
Online resources and browsing. One of the full online resource search strategies reported in supplement . “Date: 12/01/16. Portal/URL: Google. https://www.google.co.uk/webhp?hl=en . Search terms: ((Physical training) and (man or men or male or males) and (female or females or women or woman) and (military)). Notes: First 5 pages screened on title (n=50 records).” [ 89 ]
Systematic reviews and related review types rely on thorough and complex search strategies to identify literature on a given topic. The search strategies used to conduct this data gathering are essential to the transparency and reproducibility of any systematic review. Without being able to assess the quality of the search strategies used, readers are unable to assess the quality of the systematic review [ 9 , 11 , 17 ].
When space was at a premium in publications, complete reporting of search strategies was a challenge. Because it was necessary to balance the need for transparency with publication restrictions, previous PRISMA guidelines recommended including the complete search strategy from a minimum of one database searched [ 9 ]. Many systematic reviews therefore reported only the minimum necessary. However, reporting only selected search strategies can contribute to the observed irreproducibility of many systematic reviews [ 11 , 17 ].
The prior versions of PRISMA did not elaborate on methods for reporting search strategies outside of literature databases. Subsequent to its publication, many groups have begun identifying the challenges of fully documenting other types of search methods [ 90 , 91 ]. Now recommended is the explicit documentation of all of the details of all search strategies undertaken [ 91 , 92 ]. These should be reported to ensure transparency and maximum reproducibility, including searches and purposeful browsing activities undertaken in web search engines, websites, conference proceeding databases, electronic journals, and study registries.
Journal restrictions vary, but many journals now allow authors to publish supplementary materials with their manuscripts. At minimum, all search strategies, including search strategies for web search engines, websites, conference proceedings databases, electronic journals, and study registries, should be submitted as a supplement for publication. Though most supplements are appropriately accessible on journal publishers’ web sites as submitted, supplements may disappear [ 17 ]. In addition, many supplements are only available to journal subscribers [ 93 ]. Similarly, manuscripts available on public access systems like PubMed Central [ 94 ] may not have the corresponding supplemental materials properly linked. For optimal accessibility, authors should upload complete documentation to a data repository (Table (Table2), 2 ), an institutional repository, or other secure and permanent online archive instead of relying on journal publication (see Table Table3 3 for additional information).
It is important to document and report the search strategy exactly as run, typically by copying and pasting the search strategy directly as entered into the search platform. This is to ensure that information such as the fields searched, term truncation, and combinations of terms (i.e., Boolean logic or phrases) are accurately recorded. Many times, the copied and pasted version of a search strategy will also include key information such as limits (see Item 9; Table Table2) 2 ) used, databases searched within a multi-database search, and other database-specific detail that will enable more accurate reporting and greater reproducibility. This documentation must also repeat the database or resource name, database platform or web address, and other details necessary to clearly describe the resource.
Report the full search strategy in supplementary materials as described above. Describe and link to the location of the supplementary materials in the methods section.
Item 9: Limits and restrictions
Specify that no limits were used, or describe any limits or restrictions applied to a search (e.g., date or time period, language, study design) and provide justification for their use.
No limits . “We imposed no language or other restrictions on any of the searches.” [ 95 ]
Limits described without justification . “The search was limited to the English language and to human studies.” [ 96 ]
“The following search limits were then applied: randomized clinical trials (RCTs) of humans 18 years or older, systematic reviews, and meta-analyses.” [ 97 ]
Limits described with justification . “The search was limited to publications from 2000 to 2018 given that more contemporary studies included patient cohorts that are most reflective of current co-morbidities and patient characteristics as a result of the evolving obesity epidemic.” [ 98 ]
Limits described, one with justification . “Excluded publication types were comments, editorials, patient education handouts, newspaper articles, biographies, autobiographies, and case reports. All languages were included in the search result; non-English results were removed during the review process…. To improve specificity, the updated search was limited to human participants.” [ 99 ]
Many databases have features that allow searchers to quickly restrict a search using limits. What limits are available in a database are unique to both the database and the platform used to search it. Limits are dependent on the accuracy of the indexer, the timeliness of indexing, and the quality of any publisher-supplied data. For instance, using database limits to restrict searches to randomized controlled trials will only find records identified by the indexer as randomized controlled trials. Since the indexing may take 6 months or more to complete for any given article, searchers risk missing new articles when using database limits.
Using database-provided limit features should not be confused with using filters (see Item 10; Table Table2) 2 ) or inclusion criteria for the systematic review. For example, systematic review teams may choose to only include English-language randomized controlled trials. This can be done using limits, a combination of a filter (see Item 10) and screening, or screening alone. It should be clear to the reader which approach is used. For instance, in the “ Limits described, with one justification ” example above, the authors used database limits to restrict their search by publication type, but they did not use database limits to restrict by language, even though that was a component of their eligibility criteria. They also used database limits to restrict to human participants in their search update.
It is important for transparency and reproducibility that any database limits applied when running the search are reported accurately, as their use has high potential for introducing bias into a search [ 1 , 64 , 100 , 101 ]. Database limits are not recommended for use in systematic reviews, due to their fallibility [ 39 , 100 ]. If used, review teams should include a statement of justification for each use of a database limit in the methods section, the limitations section, or both [ 102 , 103 ]. In the examples above, only the last two examples provide some justification in the methods section (“to improve specificity” [ 99 ] and “contemporary studies included patient cohorts that are most reflective of current co-morbidities and patient characteristics as a result of the evolving obesity epidemic” [ 98 ]).
Report any limits or restrictions used or that no limits were used in the abstract, methods section, and in any supplementary materials, including the full search strategies (Item 8). Report the justification for any limits used within the methods section and/or in the limitations section.
Item 10. Search filters
Indicate whether published search filters were used (as originally designed or modified), and if so, cite the filter(s) used.
“For our MEDLINE search we added a highly sensitive filter for identifying randomised trials developed by the Cochrane Collaboration [38]. For Embase we used the filter for randomised trials proposed by the Scottish Intercollegiate Guidelines Network [ 104 ].” [ 105 ]
Filters are a predefined combination of search terms developed to identify references with a specific content, such as a particular type of study design (e.g., randomized controlled trials) [ 106 ], populations (e.g., the elderly), or a topic (e.g., heart failure) [ 107 ]. They often consist of a combination of subject headings, free-text terms, and publication types [ 107 ]. For systematic reviews, filters are generally recommended for use instead of limits built into databases, as discussed in Item 9, because they provide the much higher sensitivity (Table (Table2) 2 ) required for a comprehensive search [ 108 ].
Any filters used as part of the search strategy should be cited, whether published in a journal article or other source. This enables readers to assess the quality of the filter(s) used, as most published search filters are validated and/or peer reviewed [ 106 , 107 ]. Many commonly used filters are published on the InterTASC Information Specialists’ Sub-Group [ 109 ], in the Cochrane Handbook [ 4 , 39 ], and through the Health Information Research Unit of McMaster University [ 110 ].
Cite any search filter used in the methods section and describe adaptations made to any filter. Include the copied and pasted details of any search filter used or adapted for use as part of the full search strategy (Item 8).
Item 11. Prior work
Indicate when search strategies from other literature reviews were adapted or reused for a substantive part or all of the search, citing the previous review(s).
“We included [search strategies] used in other systematic reviews for research design [ 111 ], setting [ 112 , 113 ], physical activity and healthy eating [ 114 – 116 ], obesity [ 111 ], tobacco use prevention [ 117 ], and alcohol misuse [ 118 ]. We also used a search [strategy] for intervention (implementation strategies) that had been employed in previous Cochrane Reviews [ 119 , 120 ], and which was originally developed based on common terms in implementation and dissemination research.” [ 121 ]
Many authors may also examine previously published search strategies to develop the search strategies for their review. Sometimes, authors adapt or reuse these searches for different systematic reviews [ 122 ]. When basing a new search strategy on a published search strategy, it is appropriate to cite the original publication(s) consulted.
Search strategies differ from filters (Item 10) because search strategies are often developed for a specific project, not necessarily designed to be repeatedly used. Filters, on the other hand, are developed with the express purpose of reuse. Filters are often objectively derived, tested, and validated, whereas most search strategies published as part of systematic review or other evidence synthesis are “best guess,” relying on the expertise of the searcher and review team [ 107 ].
As in the example above, researchers may rely on multiple prior published searches to construct a new search for a novel review. Many times, researchers will use the same searches from a published systematic review to update the existing systematic review. In either case, it is helpful to the readers to understand whether major portions of a search are being adapted or reused.
Report any prior work consulted, adapted, or reused in the methods section. Include the copied and pasted search strategies used, including portions or the entirety of any prior work used or adapted for use, in the full search strategy (Item 8).
Item 12. Updates
Report the methods used to update the search(es) (e.g., rerunning searches, email alerts).
“Ovid Auto Alerts were set up to provide weekly updates of new literature until July 09, 2012.” [ 123 ]
“ Two consecutive searches were conducted and limited by publication type and by date, first from January 1, 1990, to November 30, 2012, and again from December 1, 2012, to July 31, 2015, in an updated search…. The original search strategy was used to model the updated search from December 1, 2012, to July 31, 2015. The updated search strategy was consistent with the original search; however, changes were required in the ERIC database search because of a change in the ERIC search algorithm. Excluded publication types were identical to the initial search. To improve specificity, the updated search was limited to human participants.” [ 99 ]
The literature search is usually conducted at the initial stage of the production of a systematic review. As a consequence, the results of a search may be outdated before the review is published [ 124 – 126 ]. The last search in a review should be conducted ideally less than 6 months before publication [ 90 , 92 , 125 ]. For this reason, authors often update searches by rerunning (Table (Table2) 2 ) the same search(es) or otherwise updating searches before the planned publication date. Updating searches differs from updating a systematic review, i.e., when the same or different authors or groups decide to redo a published systematic review to bring its findings up to date. If authors are updating a published systematic review, either authored by the same review team or another, Item 11 contains relevant guidance.
When reporting search updates, the extent of reporting depends on methods used and any changes that were made while updating the searches. If there are no changes in information sources and/or search syntax (Table (Table2), 2 ), it is sufficient to indicate the date the last search was run in the methods section and in the supplementary materials. If there are any changes in information sources and/or search syntax, the changes should be indicated (e.g., different set of databases, changes in search syntax, date restrictions) in the methods section. Authors should explain why these changes were made. When there were changes in the search strategy syntax, the original and the updated searches should both be reported as described in Item 8.
If authors use email alerts or other methods to update searches, these methods can be briefly described by indicating the method used, the frequency of any updates, the name of the database(s) used, or other relevant information that will help readers understand how the authors conducted search updates. If deduplication methods are used as part of the search update process, these methods can be described using guidance in Item 16.
Report the methods used to update the searches in the methods section and the supplementary materials, as described above.
Item 13. Dates of searches
For each search strategy, provide the date when the last search occurred.
“A comprehensive literature search was initially run on 26 February 2017 and then rerun on 5 February 2018….” [ 127 ]
Most literature databases are regularly updated with new citations as articles are published. Citations already in the database may also be updated once new information (such as indexing terms or citing articles) is available. As an example, MEDLINE added over 900,000 indexed citations (Table (Table2) 2 ) in fiscal year 2018 [ 41 ]. In addition, the information gathered by databases (such as author affiliations in MEDLINE) can change over time. Because new citations are regularly being added, systematic review guidelines recommend updating searches throughout the writing process to ensure that all relevant articles are retrieved [ 6 , 92 ].
It is necessary for authors to document the date when searches were executed, either the date the initial search was conducted, if only searched once, or the most recent date the search was rerun. This allows readers to evaluate the currency of each search and understand what literature the search could have potentially identified [ 125 ]. In addition, it supports reproducibility and updating by allowing other researchers to use date limits to view the same “slice” of the database that the original authors used or to update a systematic review by searching from the last time point searched.
Report the date of the last search of the primary information sources used in the abstract for optimal clarity for readers [ 128 ]. Report the time frame during which searches were conducted, the initial search date(s), and/or the last update search date(s) in the methods section. Report the initial and/or last update search date with each complete search strategy in the supplementary materials, as in the examples for Item 8.
Item 14. Peer review
Describe any search peer review process.
“The strategies were peer reviewed by another senior information specialist prior to execution using the PRESS Checklist [ 1 ].” [ 129 ]
Peer reviewing search strategies is an increasingly valued component of search strategy development for systematic reviews. Expert guidance recommends taking this step to help increase the robustness of the search strategy [ 6 , 74 ]. Peer reviewing (Table (Table2) 2 ) searches is useful to help to guide and improve electronic search strategies. One of peer review’s main benefits is the reduction of errors [ 23 , 130 ]. Peer review may also increase the number of relevant records found for inclusion in reviews, thus improving the overall quality of the systematic review [ 131 ].
Authors should consider using the Peer Review of Electronic Search Strategies (PRESS) Guideline statement, a practice guideline for literature search peer review outlining the major components important to review and the benefits of peer reviewing searches [ 1 ]. Authors should strongly consider having the search strategy peer reviewed by an experienced searcher, information specialist, or librarian [ 1 , 131 ]. Though peer review may be conducted generally with publication of a protocol, for example, this item is designed to document search-specific peer review.
Describe the use of peer review in the methods section.
Item 15. Total records
Document the total number of records identified from each database and other information sources.
Methods section . “A total of 3251 citations were retrieved from the six databases and four grey literature websites.” [ 133 ] Flow diagram . Fig. Fig.1 1 . Open in a separate window Fig. 1 “Figure 1. PRISMA 2009 flow diagram” [ 132 ]
Recording the flow of citations through the systematic review process is a key component of the PRISMA Statement [ 9 , 35 ]. It is helpful to identify how many records (Table (Table2) 2 ) were identified within each database and additional source. Readers can use this information to see whether databases or expert contacts constituted the majority of the records reviewed, for example. Knowing the number of records from each source also helps with reproducibility. If a reader tries to duplicate a search from a systematic review, one would expect to retrieve nearly the same results when limiting to the timeframe in the original review. If instead, the searcher locates a drastically different number of results than reported in the original review, this can be indicative of errors in the published search [ 23 ] or major changes to a database, both of which might be reasons to update a systematic review or view the systematic review’s results with skepticism.
Report the total number of references retrieved from all sources, including updates, in the results section. Report the total number of references from each database and information source in the supplementary materials. If space permits, report the total number of references from each database in the PRISMA flow diagram [ 35 ].
Item 16. Deduplication
Describe the processes and any software used to deduplicate records from multiple database searches and other information sources.
“Duplicates were removed by the librarians (LP, PJE), using EndNote's duplicate identification strategy and then manually.” [ 134 ]
Databases contain significant overlap in content. When searching in multiple databases and additional information sources, as is necessary for a systematic review, authors often employ a variety of techniques to reduce the number of duplicates within their results prior to screening [ 135 – 138 ]. Techniques vary in their efficacy, sensitivity, and specificity (Table (Table2) 2 ) [ 136 , 138 ]. Knowing which method is used enables readers to evaluate the process and understand to what extent these techniques may have removed false positive duplicates [ 138 ]. Authors should describe and cite any software or technique used, when applicable. If duplicates were removed manually, authors should include a description.
Report any deduplication method used in the methods section. The total number of references after deduplication should be reported in the PRISMA flow diagram [ 35 ].
Part 5. Discussion and conclusions
The PRISMA-S extension is designed to be used in conjunction with PRISMA 2020 [ 35 ] and PRISMA extensions including PRISMA-P for protocols [ 36 ], PRISMA-ScR for scoping reviews [ 139 ], the PRISMA Network Meta-analyses statement [ 140 ], and PRISMA-IPD for systematic reviews using individual patient data [ 141 ]. It may also be used with other reporting guidelines that relate to systematic reviews and related review types, such as RepOrting standards for Systematic Evidence Syntheses (ROSES) [ 142 ]. It provides additional guidance for systematic review teams, information specialists, librarians, and other researchers whose work contains a literature search as a component of the research methods. Though its origins are in the biomedical fields, PRISMA-S is flexible enough to be applied in all disciplines that use method-driven literature searching. Ultimately, PRISMA-S attempts to give systematic review teams a framework that helps ensure transparency and maximum reproducibility of the search component of their review.
PRISMA-S is intended to capture and provide specific guidance for reporting the most common methods used in systematic reviews today. As new methods and information sources are adopted, authors may need to adjust their reporting methods to accommodate new processes. Currently, PRISMA-S does not address using text mining or text analysis methods to create the search, for example, though this is an increasingly common way for information specialists to develop robust and objective search strategies [ 143 – 145 ]. Likewise, PRISMA-S does not require that decisions about the rationale behind choices in search terms and search construction be recorded, though this provides readers a great deal of insight. In the future, methods and rationales used to create search strategies may become more important for reproducibility.
PRISMA-S offers extensive guidance for many different types of information source and methods, many of them not described in detail in other reporting guidelines relating to literature searching. This includes detailed information on reporting study registry searches, web searches, multi-database searches, and updates. PRISMA-S can help authors report all components of their search, hopefully making the reporting process easier. As a note, PRISMA-S provides guidance on transparent reporting to authors and is not intended as a tool to either guide conduct of a systematic review or to evaluate the quality of a search or a systematic review.
The PRISMA-S checklist is available for download in Word and PDF formats from the PRISMA Statement web site [ 37 ]. The checklist should be used together with its Explanation & Elaboration documentation to provide authors with guidance for the complexities of different types of information sources and methods.
We intend to work with systematic review and information specialist organizations to broadly disseminate PRISMA-S and encourage its adoption by journals. In addition, we plan to host a series of webinars discussing how to use PRISMA-S most effectively. These webinars will also be available for later viewing and will serve as a community resource.
We hope that journal editors will recommend authors of systematic reviews and other related reviews to use PRISMA-S and submit a PRISMA-S checklist with their manuscripts. We also hope that journal editors will encourage more stringent peer review of systematic review searches to ensure greater transparency and reproducibility within the review literature.
Acknowledgements
We would like to thank all of the members of the PRISMA-S Group, which is comprised of participants in the Delphi process, consensus conference, or both. PRISMA-S Group members include Heather Blunt (Dartmouth College), Tara Brigham (Mayo Clinic in Florida), Steven Chang (La Trobe University), Justin Clark (Bond University), Aislinn Conway (BORN Ontario and CHEO Research Institute), Rachel Couban (McMaster University), Shelley de Kock (Kleijnen Systematic Reviews Ltd), Kelly Farrah (Canadian Agency for Drugs and Technologies in Health (CADTH)), Paul Fehrmann (Kent State University), Margaret Foster (Texas A & M University), Susan A. Fowler (Washington University in St. Louis), Julie Glanville (University of York), Elizabeth Harris (La Trobe University), Lilian Hoffecker (University of Colorado Denver), Jaana Isojarvi (Tampere University), David Kaunelis (Canadian Agency for Drugs and Technologies in Health (CADTH)), Hans Ket (VU Amsterdam), Paul Levay (National Institute for Health and Care Excellence (NICE)), Jennifer Lyon, Jessie McGowan (uOttawa), M. Hassan Murad (Mayo Clinic), Joey Nicholson (NYU Langone Health), Virginia Pannabecker (Virginia Tech), Robin Paynter (VA Portland Health Care System), Rachel Pinotti (Icahn School of Medicine at Mount Sinai), Amanda Ross-White (Queens University), Margaret Sampson (CHEO), Tracy Shields (Naval Medical Center Portsmouth), Adrienne Stevens (Ottawa Hospital Research Institute), Anthea Sutton (University of Sheffield), Elizabeth Weinfurter (University of Minnesota), Kath Wright (University of York), and Sarah Young (Carnegie Mellon University). We would also like to thank Kate Nyhan (Yale University), Katharina Gronostay (IQWiG), the many others who contributed to the PRISMA-S project anonymously or as draft reviewers, and our peer reviewers. We would like to give special thanks to the late Douglas G. Altman (D.G.A.; University of Oxford) for his support and guidance, and the co-chairs of the Medical Library Association’s Systematic Reviews SIG in 2016, Margaret Foster (Texas A & M University) and Susan Fowler (Washington University in St. Louis), for allowing us to use one of their meeting times for the consensus conference.
Abbreviations
Authors’ contributions.
M.L.R. conceived and designed the study, conducted the thematic and quantitative analyses, curated the data, drafted the manuscript, and reviewed and edited the manuscript. M.L.R. is the guarantor. J.B.K. and S.K. contributed to the design of the study, developed the literature search strategies, contributed to the thematic content analyses, drafted a portion of the Elaboration & Explanation, and reviewed and edited the manuscript. J.B.K. developed the survey instrument. M.L.R., J.B.K., and S.K. hosted and organized the consensus conference. S.W. and A.P.A. contributed to the thematic content analysis, drafted a portion of the Elaboration & Explanation, and reviewed and edited the manuscript. S.W. supervised the draft revision documentation. D.M. helped conceive and design the study. M.J.P. provided substantive review and editing of the checklist, Explanation & Elaboration, and final manuscript. The author (s) read and approved the final manuscript.
Melissa Rethlefsen was funded in part by the University of Utah’s Center for Clinical and Translational Science under the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR002538 in 2017–2018 . The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Shona Kirtley was funded by the Cancer Research UK (grant C49297/A27294). The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed are those of the authors and not necessarily those of the Cancer Research UK.
Matthew Page is supported by an Australian Research Council Discovery Early Career Researcher Award (DE200101618).
David Moher is supported by a University Research Chair, University of Ottawa, Ottawa, Canada.
The consensus conference was sponsored by the Systematic Reviews SIG of the Medical Library Association. There was no specific funding associated with this event.
Availability of data and materials
Ethics approval and consent to participate.
This study was declared exempt by the University of Utah Institutional Review Board (IRB_00088425). Consent was received from all survey participants.
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests. MJP and DM are leading the PRISMA 2020 update.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Melissa L. Rethlefsen, Email: moc.liamg@nesfelhterlm .
Shona Kirtley, Email: [email protected] .
Siw Waffenschmidt, Email: [email protected] .
Ana Patricia Ayala, Email: [email protected] .
David Moher, Email: ac.irho@rehomd .
Jonathan B. Koffel, Email: ude.nmu@leffokbj .
PRISMA-S Group: Heather Blunt , Tara Brigham , Steven Chang , Justin Clark , Aislinn Conway , Rachel Couban , Shelley de Kock , Kelly Farrah , Paul Fehrmann , Margaret Foster , Susan A. Fowler , Julie Glanville , Elizabeth Harris , Lilian Hoffecker , Jaana Isojarvi , David Kaunelis , Hans Ket , Paul Levay , Jennifer Lyon , Jessie McGowan , M. Hassan Murad , Joey Nicholson , Virginia Pannabecker , Robin Paynter , Rachel Pinotti , Amanda Ross-White , Margaret Sampson , Tracy Shields , Adrienne Stevens , Anthea Sutton , Elizabeth Weinfurter , Kath Wright , and Sarah Young
Proceedings of the International Conference on Economic Management, Accounting and Tourism (ICEMAT 2023)
A Systematic Literature Review on Entrepreneurial Intention: PRISMA method
Several studies use Entrepreneurial Intention as a theoretical framework. Thus, it requires a more extension of Entrepreneurial Intention research to be investigated. The Systematic Literature Review (SLR) is one of the methods to find the research gaps. However, none of the researchers comprehensively conducted a Systematic Literature Review on Entrepreneurial Intention. Therefore, this study aims to review the main finding, database, citation, study setting, methodology, unit of analysis, field of study analysis, and underpinning theory of previous studies on Entrepreneurial Intention. A total of 152 articles addressing Entrepreneurial Intention published between 2000 and 2021 have been analyzed. The present study identifies Self-Efficacy as the most often used by authors as an antecedent of Entrepreneurial Intention. Most of the Entrepreneurial Intention research was conducted in a quantitative method specified in the survey method and most of the researchers used Structural Equation Modelling and Multiple Regression for data analysis. Most study has been done in multi-country, dominated by students as a unit of analysis and management, education, business, and economics are the fields of study that frequently use Entrepreneurial Intention as the topic of research. Almost half of the Entrepreneurial Intention research uses the Theory of Planned Behaviour (TPB) as the underpinning theory of the research.
Download article (PDF)
Cite this article
To read this content please select one of the options below:
Please note you do not have access to teaching notes, regulatory framework on governing equity crowdfunding: a systematic literature review and future directions.
Journal of Financial Regulation and Compliance
ISSN : 1358-1988
Article publication date: 16 May 2024
The purpose of this study is to comprehensively analyse and compare equity crowdfunding (ECF) regulations across 26 countries, shedding light on the diverse regulatory frameworks, investor and issuer limits and the evolution of ECF globally. By addressing this research gap and providing consolidated insights, the study aims to inform policymakers, researchers and entrepreneurs about the regulatory landscape of ECF, fostering a deeper understanding of its potential and challenges in various economies. Ultimately, the study contributes to the advancement of ECF as an alternative financing method for small and medium enterprises (SMEs) and startups, empowering them to access much-needed capital for growth.
Design/methodology/approach
The study used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) model for a systematic literature review on global ECF regulations. Starting with 74 initial articles from Web of Sciences and Scopus databases, duplicates were removed and language criteria applied, leaving 42 articles. After a thorough full-text screening, 20 articles were excluded, resulting in the review of 22 papers from 2016 to 2022. PRISMA’s structured framework enhances the quality of systematic reviews, ensuring transparency and accessibility of findings for various stakeholders, including researchers, practitioners and policymakers, in the field of ECF regulations.
This study examines ECF regulations across various countries. Notably, the UK has advanced regulations, while the USA adopted them later through the Jumpstart Our Business Startups Act. Canada regulates at the provincial level. Malaysia and China were early adopters in Asia, but Hong Kong, Japan, Israel and India have bans. Turkey introduced regulations in 2019. New Zealand and Australia enacted laws, with Australia referring to it as “crowd-sourced equity funding”. Italy, Austria, France, Germany and Belgium have established regulations in Europe. These regulations vary in investor and issuer limits, disclosure requirements and anti-corruption measures, impacting the growth of ECF markets.
Research limitations/implications
This study’s findings underscore the diverse regulatory landscape governing ECF worldwide. It reveals that regulatory approaches vary from liberal to protectionist, reflecting each country’s unique economic and political context. The implications of this research highlight the need for cross-country analysis to inform practical implementation and the effectiveness of emerging ECF ecosystems. This knowledge can inspire regulatory adjustments, support startups and foster entrepreneurial growth in emerging economies, ultimately reshaping early-stage funding for new-age startups and SMEs on a global scale.
Originality/value
This study’s originality lies in its comprehensive analysis of ECF regulations across 26 diverse countries, shedding light on the intricate interplay between regulatory frameworks and a nation’s political-economic landscape. By delving into the nuanced variations in investor limits, investment types and regulatory strategies, it unveils the multifaceted nature of ECF regulation globally. Furthermore, this research adds value by comparing divergent perspectives on investment constraints and offering an understanding of their impact on ECF efficacy. Ultimately, the study’s unique contribution lies in its potential to inform practical implementation, shape legislative frameworks and catalyse entrepreneurial ecosystems in emerging economies, propelling the evolution of early-stage funding practices.
- Systematic literature review
- Crowdfunding
- Equity crowdfunding
- Regulations
- Entrepreneurial finance
Gupta, P. , Singh, S. , Ghosh, R. , Kumar, S. and Jain, C. (2024), "Regulatory framework on governing equity crowdfunding: a systematic literature review and future directions", Journal of Financial Regulation and Compliance , Vol. ahead-of-print No. ahead-of-print. https://doi.org/10.1108/JFRC-10-2023-0160
Emerald Publishing Limited
Copyright © 2024, Emerald Publishing Limited
Related articles
We’re listening — tell us what you think, something didn’t work….
Report bugs here
All feedback is valuable
Please share your general feedback
Join us on our journey
Platform update page.
Visit emeraldpublishing.com/platformupdate to discover the latest news and updates
Questions & More Information
Answers to the most commonly asked questions here
- Open access
- Published: 16 May 2024
DNA methylation and type 2 diabetes: a systematic review
- Nikhil Nadiger 1 , 2 ,
- Jyothisha Kana Veed 2 ,
- Priyanka Chinya Nataraj 2 nAff3 &
- Arpita Mukhopadhyay 2
Clinical Epigenetics volume 16 , Article number: 67 ( 2024 ) Cite this article
Metrics details
DNA methylation influences gene expression and function in the pathophysiology of type 2 diabetes mellitus (T2DM). Mapping of T2DM-associated DNA methylation could aid early detection and/or therapeutic treatment options for diabetics.
A systematic literature search for associations between T2DM and DNA methylation was performed. Prospero registration ID: CRD42020140436.
PubMed and ScienceDirect databases were searched (till October 19, 2023). Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines and New Castle Ottawa scale were used for reporting the selection and quality of the studies, respectively.
Thirty-two articles were selected. Four of 130 differentially methylated genes in blood, adipose, liver or pancreatic islets ( TXNIP , ABCG1 , PPARGC1A , PTPRN2 ) were reported in > 1 study. TXNIP was hypomethylated in diabetic blood across ethnicities. Gene enrichment analysis of the differentially methylated genes highlighted relevant disease pathways (T2DM, type 1 diabetes and adipocytokine signaling). Three prospective studies reported association of methylation in IGFBP2 , MSI2 , FTO , TXNIP , SREBF1 , PHOSPHO1 , SOCS3 and ABCG1 in blood at baseline with incident T2DM/hyperglycemia. Sex-specific differential methylation was reported only for HOOK2 in visceral adipose tissue (female diabetics: hypermethylated, male diabetics: hypomethylated). Gene expression was inversely associated with methylation status in 8 studies, in genes including ABCG1 (blood), S100A4 (adipose tissue), PER2 (pancreatic islets), PDGFA (liver) and PPARGC1A (skeletal muscle).
This review summarizes available evidence for using DNA methylation patterns to unravel T2DM pathophysiology. Further validation studies in diverse populations will set the stage for utilizing this knowledge for identifying early diagnostic markers and novel druggable pathways.
Introduction
Type 2 diabetes mellitus (T2DM) is a disorder of genetic and environmental factors. It is projected to affect 693 million people worldwide by 2045 [ 1 ]. DNA methylation had been proposed as one of the epigenetic phenomena for explaining the missing heritability of T2DM, as multiple, large genome-wide association studies have been able to account for only < 20% of the estimated T2DM heritability [ 2 ]. DNA methylation is an epigenetic phenomenon in which the C5 carbon of the cytosine residue is attached to a methyl group, predominantly in cytosine-phosphate-guanine (CpG) sites [ 3 , 4 , 5 ]. This epigenetic alteration influences gene expression, and thereby, gene function [ 6 , 7 ].
DNA methylation has been studied extensively in relation to T2DM, and 3 systematic reviews have summarized the findings a few years back [ 8 , 9 , 10 ]. From systematic literature done till August 2015, Muka et al. [ 10 ] could not find any consistent association between global DNA methylation with T2DM, glucose, insulin and insulin resistance and reported epigenetic regulation of few candidate genes in blood cells, muscle, adipose tissue and placenta without any overlap between them. Walaszczyk et al . [ 9 ] could replicate association of methylation with T2DM in blood samples from the Lifelines study at 5 CpGs (in ABCG1 , LOXL2 , TXNIP , SLC1A5 and SREBF1 ) out of the 52 CpGs they identified as reported to be differentially methylated in T2DM through a systematic review of the literature done till April 2017. Willmer et al . [ 8 ] also focused on differential methylation signatures in blood samples and reported TCF7L2 , KCNQ1 , ABCG1 , TXNIP , PHOSPHO1 , SREBF1 , SLC30A8 and FTO genes to be reproducibly associated with T2DM across multiple population groups in the literature reviewed between January 2002 and July 2018.
DNA methylation has been touted as a strong candidate biological process for identification of diagnostic and therapeutics for T2DM [ 11 ]. While the available systematic reviews have looked at DNA methylation associated with T2DM [ 8 , 9 , 10 ], they have not evaluated T2DM-associated DNA methylation comprehensively in all available human tissue and cell types. We set out to fill this research gap with the no time period cutoff until October 19, 2023, and including all available human tissue and cell types. We also report associated gene expression data, role of sex and ethnicity, in relation to DNA methylation in our review.
PubMed and Science Direct databases were independently searched by authors (NN, PN and JKV) using the key terms “type 2 diabetes mellitus” and “DNA methylation,” and their associated terms for all studies published up to October 19, 2023. All articles from the time of publication listing were considered, and as such no start date was set. No filters were applied during the search using the keywords, so as to not exclude any mislabeled/mis-annotated article type. The detailed search strategy is given in Additional file 1 : Table S1.
Study inclusion and exclusion criteria
The inclusion criteria were full-text English language articles on DNA methylation associated with T2DM in human subjects. Case–control and prospective studies investigating genome-wide methylation were included. Reviews, animal model studies, in vitro studies, irrelevant articles and articles published in other languages were excluded.
All participants, regardless of gender and ethnicity, classified as adults aged 18 years and above were included. All individuals who did not satisfy these criteria—children and adolescents under 18 years of age; as well as subjects with type 1 diabetes (T1DM) or gestational diabetes were excluded. As the association of DNA methylation with T2DM was the focus of this systematic review, intervention studies and clinical trials were excluded. Studies reporting association of DNA methylation with diabetes-related traits (hyperglycemia and insulin resistance) were retained.
All the articles were assessed for their eligibility based on their abstract or full text.
Disagreements between the authors, such as categorization and selection of articles, and data extraction, were resolved through discussion with AM. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklist was followed to represent the method used [ 12 ]. A total of 32 full-text articles are included in this systematic review.
The assessment of quality of the studies was done by adapting the New Castle Ottawa scale (NOS) [ 13 ]. The parameters used for the assessment are adequacy of case definition, representativeness of cases, selection of controls, definition of controls, comparability of cases and controls, ascertainment of exposure and method used for ascertainment of cases and controls. Scores were given to each of the included studies, and the total score was calculated according to the score sheet (NOS).
This review protocol was registered with the International Prospective Register of Systematic Reviews (PROSPERO) database ( https://www.crd.york.ac.uk/prospero/ ) [ 14 ] (accessed April 18, 2023) (registration ID: CRD42020140436).
Pathologically connected pathways with differentially methylated genes in T2DM were analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Jensen Disease database via Enrichr-KG [ 15 ].
We identified a total of 5819 articles during the initial search. Duplicates, irrelevant articles based on the study design, publication language, article type, and other articles not within our scope of review were removed. Thirty-two full-text articles were finally selected (Fig. 1 ).
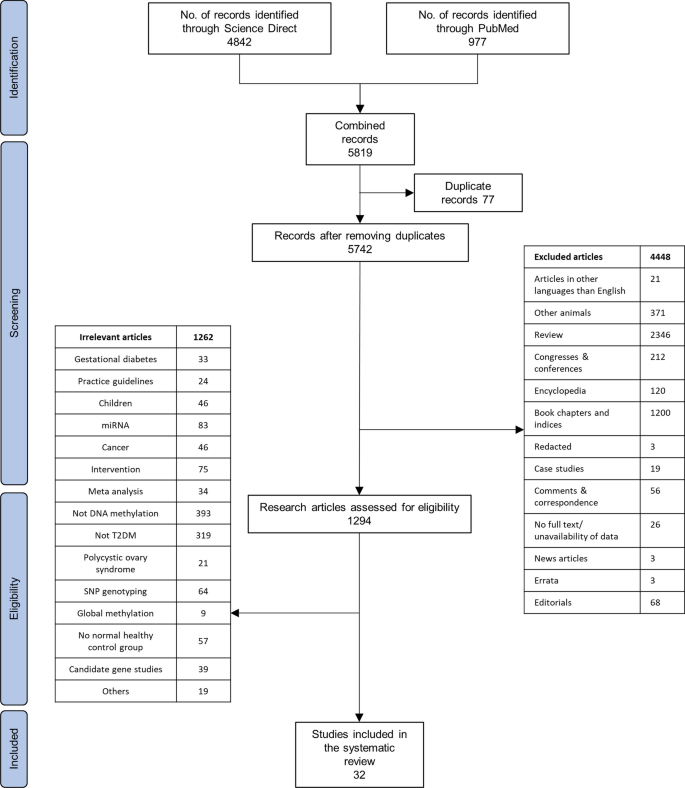
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [ 12 ] flowchart for the literature search process, performed up to October 19, 2023
NOS was used to access the quality of the articles. Of the 32 studies, 16 were assigned a score of more than 5, indicating high quality (Additional file 2 : Table S2). As all the studies have used the same method of ascertainment for cases and controls, and the authors are not blinded to case–control status, these redundant scores are not presented. As the nonresponse rate was not available for any of the studies, this also has been omitted from the quality assessment table.
Case–control studies that reported differential DNA methylation between T2DM (case) and normoglycemic (control) subjects or reported associations between DNA methylation and clinical parameters related to glycemic control of the subjects (HbA1c, fasting blood glucose) and prospective nested case–control studies that reported differential DNA methylation measured at baseline/recruitment between subjects who developed T2DM (incident cases) and those that remained normoglycemic (control) during the follow-up period were finally included.
Participant details such as number of cases and controls and location of the study are also included. Details of the study participants who do not explicitly belong to either case or control group are also presented. The tissue source of the gene/loci identified in; method used for determining methylation status; and the validation method used for confirming the methylation status are tabulated in Table 1 .
The loci/genes reported to be differentially methylated are tabulated in Table 2 , where their methylation status is represented as downward arrow (hypomethylation) or upward arrow (hypermethylation). Wherever reported, the statistical significance of methylation ( P value) is also mentioned. For studies reporting more than 10 differentially methylated genes, the top 5 hypo- and hypermethylated genes are listed.
Methods of DNA methylation analysis
Majority of the evaluated studies had employed array-based techniques to assess DNA methylation levels. Eighteen of 32 studies used Illumina 450 k array. Other array-based studies used Illumina 27 k array (2 studies), Illumina EPIC BeadChip array (4 studies; of which 2 studies specifically mentioned using the 850 k array—EPIC v1 array targeting 850 k probes), Affymetrix SNP6 microarray (1 study), Affymetrix GeneChip promoter 1.0R array (1 study) or Affymetrix axiom genome-wide Taiwan BioBank (TWB) array (1 studies). Rest of the studies used techniques such as methylated DNA immunoprecipitation (MEDIP) (2 studies), MEDIP-chromatin immune precipitation (1 study), reduced representation bisulfite sequencing (1 study) or next-generation sequencing (1 study) to measure DNA methylation levels.
Tissues used in DNA methylation analyses
Of the 32 articles retrieved, 17 (53%) studies used blood samples, 4 (13%) studies used pancreatic islet samples, 5 (16%) studies used adipose tissue samples, 2 (6%) studies used liver samples, 1 (3%) study used spermatozoa samples and 3 (9%) used skeletal muscle samples for their DNA methylation analyses. None of the 32 studies reviewed here utilized more than one tissue from the same subjects for DNA methylation analyses.
Genome-wide methylation analysis for T2DM
Of the 32 genome-wide methylation studies reviewed here, we identified a total of 130 loci that were differentially methylated between T2DM cases and controls across. In an instance where a study reports < 10 differentially methylated genes/loci, they are presented individually. However, in the case of a study which reports > 10 genes/loci, only the top 5 hypo- and 5 hypermethylated genes are highlighted for brevity and reported in Table 2 . The direction of methylation (hyper- or hypomethylated in T2DM, compared to controls) and the reported P values (both unadjusted, and after multiple testing correction) have been included.
We identified genes such as ABCG1, PPARGC1A , PTPRN2 and TXNIP with well-known T2DM genetic risk variants, which were consistently reported to be differentially methylated in more than one study (Fig. 2 ). Tissues used in identification of these gene were blood cells, liver, pancreatic islets and adipose tissue. TXNIP (cg19693031) was the most common gene identified consistently as hypomethylated in diabetic blood (9 studies). TXNIP also harbors established T2DM genetic risk variants [ 16 , 17 ].
A pie chart depicting the genes that were consistently reported to be differentially methylated in ≥ 2 studies in various tissues from T2DM subjects. ↑: Hypomethylation, ↓: Hypermethylation in T2DM individuals compared to normoglycemics. PPARGC1A (chr4: 24,024,251–500) hypomethylated, (chr4: 24,111,501–750) hypermethylated in spermatozoa [ 57 ]
Although blood is not an insulin-responsive tissue, it is the prime minimally invasive tissue available for investigating T2DM-associated epigenetic markers. With the bulk (50%) of the studies coming from Europe, ABCG1 [ 18 , 19 ] and TXNIP [ 16 , 17 , 19 , 20 , 21 , 22 , 23 , 24 , 25 ] were some of the blood-based epigenetic markers which were reported to be associated with T2DM in more than one study. We were unable to find any study where differential methylation was investigated simultaneously in blood and other tissues from the same subjects.
Pancreatic islets
Insufficient secretion of insulin from pancreatic beta cells and increased secretion of glucagon from pancreatic alpha cells leads to development of T2DM and is known to be regulated by DNA methylation [ 26 ]. Three of the 32 studies, from Italy, South Korea and Sweden, included in this review have interrogated DNA methylation in pancreatic islets from T2DM individuals, donated after their death. Regions in SFRS2IP [ 3 ], MSI2 [ 27 ], which are known to be associated with critical roles in nucleic acid binding, and B3GNT7 [ 28 ] that is involved in synthesis of glycoprotein, were reported to be hypomethylated in pancreatic islets from T2DM individuals. Considering that DNA methylation can change based on the time of collection of tissue after death [ 29 , 30 ], findings from these studies need to be interpreted in cognizance of the lack of details available in these studies about the cause of death or collection and storage of pancreatic islet tissue after death.
Adipose tissue
Adipose tissue is known to play a critical role in regulating body metabolism and energy homeostasis [ 31 ]. Dysregulation in adipose biology imposes serious health complications such as obesity and development of T2DM [ 31 ]. DNA methylation is an important regulator factor in development [ 32 , 33 ] and dysfunction [ 34 , 35 ] of adipose tissue. Five studies—4 of these representing the European population—included in this review have dissected whether T2DM, and related risk factors are associated with epigenetic modifications in human adipose tissue [ 36 , 37 , 38 , 39 , 40 ]. It is possible that DNA methylation alterations in these reported genes including C1orf52 [ 36 ], HOOK2 [ 37 ], MFSD1 [ 38 ], HNF4A [ 39 ] and L1TD1 [ 40 ] contribute to or are caused by T2DM.
C1orf52 is involved in RNA binding in adipose tissue [ 41 ], and HOOK2 is responsible for cytoskeleton maintenance via regulation of microtubules [ 42 ], while MSFD1 regulates lysosome transport [ 43 ]. Epigenetic alterations in such genes involved in cell structure and function can cause dysfunction in adipose tissue, thereby leading to insulin resistance. While HNF4A mainly regulates transcription in hepatocytes and is associated with Fanconi renotubular syndrome 4 with maturity-onset diabetes of the young [ 44 ] and maturity-onset diabetes of the young, type 1 [ 45 ], it is also known to play a role in lipid and glucose metabolism [ 46 , 47 ]. L1TD1 is predicted to be involved in single-stranded RNA-binding activity [ 48 ].
Liver is known to be involved in regulating glucose level by storing and releasing glycogen in response to insulin and glucagon [ 49 ]. Impaired hepatic gluconeogenesis, glycogenolysis and insulin sensitivity are known to play an important role in T2DM development and other risk factors. Altered hepatic metabolism could be the cause or consequence of DNA methylation modification. Genes involved in intracellular tyrosine kinase activity— PDGFA [ 50 ], transferring phosphorus-containing groups and protein tyrosine kinase activity— RIPK4 [ 51 ], heme binding and oxidoreductase activity— CYB561D1 [ 51 ], were found to be hypomethylated in the diabetic groups. However, the gene involved in inflammation— IL23Ap19 [ 51 ] was identified to be hypermethylated in the diabetic group. Of the two studies reported here, one was from France and the other from Finland.
Gene expression studies
Out of the 32 studies reviewed, 8 had also examined differences in gene expression between T2DM and normoglycemic individuals. To examine if increase in methylation of a gene causes decrease in expression of that gene, we analyzed the studies that report both differentially methylated genes and gene expression, in the same population and study setting, using tissues from the same study participants (Table 3 ). For most of the loci with both DNA methylation and gene expression data available, we found that increase in methylation was associated with decrease in expression, concurrent to the current understanding [ 6 ]. Hypermethylation of PPARGC1A in skeletal muscles [ 52 ], ABCG1 in blood [ 18 ] and PER2 in pancreatic islets [ 3 ] was associated with lower expression of the corresponding genes.
Twin studies
Five of the 32 studies reviewed here have investigated DNA methylation in monozygotic twin cohorts [ 17 , 21 , 28 , 36 , 53 ] (Table 4 ). MALTI [ 53 ] which is known to be involved in energy and insulin signaling pathways [ 54 ], PTBP1 [ 36 ] that is involved in nucleic acid binding, and ANO8 [ 28 ] that is involved in calcium transport, were hypermethylated in diabetic twins in peripheral blood, adipose tissue and pancreatic islets, respectively. TXNIP [ 17 , 21 ], COL21A1 [ 36 ] and B3GNT7 [ 28 ] were hypomethylated in blood cells, adipose tissue and pancreatic islets, respectively, from the diabetic twins. Dayeh et al . reported differential methylation of ABCG1 (hypermethylated in blood and adipose tissue) and PHOSPHO1 (hypomethylated in skeletal muscle) in monozygotic twins discordant for T2DM [ 55 ].
Association between diabetes related traits and DNA methylation
Only 4 of the 32 studies reported association between diabetes-related traits (hyperglycemia and insulin resistance) and DNA methylation [ 17 , 18 , 19 , 22 ]. Kriebel et al . reported significant association between measures of glucose metabolism phenotypic traits and methylation levels of 31 CpG sites in PBMCs [ 18 ]. Five CpGs were found to be associated with fasting glucose, 1 CpG with 2-h glucose, 8 with fasting insulin and 26 with Homeostatic Model Assessment of Insulin Resistance (HOMA-IR) in model 1 (Table 2 ) [ 18 ]. There was no significant association between HbA1c and DNA methylation levels in model 1; in model 2, after adjustment for body mass index (BMI), the effect strength was reduced by 30% for DNA methylation associations with fasting glucose suggesting that the associations between DNA methylation and diabetes-related traits are partially mediated by BMI [ 18 ].
Kulkarni et al . investigated association between 446,356 autosomal CpG sites and phenotypic traits in PBMCs, of which a total of 51 CpG sites were significantly associated with T2DM, 19 with FBG and 24 with HOMA-IR (Table 2 ) [ 19 ].
Wang et al . report association between 63 differential methylated loci and fasting blood glucose and association between 6 differentially methylated loci with HbA1c in blood samples from twins discordant for diabetes [ 17 ]. Among these, hypomethylation of TXNIP [ 17 , 19 ] and hypermethylation of ABCG1 [ 18 , 19 ] were positively associated with fasting blood glucose (FBG), and hypermethylation of SAMD12 was negatively associated with FBG [ 19 ]. TXNIP hypomethylation in blood cells was found to be associated with hyperglycemia in individuals from Taiwan [ 23 ], France [ 24 ], the USA [ 21 ] and China [ 17 ].
Dawes et al . performed genome-wide DNA methylation on blood samples from normoglycemic (n = 142), pre-diabetic (n = 274) and diabetic (n = 90) individuals [ 22 ]. They identified HbA1c-associated DNA methylation loci by regressing the probes against HbA1c values, while controlling for age, sex and BMI [ 22 ]. They report cg19693031 ( TXNIP ) as the locus most highly associated with HbA1c [ 22 ].
Enrichment analysis of genes differentially methylated in T2DM
Enrichment analysis of signaling pathways relevant to the pathophysiology of T2DM using Enrichr-KG [ 15 ] was done in two steps. Initially, all 130 genes differentially methylated in T2DM in all 32 studies reviewed were included (Fig. 3 ). To take into account reproducibility of these findings, enrichment analysis was separately done specifically for the genes ( ABCG1 , TXNIP , PTPRN2 , PPARGC1A ) that were reported to be differentially methylated in T2DM in more than one study (Fig. 4 ). TXNIP hypomethylation in blood was linked to hyperglycemia. PPARGC1A hypermethylation in skeletal muscles, and two CpG sites that were hyper- and hypomethylated, respectively, in spermatozoa, was linked to hyperglycemia and adipocytokine signaling pathway. PTPRN2 that was reported to be hypermethylated in blood and hypomethylated in adipose tissue was associated with T2DM and T1DM.
Gene enrichment analysis of 17 of the 130 genes reported to be differentially methylated in T2DM subjects in the 32 studies included for review using Enrichr-KG. These genes were mapped to diabetes and related disorders. Insulin resistance, glucagon signaling pathway, glaucoma, AMPK signaling pathway, cholinergic synapse, ovarian cancer, amphetamine addiction and Huntington’s disease were found to be associated with KCNQ1 , FTO , PPARGC1A , PTPRN2 , ELOVL5 , HNF1B , HNF4A , VPS13A , MAEA , CREB1 , CPT1A , PRKCZ , PRKCB , CREB3L2 , CDKN2A and TGFBR3
Gene enrichment analysis of 4 genes reported to be differentially methylated in T2DM subjects in > 1 study from among the 32 studies included for review using Enrichr-KG. Hyperglycemia, type 1 diabetes, adipocytokine signaling pathway, glucagon signaling pathway, longevity regulating pathway and ABC transporters were found to be associated with PPARGC1A , TXNIP , PTPRN2 and ABCG1
Subgroup analysis based on ethnicity
Out of the 32 studies, 16 (50%) were from Europe, 4 (13%) were from North America, 8 (25%) were from Asia and 1 (3%) from Africa. Three studies (9%) did not report their subjects’ ethnicity/demography.
TXNIP was the most commonly reported hypomethylated gene in blood cells of T2DM individuals from all the geographic locations [ 16 , 17 , 19 , 20 , 21 , 22 , 23 , 24 ]. ABCG1 was found be to hypermethylated in blood cells of type 2 diabetics in studies from Europe [ 18 ] and the USA [ 19 ]. PTPRN2 was reported to be hypermethylated in peripheral blood in studies from China [ 56 ] and France [ 24 ]. Conversely, PTPRN2 was reported to be hypomethylated in adipose tissue from a Spanish study [ 37 ].
Subgroup analysis based on sex
PPARGC1A was assessed for differential methylation in two studies which had only male participants [ 52 , 57 ]. PPARGC1A was hypermethylated in skeletal muscle of T2DM men [ 52 ]. Of the two differentially methylated regions in PPARGC1A identified in sperm, chr4: 24,111,501–750 was reported to be hypermethylated, and chr4: 24,024,251–500 was reported to be hypomethylated [ 57 ]. We did not find other epigenome-wide studies that report differential methylation of PPARGC1A in female-only or mixed-sex populations.
PDGFA was found to be hypomethylated in hepatocytes from liver biopsies of female T2DM participants of the discovery group and was later confirmed in both men and women by Abderrahmani et al . [ 50 ]. Similarly, hypomethylation of MSI2 in blood cells was first observed in a discovery group comprised of only men, and then in a replication group of both men and women by Jeon et al . [ 27 ].
In the cg 11,738,485-region (5 CpG nucleotides) of HOOK2 , female T2DM visceral adipose tissue samples were hypermethylated, while male T2DM samples were hypomethylated, compared to non-diabetic sex-matched control samples [ 37 ]. None of the other loci/genes were reported to be differentially methylated in a sex-specific manner.
Internal and/or external validation
Only 22% of the studies reviewed (7 out of 32) validated their findings in an independent set of subjects using the same DNA methylation measurement method that they had used for the discovery set of samples [ 17 , 25 , 27 , 36 , 37 , 50 , 53 ]. Others used either bisulfite pyrosequencing/sequencing (10 studies) [ 3 , 19 , 27 , 28 , 37 , 39 , 52 , 58 , 59 , 60 ], qPCR (1 study) [ 51 ], EpiTYPER (1 study) [ 16 ], Illumina 450 k (3 studies) [ 36 , 50 , 53 ] or MEDIP (1 study) [ 61 ] for their internal validation. Sixteen studies (50%) did not perform any validation for their findings.
Replication for case–control studies
We later looked for candidate-gene DNA methylation studies to see if the differentially methylated genes found in genome-wide studies have been confirmed in them. The following genes were reported to be differentially methylated in T2DM compared to normoglycemic controls in independent candidate-gene DNA methylation studies in the same tissue as the initial discovery group— ABCG1 [ 62 , 63 ], FTO [ 64 , 65 , 66 ], TXNIP [ 67 ] and KCNQ1 [ 64 , 68 ] in PBMCs, and PPARGC1A in pancreatic islets [ 69 ].
Prospective studies
As prospective studies observe the disease condition over a long period, they help in better understanding the role of a gene/set of genes toward pathogenesis. In our review, we came across three such studies that looked at incidence of T2DM and epigenetic modifications in genes associated with this incidence (Table 5 ).
In a 1:1 matched nested case–control study of 290 incident diabetics, who developed T2DM and 290 controls, who remained normoglycemic during the 4-year follow-up, baseline methylation at 7 CpG sites of IGFBP2 in blood cells (4 hypermethylated and 3 hypomethylated in cases) was associated with increased risk of incident T2DM [ 70 ].
Jeon et al . reported that differential methylation of three CpG sites in blood cells at baseline was associated with T2DM/hyperglycemia after a 10-year follow-up [ 27 ]. These CpG sites were cg23586172 (annotated to MSI2 , hypomethylated), cg22604213 (annotated to CXXC4, hypomethylated) and cg25290098 (hypomethylated) in T2DM [ 27 ]. They further reported MSI2 hypomethylation in a replication group of 220 normoglycemic and 220 T2DM individuals [ 27 ]. Furthermore, whole-genome bisulfite sequencing of pancreatic islets of 2 T2DM and 16 normoglycemic individuals revealed that chr17:55,484,635 in MSI2 was hypomethylated in T2DM [ 27 ]. While MSI2 hypomethylation was seen in both pancreatic islets and PBMCs, pancreatic islets showed increased difference of 16% methylation versus 3% in PBMCs of MSI2 in T2DM when compared to normoglycemics [ 27 ]. MSI2 differential methylation was not found to be replicated in locus-specific case–control studies.
From the Jerusalem LRC longitudinal study, Toperoff et al . selected 58 individuals who developed impaired glucose metabolism over a 13-year follow-up and reported hypomethylation of a single CpG site in the first intron of FTO in peripheral blood samples collected at baseline [ 58 ]. Chen et al . similarly reported hypomethylation of FTO in their case–control study [ 57 ].
In a longitudinal study of Indian Asians living in London, UK (1074 incident T2DM and 1590 normoglycemic controls), over 8 years of follow-up, Chambers et al . reported that DNA methylation levels of TXNIP , PROC , C7orf29 , SREBF1 , PHOSPHO1 , SOCS3 and ABCG1 in blood cells were positively associated with future T2DM incidence [ 71 ]. Of these, higher baseline methylation levels in TXNIP , SREBF1 , PHOSPHO1 , SOCS3 and ABCG1 were also associated with incident T2DM in an European cohort of 377 incident T2DM and 764 normoglycemic individuals [ 71 ].
Differential methylation in animal models
To check if animal model studies exist that have investigated or reported differential methylation in the genes identified as differentially methylated in the human case–control studies as playing causal or mechanistic role in the development of T2DM, a simple literature search was done using PubMed and bibliography search. A study in rat pancreatic islets reported Kcnq1 was hypomethylated in older rats (15 months of age) when compared to younger rats (3 months of age), but this difference was not statistically significant, while there was no comparison done with a rat T2DM model [ 72 ]. Though Toperoff et al . reported hypomethylation of KCNQ1 in blood cells [ 58 ], there are no human pancreatic islet studies reporting hypomethylation of KCNQ1 . Identification of multiple variants in genome-wide association studies [ 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 , 81 ] points toward the likely importance of KCNQ1 in T2DM pathophysiology.
High-fat diet was shown to induce hypermethylation of Tcf7l2, and subsequently, gene expression was decreased in mouse islets [ 82 ]. This is in contrast to the findings where TCF7L2 is hypomethylated in T2DM human blood cells [ 58 ] and pancreatic islets [ 59 ]. It is to be noted that the mice used were non-diabetic adult males aged 8 weeks (equivalent to middle-aged humans [ 83 ]) [ 82 ], while the human study group were a mix of men and women aged about 58–65 years, and for the human pancreatic islet study, the samples had been collected post-mortem [ 58 , 59 ]. Although there is an inverse differential methylation status among mice and humans, it is important to note that a high-fat diet caused suppression of Tcf7l2 gene expression and thus decreases pancreatic beta-cell survival (mediated via the transcription of Wnt/Beta-catenin signaling pathway [ 84 ]) [ 82 ].
From the 32 studies finally included for this systematic review, we identified 130 genes with T2DM-associated differential methylation across all tissues analyzed. These comprise of the top 5 hypo- and hypermethylated genes for studies reporting more than 10 differentially methylated genes/loci. Of these 130 genes, 4 (3%; ABCG1, PPARGC1A , PTPRN2 and TXNIP ) were reported in > 1 studies. The genes and associated pathways with altered DNA methylation in T2DM are conceptually summarized in Fig. 3 (for 16 of the 130 genes, for which pathway analysis could be conducted) and Fig. 4 (for the 4 genes reported to be differentially methylated in > 1 studies).
Previous systematic reviews [ 8 , 9 ] have reported differentially methylated loci in genes in T2DM blood cells including ABCG1 , TXNIP , KCNQ1 . While another such review by Muka et al . reported several epigenetically regulated genes from blood cells, adipose tissue, muscle and placenta, there was no overlap between them, and no association was found between global DNA methylation and T2DM/hyperglycemic markers [ 10 ].
We did not limit our search to a particular method used to identify DNA methylation, and several studies included have used Illumina’s 450 k array. The common method of validation/replication in the studies reviewed here was bisulfite pyrosequencing. We also looked at candidate-gene DNA methylation studies which aimed to replicate/validate the epigenome-wide studies reviewed here and found that in blood cells, ABCG1 [ 62 ], FTO [ 64 ] and KCNQ1 [ 64 ] were hypermethylated, while TXNIP was hypomethylated [ 67 ]. TXNIP codes for thioredoxin-interacting protein, and this protein plays a major role in pathways generating reactive oxygen species [ 85 ], regulating redox-dependent signaling pathways, mediating oxidative stress, suppressing cell growth and inducing pancreatic beta-cell apoptosis [ 86 ]. ABCG1 codes for the protein responsible for intracellular sterol transport [ 87 ], and it regulates cholesterol efflux from macrophages to high-density lipoprotein in diabetics [ 88 ], indicated by altered lipid levels [ 89 ]. While genetic variants and epigenetic modification of KCNQ1 have been linked with T2DM via whole body insulin sensitivity [ 90 ], there is no clear evidence for the mechanistic link. Likewise, there has been no clear evidence of FTO link with T2DM.
As gene expression is known to be regulated by DNA methylation, it is important to validate this claim in the epigenome-wide association studies. We were able to report the relation between DNA methylation in the promoter region and expression of the corresponding gene, as none of the studies had mentioned methylation status of other regions of the genes. Of the studies reviewed here, we found that DNA methylation of genes was inversely related to gene expression. For example, hypomethylation of S100A4 in adipose tissue [ 36 ] and PDGFA in hepatocytes [ 50 ] was associated with increased expression of these genes, and hypermethylation of PPARGC1A in skeletal muscles [ 52 ], ABCG1 in blood [ 18 ] and PER2 in pancreatic islets [ 3 ] was associated with lower expression of the corresponding genes. Even though we observed DNA methylation being related inversely with expression of the corresponding gene in the studies reviewed, this is not a rule as has been reported repeatedly [ 91 ]. It is also important to note that there have been reports of methylation levels differing between different regions of the gene that influence gene expression; for instance, Anastasiadi et al . recently reported that gene expression is dependent on methylation of the first exon, more than methylation of the promoter region [ 92 ]. Moreover, in other studies such as one by Ball and colleagues, highly expressed genes have been reported to have low methylation levels in the promoter region and high methylation levels in rest of the gene body [ 93 ]. We could not, however, evaluate the relations between DNA methylation in various regions of a gene and its corresponding expression in this study since the studies reviewed by us have reported DNA methylation specifically in the promoter region.
Epigenetic studies on twins discordant for disease status are crucial in understanding the genetic basis of epigenetic differences observed in cross-sectional studies. Of the 5 studies included in our search, 3 did not have any common differentially methylated genes among them, while the other two studies that used blood cells as the source tissue had TXNIP as the common differentially methylated gene between them, with hypomethylation of TXNIP in diabetic blood samples observed in both these studies [ 17 , 21 ]. TXNIP is the only gene reported to be hypomethylated in diabetic blood in both case–control studies [ 55 ] and in twin studies [ 17 , 21 ] where the influence of underlying genetic factors is not masked. TXNIP has also been reported to be hypomethylated in diabetic pancreatic islets [ 55 ] and skeletal muscle [ 55 ], making it a potentially important causal gene in the pathophysiology of T2DM.
T2DM is known to be associated with other comorbidities such as obesity and cardiovascular complication. These comorbidities share some common risk factors like age, BMI and cholesterol content in blood. These risk factors are influenced by genes such as KCNQ1 , TCF7L2 and FTO [ 94 ]. Other systematic reviews have looked at epigenetic changes in obesity [ 95 ], aging [ 96 , 97 ] and cardiovascular complications [ 98 ]. Andrade et al . aimed to identify epigenetic changes in human adipose tissue from obese/overweight individuals with and without metabolic disorders like T2DM [ 95 ]. They also report differentially methylated genes that we have been reported in this review, such as KCNQ1 , FASN , MFSD1 , TXNIP , PPARG , IRS1 and TCF7L2 , from the same studies [ 95 ]. Krolevets et al . report that in addition to about 75,000 CpG sites and 19,000 genes, PTPRN2 was among the most frequently reported gene that was associated with cardiac disorders, although the direction of methylation is not specified [ 98 ]. Of the two studies that investigated DNA methylation in aging [ 96 , 97 ], no genes/CpG sites/studies were common with the ones mentioned in our review.
One of the most important factors in looking at T2DM as an epidemic is the geographic location of the site of reported data. With a large amount of data coming in from Europe alone, it is important to perform similar studies in other parts of the world and including various other ethnic groups to validate these reports and also help in mapping the genetic diversity to be able to tackle T2DM. India being the most populous country [ 99 ] with about 11% of Indians suffering from T2DM (in 2020) [ 100 ], it is imperative to study this population to uncover T2DM susceptible loci/genes. Of note, Chambers et al . have followed up London resident Indian Asians, for 8 years, and found that DNA methylation levels of TXNIP , PROC , C7orf29 , SREBF1 , PHOSPHO1 , SOCS3 and ABCG1 were positively associated with future T2DM incidence [ 71 ], but similar studies are lacking in Indians living in India, where exposure to pollution and availability and consumption of healthy diet are vastly different.
As for sex-specific methylation signatures of T2DM, differences were not seen between men and women except in genes HOOK2 [ 37 ] and MSI2 [ 27 ], which were hypermethylated in adipose tissue, and hypomethylated in blood, respectively . Finally, we searched if the genes which we found to be highly reported to be differentially methylated in human were also reported to be differentially methylated in animal models. KCNQ1 was reported to be hypomethylated in both T2DM human [ 58 ], and older mice model compared with younger mice [ 72 ] suggesting age-related methylation changes across species. In both humans [ 58 ], and mice fed with a high-fat diet, TCF7L2 was hypomethylated, and this DNA methylation change in mice was induced because of their diet [ 82 ], suggesting that nutrient consumption plays a role in epigenetic modification of genes involved in beta-cell function, and a healthy diet can have a protective role in maintaining homeostasis.
Although we did not look at clinical trials and candidate-gene studies that report differential DNA methylation, our review is an up-to-date report of epigenome-wide studies that includes prospective studies. We also report gene expression data in comparison with DNA methylation. Furthermore, a systematic report of differentially methylated gene/loci in tissues including blood cells, adipose tissue, pancreatic islet, skeletal muscles, liver and spermatozoa is included. While sex and ethnicity play a major role in pathology, we have tried to highlight these effects.
As with previous reviews, we emphasize the need for more prospective studies and replication of genome-wide association studies in different tissue types and populations.
From the 32 studies that report differentially methylated genes/loci between T2DM and normoglycemic individuals, ABCG1 (hypermethylated in blood), FTO (hypermethylated in blood and spermatozoa), KCNQ1 (hypermethylated in blood and hypomethylated in spermatozoa), TXNIP (hypomethylated in blood), PPARGC1A loci at chr4: 24,111,501–750 (hypermethylated in skeletal muscle and spermatozoa) and loci at chr4: 24,024,251–500 (hypomethylated in spermatozoa), PTPRN2 (hypermethylated in blood, hypomethylated in adipose tissue) were reported in more than one study. We found reports of hypermethylation of these genes that were associated with decreased gene expression, and vice versa. We also report findings from studies done on monozygotic twins. Various traits that can affect T2DM such as sex, glucose levels, BMI and ethnicity were also taken into consideration.
As there were multiple methods that were used to measure DNA methylation, internal and external validation of these results is also reported. Finally, animal model studies that have reported differential DNA methylation of the genes that were found to be differentially methylated in human studies were looked at to get an understanding of the likely mechanisms linking epigenetic dysregulation of these genes in T2DM to its pathophysiology.
Although the majority of the top differentially methylated genes are well known, other more recent genes reported here should be investigated further to understand their role in pathogenesis of T2DM.
Data availability statement
All relevant data are presented as tables and/or figures.
Abbreviations
ATP-Binding Cassette Subfamily G Member 1
Anoctamin 8
Beta 1,3-N-Acetylglucosaminyltransferase 7
Chromosome 1 Open Reading Frame 52
Chromosome 7 Open Reading Frame 29
Collagen Type XXI Alpha 1
Cytochrome B561 Family Member D1
CXXC Finger Protein 4
Alpha-Ketoglutarate Dependent Dioxygenase
Glucagon Like Peptide 1 Receptor
Glutathione Peroxidase 6
Hepatocyte Nuclear Factor 4 Alpha
Hook Microtubule Tethering Protein 2
Insulin-Like Growth Factor-Binding Protein 2
Interleukin-23 Subunit Alpha
Potassium Voltage-Gated Channel Subfamily Q Member 1
LINE1 Type Transposase Domain Containing 1
Lysyl Oxidase Homolog 2
Mucosa-Associated Lymphoid Tissue Lymphoma Translocation Protein 1
Major Facilitator Superfamily Domain Containing 1
Musashi RNA-Binding Protein 2
Platelet Derived Growth Factor Subunit A
Pancreatic and Duodenal Homeobox 1
Period Circadian Regulator 2
Phosphoethanolamine/Phosphocholine Phosphatase 1
Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1-Alpha
Protein C, Inactivator Of Coagulation Factors Va And VIIIa
Polypyrimidine Tract-Binding Protein 1
Protein Tyrosine Phosphatase Receptor Type N2
Receptor Interacting Serine/Threonine Kinase 4
S100 Calcium-Binding Protein A4
Sterile Alpha Motif Domain Containing 12
Solute Carrier Family 1 Member 5
Solute Carrier Family 22 Member 1
Solute Carrier Family 22 Member 3
Solute Carrier Family 30 Member 8
Sterol Regulatory Element-Binding Transcription Factor 1
Suppressor Of Cytokine Signaling 3
Transcription Factor 7-Like 2
Thioredoxin-Interacting Protein
Cho NH, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, et al. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–81.
Article CAS PubMed Google Scholar
Prasad R, Groop L. Genetics of type 2 diabetes—pitfalls and possibilities. Genes. 2015;6(1):87–123.
Article CAS PubMed PubMed Central Google Scholar
Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31(6):1405–26.
Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2(6):607–17.
Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597–610.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38.
Hall E, Dayeh T, Kirkpatrick CL, Wollheim CB, Dekker Nitert M, Ling C. DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BMC Med Genet. 2013;14:76–76.
Willmer T, Johnson R, Louw J, Pheiffer C. Blood-based DNA methylation biomarkers for type 2 diabetes: potential for clinical applications. Front Endocrinol. 2018;4(9):744.
Article Google Scholar
Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H, et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case–control sample of the Lifelines study. Diabetologia. 2018;61(2):354–68.
Muka T, Nano J, Voortman T, Braun KVE, Ligthart S, Stranges S, et al. The role of global and regional DNA methylation and histone modifications in glycemic traits and type 2 diabetes: a systematic review. Nutr Metab Cardiovasc Dis. 2016;26(7):553–66.
Rönn T, Ling C. DNA methylation as a diagnostic and therapeutic target in the battle against Type 2 diabetes. Epigenomics. 2015;7(3):451–60.
Article PubMed Google Scholar
Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;1(4):1.
Wells G, Shea B, O’Connell D, Peterson J, Welch, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. 2014. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
PROSPERO. [cited 2023 Apr 18]. https://www.crd.york.ac.uk/prospero/
Evangelista JE, Xie Z, Marino GB, Nguyen N, Clarke DJB, Ma’ayan A. Enrichr-KG: bridging enrichment analysis across multiple libraries. Nucl Acids Res. 2023;gkad393.
Soriano-Tárraga C, Jiménez-Conde J, Giralt-Steinhauer E, Mola-Caminal M, Vivanco-Hidalgo RM, Ois A, et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet. 2016;25(3):609–19.
Wang Z, Peng H, Gao W, Cao W, Lv J, Yu C, et al. Blood DNA methylation markers associated with type 2 diabetes, fasting glucose, and HbA1c levels: an epigenome-wide association study in 316 adult twin pairs. Genomics. 2021;113(6):4206–13.
Kriebel J, Herder C, Rathmann W, Wahl S, Kunze S, Molnos S, et al. Association between DNA methylation in whole blood and measures of glucose metabolism: KORA F4 study. PLoS ONE. 2016;11(3):e0152314–e0152314.
Article PubMed PubMed Central Google Scholar
Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JWJ, Göring HHH, et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet. 2015;24(18):5330–44.
Meeks KAC, Henneman P, Venema A, Addo J, Bahendeka S, Burr T, et al. Epigenome-wide association study in whole blood on type 2 diabetes among sub-Saharan African individuals: findings from the RODAM study. Int J Epidemiol. 2019;48(1):58–70.
Xiang Y, Wang Z, Hui Q, Gwinn M, Vaccarino V, Sun YV. DNA Methylation of TXNIP independently associated with inflammation and diabetes mellitus in twins. Twin Res Hum Genet. 2021;24(5):273–80.
Dawes K, Philibert W, Darbro B, Simons RL, Philibert R. Additive and interactive genetically contextual effects of HbA1c on cg19693031 methylation in type 2 diabetes. Genes (Basel). 2022;13(4):683.
Tsai HH, Shen CY, Ho CC, Hsu SY, Tantoh DM, Nfor ON, et al. Interaction between a diabetes-related methylation site (TXNIP cg19693031) and variant (GLUT1 rs841853) on fasting blood glucose levels among non-diabetics. J Transl Med. 2022;20(1):87.
Khamis A, Ning L, Balkau B, Bonnefond A, Canouil M, Roussel R, et al. Epigenetic changes associated with hyperglycaemia exposure in the longitudinal DESIR cohort. Diabetes Metab. 2022;48(4):101347.
Florath I, Butterbach K, Heiss J, Bewerunge-Hudler M, Zhang Y, Schöttker B, et al. Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia. 2016;59(1):130–8.
Kuroda A, Rauch TA, Todorov I, Ku HT, Al-Abdullah IH, Kandeel F, et al. Insulin gene expression is regulated by DNA methylation. PLoS ONE. 2009;4(9):6953.
Jeon JP, Koh IU, Choi NH, Kim BJ, Han BG, Lee S. Differential DNA methylation of MSI2 and its correlation with diabetic traits. PLoS ONE. 2017;12(5):e0177406–e0177406.
Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10(3):e1004160–e1004160.
Sjöholm LK, Ransome Y, Ekström TJ, Karlsson O. Evaluation of post-mortem effects on global brain DNA methylation and hydroxymethylation. Basic Clin Pharmacol Toxicol. 2018;122(2):208–13.
Vilahur N, Baccarelli AA, Bustamante M, Agramunt S, Byun HM, Fernandez MF, et al. Storage conditions and stability of global DNA methylation in placental tissue. Epigenomics. 2013;5(3):341–8.
Makki K, Froguel P, Wolowczuk I. Adipose tissue in obesity-related inflammation and insulin resistance: cells, cytokines, and chemokines. ISRN Inflammation. 2013;22(2013):1–12.
Dahlman I, Sinha I, Gao H, Brodin D, Thorell A, Rydén M, et al. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. Int J Obes. 2015;39(6):910–9.
Article CAS Google Scholar
Fujiki K, Shinoda A, Kano F, Sato R, Shirahige K, Murata M. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat Commun. 2013;4(1):2262.
Pfeiffer S, Krüger J, Maierhofer A, Böttcher Y, Klöting N, El Hajj N, et al. Hypoxia-inducible factor 3A gene expression and methylation in adipose tissue is related to adipose tissue dysfunction. Sci Rep. 2016;6(1):27969.
Wang X, Cao Q, Yu L, Shi H, Xue B, Shi H. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight. 2016 Nov 17 [cited 2023 Jun 17]; 1(19). https://insight.jci.org/articles/view/87748
Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63(9):2962–76.
Rodríguez-Rodero S, Menéndez-Torre E, Fernández-Bayón G, Morales-Sánchez P, Sanz L, Turienzo E, et al. Altered intragenic DNA methylation of HOOK2 gene in adipose tissue from individuals with obesity and type 2 diabetes. PLoS ONE. 2017;12(12):e0189153–e0189153.
Wang C, Ha X, Li W, Xu P, Zhang Z, Wang T, et al. Comparative gene expression profile and DNA methylation status in diabetic patients of Kazak and Han people. Medicine. 2018;97(36):e11982–e11982.
Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS ONE. 2012;7(12):e51302–e51302.
Andersen E, Ingerslev LR, Fabre O, Donkin I, Altıntaş A, Versteyhe S, et al. Preadipocytes from obese humans with type 2 diabetes are epigenetically reprogrammed at genes controlling adipose tissue function. Int J Obes. 2019;43(2):306–18.
Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–903.
Walenta JH, Didier AJ, Liu X, Krämer H. The golgi-associated Hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol. 2001;152(5):923–34.
Massa López D, Thelen M, Stahl F, Thiel C, Linhorst A, Sylvester M, et al. The lysosomal transporter MFSD1 is essential for liver homeostasis and critically depends on its accessory subunit GLMP. Elife. 2019;8:e50025.
Kashoor I, Batlle D. Proximal renal tubular acidosis with and without Fanconi syndrome. Kidney Res Clin Pract. 2019;38(3):267–81.
Yamagata K. Roles of HNF1α and HNF4α in Pancreatic β-Cells. In: Vitamins & Hormones [Internet]. Elsevier; 2014 [cited 2023 Aug 5]. pp. 407–23. https://linkinghub.elsevier.com/retrieve/pii/B9780128001745000168
Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4α (Nuclear Receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21(4):1393–403.
Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA. 1997;94(24):13209–14.
Vedi M, Smith JR, Thomas Hayman G, Tutaj M, Brodie KC, De Pons JL, et al. 2022 updates to the rat genome database: a findable, accessible, interoperable, and reusable (FAIR) resource. Genetics. 2023;224(1):1042.
Han HS, Kang G, Kim JS, Choi BH, Koo SH. Regulation of glucose metabolism from a liver-centric perspective. Exp Mol Med. 2016;48(3):e218–e218.
Abderrahmani A, Yengo L, Caiazzo R, Canouil M, Cauchi S, Raverdy V, et al. Increased hepatic PDGF-AA signaling mediates liver insulin resistance in obesity-associated type 2 diabetes. Diabetes. 2018;67(7):1310–21.
Nilsson E, Matte A, Perfilyev A, de Mello VD, Käkelä P, Pihlajamäki J, et al. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J Clin Endocrinol Metab. 2015;100(11):E1491-1501.
Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10(3):189–98.
Yuan W, Xia Y, Bell CG, Yet I, Ferreira T, Ward KJ, et al. An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat Commun. 2014;5:5719–5719.
Kiechl S, Wittmann J, Giaccari A, Knoflach M, Willeit P, Bozec A, et al. Blockade of receptor activator of nuclear factor-κB (RANKL) signaling improves hepatic insulin resistance and prevents development of diabetes mellitus. Nat Med. 2013;19(3):358–63.
Dayeh T, Tuomi T, Almgren P, Perfilyev A, Jansson PA, De Mello VD, et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics. 2016;11(7):482–8.
Chen YT, Lin WD, Liao WL, Tsai YC, Liao JW, Tsai FJ. NT5C2 methylation regulatory interplay between DNMT1 and insulin receptor in type 2 diabetes. Sci Rep. 2020;10(1):16087.
Chen X, Lin Q, Wen J, Lin W, Liang J, Huang H, et al. Whole genome bisulfite sequencing of human spermatozoa reveals differentially methylated patterns from type 2 diabetic patients. Journal of diabetes investigation. 2019
Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21(2):371–83.
Volkov P, Bacos K, Ofori JK, Esguerra JLS, Eliasson L, Rönn T, et al. Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes. 2017;66(4):1074–85.
García-Calzón S, Perfilyev A, Männistö V, de Mello VD, Nilsson E, Pihlajamäki J, et al. Diabetes medication associates with DNA methylation of metformin transporter genes in the human liver. Clin Epigenetics. 2017;9:102–102.
Zampieri M, Bacalini MG, Barchetta I, Scalea S, Cimini FA, Bertoccini L, et al. Increased PARylation impacts the DNA methylation process in type 2 diabetes mellitus. Clin Epigenet. 2021;13(1):114.
Qie R, Chen Q, Wang T, Chen X, Wang J, Cheng R, et al. Association of ABCG1 gene methylation and its dynamic change status with incident type 2 diabetes mellitus: the Rural Chinese Cohort Study. J Hum Genet. 2021;66(4):347–57.
Krause C, Sievert H, Geißler C, Grohs M, El Gammal AT, Wolter S, et al. Critical evaluation of the DNA-methylation markers ABCG1 and SREBF1 for Type 2 diabetes stratification. Epigenomics. 2019;11(8):885–97.
van Otterdijk SD, Binder AM, Szarc Vel Szic K, Schwald J, Michels KB. DNA methylation of candidate genes in peripheral blood from patients with type 2 diabetes or the metabolic syndrome. PLoS ONE. 2017;12(7):e0180955–e0180955.
Toperoff G, Kark JD, Aran D, Nassar H, Ahmad WA, Sinnreich R, et al. Premature aging of leukocyte DNA methylation is associated with type 2 diabetes prevalence. Clin Epigenetics. 2015;7(1):35–35.
Huang S, Qin P, Chen Q, Zhang D, Cheng C, Guo C, et al. Association of FTO gene methylation with incident type 2 diabetes mellitus: A nested case–control study. Gene. 2021;786: 145585.
Zhang D, Cheng C, Cao M, Wang T, Chen X, Zhao Y, et al. TXNIP hypomethylation and its interaction with obesity and hypertriglyceridemia increase type 2 diabetes mellitus risk: a nested case-control study. J Diabetes. 2020;12(7):512–20.
Hu F, Zhang Y, Qin P, Zhao Y, Liu D, Zhou Q, et al. Integrated analysis of probability of type 2 diabetes mellitus with polymorphisms and methylation of KCNQ1 gene: a nested case-control study. J Diabetes. 2021;13(12):975–86.
Ling C, Del Guerra S, Lupi R, Rönn T, Granhall C, Luthman H, et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51(4):615–22.
Wittenbecher C, Ouni M, Kuxhaus O, Jähnert M, Gottmann P, Teichmann A, et al. Insulin-like growth factor binding protein 2 (IGFBP-2) and the risk of developing type 2 diabetes. Diabetes. 2019;68(1):188–97.
Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3(7):526–34.
Sandovici I, Hammerle CM, Cooper WN, Smith NH, Tarry-Adkins JL, Dunmore BJ, et al. Ageing is associated with molecular signatures of inflammation and type 2 diabetes in rat pancreatic islets. Diabetologia. 2016;59(3):502–11.
Lee YH, Kang ES, Kim SH, Han SJ, Kim CH, Kim HJ, et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Hum Genet. 2008;53(11–12):991–8.
Liu Y, Zhou DZ, Zhang D, Chen Z, Zhao T, Zhang Z, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia. 2009;52(7):1315–21.
Hu C, Wang C, Zhang R, Ma X, Wang J, Lu J, et al. Variations in KCNQ1 are associated with type 2 diabetes and beta cell function in a Chinese population. Diabetologia. 2009;52(7):1322–5.
Jonsson A, Isomaa B, Tuomi T, Taneera J, Salehi A, Nilsson P, et al. A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes. 2009;58(10):2409–13.
Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092–7.
Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40(9):1098–102.
Müssig K, Staiger H, Machicao F, Kirchhoff K, Guthoff M, Schäfer SA, et al. Association of Type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes. 2009;58(7):1715–20.
Qian Y, Dong M, Lu F, Li H, Jin G, Hu Z, et al. Joint effect of CENTD2 and KCNQ1 polymorphisms on the risk of type 2 diabetes mellitus among Chinese Han population. Mol Cell Endocrinol. 2015;407:46–51.
Been LF, Ralhan S, Wander GS, Mehra NK, Singh J, Mulvihill JJ, et al. Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: a study of 3,310 subjects from India and the US. BMC Med Genet. 2011;12(1):18.
Hu Y, Shi P, He K, Zhu YQ, Yang F, Yang M, et al. Methylation of Tcf712 promoter by high-fat diet impairs β-cell function in mouse pancreatic islets. Diabetes Metab Res Rev. 2018;34(4): e2980.
Flurkey K, Currer JM, Harrison D. Mouse models in aging research. In: The mouse in biomedical research. Elsevier; 2007, pp. 637–72.
Papadopoulou S, Edlund H. Attenuated Wnt signaling perturbs pancreatic growth but not pancreatic function. Diabetes. 2005;54(10):2844–51.
Kumar A, Mittal R. Mapping Txnip: key connexions in progression of diabetic nephropathy. Pharmacol Rep. 2018;70(3):614–22.
Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces β-cell apoptosis. Endocrinology. 2005;146(5):2397–405.
Tarling EJ, Edwards PA. ATP binding cassette transporter G1 (ABCG1) is an intracellular sterol transporter. Proc Natl Acad Sci USA. 2011;108(49):19719–24.
Westerterp M, Bochem AE, Yvan-Charvet L, Murphy AJ, Wang N, Tall AR. ATP-binding cassette transporters, atherosclerosis, and inflammation. Circ Res. 2014;114(1):157–70.
Wilson PW, McGEE DL, Kannel WB. Obesity, very low density lipoproteins, and glucose intolerance over fourteen years: The Framingham Study. Am J Epidemiol. 1981;114(5):697–704.
Shah UJ, Xie W, Flyvbjerg A, Nolan JJ, Højlund K, Walker M, et al. Differential methylation of the type 2 diabetes susceptibility locus KCNQ1 is associated with insulin sensitivity and is predicted by CpG site specific genetic variation. Diabetes Res Clin Pract. 2019;148:189–99.
Mattei AL, Bailly N, Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38(7):676–707.
Anastasiadi D, Esteve-Codina A, Piferrer F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin. 2018;11(1):37.
Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–8.
Berumen J, Orozco L, Betancourt-Cravioto M, Gallardo H, Zulueta M, Mendizabal L, et al. Influence of obesity, parental history of diabetes, and genes in type 2 diabetes: a case-control study. Sci Rep. 2019;9(1):2748.
Andrade S, Morais T, Sandovici I, Seabra AL, Constância M, Monteiro MP. Adipose tissue epigenetic profile in obesity-related dysglycemia: a systematic review. Front Endocrinol. 2021;29(12): 681649.
Ryan J, Wrigglesworth J, Loong J, Fransquet PD, Woods RL. A systematic review and meta-analysis of environmental, lifestyle, and health factors associated with DNA methylation age. J Gerontol Ser A. 2020;75(3):481–94.
Oblak L, Van Der Zaag J, Higgins-Chen AT, Levine ME, Boks MP. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res Rev. 2021;69: 101348.
Krolevets M, Cate VT, Prochaska JH, Schulz A, Rapp S, Tenzer S, et al. DNA methylation and cardiovascular disease in humans: a systematic review and database of known CpG methylation sites. Clin Epigenet. 2023;15(1):56.
Department of Economic and Social Affairs UN. India to overtake China as world’s most populous country in April 2023, United Nations projects. [cited 2023 Jun 8]. https://www.un.org/en/desa/india-overtake-china-world-most-populous-country-april-2023-united-nations-projects
Anjana RM, Unnikrishnan R, Deepa M, Pradeepa R, Tandon N, Das AK, et al. Metabolic non-communicable disease health report of India: the ICMR-INDIAB national cross-sectional study (ICMR-INDIAB-17). Lancet Diabetes Endocrinol. 2023;11(7):474–89.
Hwang JY, Lee HJ, Go MJ, Jang HB, Choi NH, Bae JB, et al. Genome-wide methylation analysis identifies ELOVL5 as an epigenetic biomarker for the risk of type 2 diabetes mellitus. Sci Rep. 2018;8(1):14862–14862.
Zou L, Yan S, Guan X, Pan Y, Qu X. Hypermethylation of the PRKCZ Gene in Type 2 diabetes mellitus. J Diabetes Res. 2013;2013:721493–721493.
Chen YT, Liao JW, Tsai YC, Tsai FJ. Inhibition of DNA methyltransferase 1 increases nuclear receptor subfamily 4 group A member 1 expression and decreases blood glucose in type 2 diabetes. Oncotarget. 2016;7(26):39162–70.
Davegårdh C, Säll J, Benrick A, Broholm C, Volkov P, Perfilyev A, et al. VPS39-deficiency observed in type 2 diabetes impairs muscle stem cell differentiation via altered autophagy and epigenetics. Nat Commun. 2021;12(1):2431.
Whytock KL, Pino MF, Sun Y, Yu G, De Carvalho FG, Yeo RX, et al. Comprehensive interrogation of human skeletal muscle reveals a dissociation between insulin resistance and mitochondrial capacity. Am J Physiol-Endocrinol Metabol. 2023;325(4):E291-302.
Download references
Acknowledgements
AM is supported by the Wellcome Trust/DBT India Alliance Fellowship [Grant Number IA/CPHI/19/1/504593]. We thank Ms. Ramya for her insightful comments.
This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
Author information
Priyanka Chinya Nataraj
Present address: Vedantu, Bangalore, India
Authors and Affiliations
Research Scholar, Manipal Academy of Higher Education, Manipal, India
Nikhil Nadiger
Division of Nutrition, St. John’s Research Institute, St. John’s Medical College, St Johns National Academy of Health Sciences, Sarjapura Road, Koramangala, Bangalore, 560034, India
Nikhil Nadiger, Jyothisha Kana Veed, Priyanka Chinya Nataraj & Arpita Mukhopadhyay
You can also search for this author in PubMed Google Scholar
Contributions
AM was involved in conceptualization, review and editing of the manuscript. NN, JKV and PCN were involved in data extraction, formal analysis, investigation and writing the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Correspondence to Arpita Mukhopadhyay .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Conflict of interest.
The authors declare no conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1 search strategy for the systematic review of dna methylation association with t2dm, 13148_2024_1670_moesm2_esm.docx.
Additional file 2 Qualitative assessment of research articles included in the review based on the New Castle Ottawa Scale (NOS)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Nadiger, N., Veed, J.K., Chinya Nataraj, P. et al. DNA methylation and type 2 diabetes: a systematic review. Clin Epigenet 16 , 67 (2024). https://doi.org/10.1186/s13148-024-01670-6
Download citation
Received : 20 January 2024
Accepted : 11 April 2024
Published : 16 May 2024
DOI : https://doi.org/10.1186/s13148-024-01670-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Type 2 diabetes
- DNA methylation
- Epigenome-wide association studies
- Epigenetics
Clinical Epigenetics
ISSN: 1868-7083
- Submission enquiries: [email protected]
- General enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
Here you can access information about the PRISMA reporting guidelines, which are designed to help authors transparently report why their systematic review was done, what methods they used, and what they found. The main PRISMA reporting guideline (the PRISMA 2020 statement) primarily provides guidance for the reporting of systematic reviews ...
The PRISMA 2020 items are relevant for mixed-methods systematic reviews (which include quantitative and qualitative studies), but reporting guidelines addressing the presentation and synthesis of qualitative data should also be consulted. 14 15 PRISMA 2020 can be used for original systematic reviews, updated systematic reviews, or continually ...
Development of PRISMA 2020. A complete description of the methods used to develop PRISMA 2020 is available elsewhere.35 We identified PRISMA 2009 items that were often reported incompletely by examining the results of studies investigating the transparency of reporting of published reviews.17 21 36 37 We identified possible modifications to the PRISMA 2009 statement by reviewing 60 documents ...
It has been more than a decade since the original publication of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement [], and it has become one of the most cited reporting guidelines in biomedical literature [2, 3].Since its publication, multiple extensions of the PRISMA Statement have been published concomitant with the advancement of knowledge synthesis ...
2. Development of PRISMA 2020. A complete description of the methods used to develop PRISMA 2020 is available elsewhere [35].We identified PRISMA 2009 items that were often reported incompletely by examining the results of studies investigating the transparency of reporting of published reviews [17, 21, 36, 37].We identified possible modifications to the PRISMA 2009 statement by reviewing 60 ...
PRISMA is an evidence-based minimum set of items for reporting in systematic reviews and meta-analyses. PRISMA focuses on the reporting of reviews evaluating randomized trials, but can also be used as a basis for reporting systematic reviews of other types of research, particularly evaluations of interventions.
Using statistical methods for the interpretation of the results implies a systematic review containing meta-analysis ( 6 ). The PRISMA guidelines consist of a four-phase flow diagram and a 27-item checklist. The flow diagram describes the identification, screening, eligibility and inclusion criteria of the reports that fall under the scope of a ...
To document your grey literature search, download the flow diagram template version 1 PRISMA 2020 flow diagram for new systematic reviews which included searches of databases, registers and other sources or the version 2 PRISMA 2020 flow diagram for updated systematic reviews which included searches of databases, registers and other sources.
Documenting grey literature and/or hand searches. If you have also searched additional sources, such as professional organization websites, cited or citing references, etc., document your grey literature search using the flow diagram template version 1 PRISMA 2020 flow diagram for new systematic reviews which included searches of databases, registers and other sources or the version 2 PRISMA ...
views and Meta-Analyses (PRISMA) group, which. mainly consists of Cochrane authors, has developed. the PRISMA guidelines in 2009 (6). A systematic. review will extensively scan all reports ...
The Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement published in 2009 (hereafter referred to as PRISMA 2009) [4,5,6,7,8,9,10] is a reporting guideline designed to address poor reporting of systematic reviews [].The PRISMA 2009 statement comprised a checklist of 27 items recommended for reporting in systematic reviews and an "explanation and elaboration ...
PRISMA stands for Preferred Reporting Items for Systematic Reviews and Meta-Analyses. It is an evidence-based minimum set of items for reporting in systematic reviews and meta-analyses. The PRISMA statement consists of a 27-item checklist and a 4-phase flow diagram. These items have been adapted for use by students conducting systematic reviews ...
Development of PRISMA 2020. A complete description of the methods used to develop PRISMA 2020 is available elsewhere. 35 We identified PRISMA 2009 items that were often reported incompletely by examining the results of studies investigating the transparency of reporting of published reviews. 17 21 36 37 We identified possible modifications to the PRISMA 2009 statement by reviewing 60 documents ...
Systematic reviews in the literature have become more important as a means of keeping clinicians up to date on advances in the field and as a method of developing clinical practice guidelines. ... been made in the science of systematic reviews. PRISMA also has adopted the definition used by the Cochrane Collaboration: "a systematic review is ...
The PRISMA flow diagram, depicting the flow of information through the different phases of a systematic review. PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) is an evidence-based minimum set of items aimed at helping scientific authors to report a wide array of systematic reviews and meta-analyses, primarily used to assess the benefits and harms of a health care ...
Systematic reviews and meta-analyses are essential to summarise evidence relating to efficacy and safety of healthcare interventions accurately and reliably. The clarity and transparency of these reports, however, are not optimal. Poor reporting of systematic reviews diminishes their value to clinicians, policy makers, and other users. Since the development of the QUOROM (quality of reporting ...
Systematic reviews and Meta-Analyses (PRISMA). Using the chart flow and the list of items of this method, a literature search was conducted in Science Direct Freedom Collection, Elsevier database, Web of Science - Core Collection, Springer Link Journals. In this paper are given 2 examples of systematic reviews that were obtained using PRISMA ...
PDF | On May 14, 2024, Antonio E. L. Nyoko and others published A Systematic Literature Review on Entrepreneurial Intention: PRISMA method | Find, read and cite all the research you need on ...
Indicate when search strategies from other literature reviews were adapted or reused for a substantive part or all of the search, citing the previous review(s). Updates: 12: Report the methods used to update the search(es) (e.g., rerunning searches, email alerts). Dates of searches: 13
TY - CONF AU - Antonio E. L. Nyoko AU - Mohd Hizam Hanafiah PY - 2024 DA - 2024/05/14 TI - A Systematic Literature Review on Entrepreneurial Intention: PRISMA method BT - Proceedings of the International Conference on Economic Management, Accounting and Tourism (ICEMAT 2023) PB - Atlantis Press SP - 254 EP - 273 SN - 2352-5428 UR - https://doi ...
Methods: A systematic literature search following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines was conducted on 22nd of February 2022 using the Ovid MEDLINE and Web of Science databases. Three categories of search terms were used: (1) self-harm, self-injury, self-mutilation, suicide, self-poisoning; (2 ...
The study used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) model for a systematic literature review on global ECF regulations. Starting with 74 initial articles from Web of Sciences and Scopus databases, duplicates were removed and language criteria applied, leaving 42 articles.
Methods. PubMed and ScienceDirect databases were searched (till October 19, 2023). Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines and New Castle Ottawa scale were used for reporting the selection and quality of the studies, respectively. Result. Thirty-two articles were selected.
Over the past decade, advances in systematic review methodology and terminology have necessitated an update to the guideline. The PRISMA 2020 statement replaces the 2009 statement and includes new reporting guidance that reflects advances in methods to identify, select, appraise, and synthesise studies. The structure and presentation of the ...
We focused on literature related to academic advising for non-traditional students in formal HE, restricting our search to both empirical and non-empirical articles published in peer-reviewed journals between 2010 and 2022. ... Employing Arksey and O'Malley's scoping review method and the PRISMA-ScR Checklist, we searched four databases ...
Volcanic clouds pose significant threats to air traffic, human health, and economic activity, making early detection and monitoring crucial. Accurate determination of eruptive source parameters is crucial for forecasting and implementing preventive measures. This review article aims to identify the most common remote sensing methods for monitoring volcanic clouds. To achieve this, we conducted ...