
New consortium to advance research on infectious disease prevention, treatment
May 29, 2024.
A new partnership between researchers at the University of Kentucky will combat the threat of infectious diseases through research directed at prevention and treatment.
The Consortium for Understanding and Reducing Infectious Diseases in Kentucky (CURE-KY) will foster multidisciplinary collaborations to address the burden of infectious diseases in the Commonwealth and beyond.
“I am thrilled by the boundless possibilities our collaborative efforts promise at the University of Kentucky. With a steadfast commitment to advancing knowledge and fostering transdisciplinary alliances, we aim to propel infectious disease research to new heights, creating a healthier Kentucky and nurturing future scientific leaders,” said Ilhem Messaoudi, PhD, CURE-KY founder and chair of the Department of Microbiology, Immunology, and Molecular Genetics in the College of Medicine.
CURE-KY grew out of the Emerging Themes for Research Program, which is part of the Research Leadership Academy (RLA) supported by the Office of the Vice President for Research and was built on the heels of UK’s COVID-19 Unified Research Experts (CURE) Alliance that was quickly assembled to support a full range of COVID-related research.
“A key facet of the Research Leadership Academy is to empower our researchers to tackle research questions that are critical to Kentucky and do so with a collaborative and innovative approach,” said Lisa Cassis, UK’s vice president for research. “CURE-KY is an exemplary model of this strategy. I am excited to see the range of this team’s scientific expertise and their dedication to advancing the health of Kentucky through vital research.”
CURE-KY brings together a diverse group of experts from the College of Medicine, College of Public Health, College of Pharmacy, Martin-Gatton College of Agriculture, Food and Environment and Markey Cancer Center.
CURE-KY will focus on research in five areas: microbial pathogenesis; vaccines, therapeutics and antimicrobial resistance; public health and community engagement; animal health; and engineering and systems.
The consortium will also work to address vaccine hesitancy and misinformation to foster trust between Kentuckians and the scientific community.
“CURE-KY leverages the expertise of some of the best and brightest physicians and scientists specializing in infectious disease treatment and prevention. With the power of this unified team, the University of Kentucky will equip the Commonwealth with the information, resources and health care solutions necessary to combat dangerous diseases,” said Charles “Chipper” Griffith III, MD, dean of the College of Medicine .
“The College of Public Health is dedicated to bettering public health, which is tightly integrated with the mission of CURE-KY. From studying prevention, control and understanding transmission dynamics to assessing risk, promoting health equity and engaging communities in disease control efforts, this work is critical toward our goal of creating a healthier Kentucky,” said Heather Bush, PhD, dean of the College of Public Health .
"We are proud to be a part of the CURE-KY consortium, which represents a pivotal step forward in our ongoing commitment to improve public health in Kentucky,” said R. Kip Guy, PhD, dean of the College of Pharmacy . “The collective expertise on our campus in drug discovery, vaccine development and infectious disease research is essential for addressing the complex challenges presented in Kentucky and beyond.”
“From Dr. Li’s groundbreaking research on infectious diseases in animals to the breadth of research in plant sciences, engineering and biotechnology, our college is a powerhouse of expertise that propels transdisciplinary projects like CURE-KY, fostering innovation and collaboration at every turn," said Nancy Cox, vice president for land-grant engagement and dean of the UK Martin-Gatton College of Agriculture, Food and Environment .
“Kentucky is home to the highest cancer rates in the country, including several cancers caused by infectious diseases such as HPV and hepatitis C. Cancer prevention is a key component of Markey’s mission to significantly reduce Kentucky’s cancer burden. By better understanding and combating diseases that can lead to cancer, CURE-KY will contribute to this mission and make progress toward our ultimate goal of conquering cancer in the Commonwealth,” said B. Mark Evers, MD, director of the Markey Cancer Center .
“We have an excellent team of scientists here at UK and on our leadership team. Together we can tackle infectious diseases from an array of perspectives to make a difference in the health of our community,” said Messaoudi.
You can find more information about CURE-KY and upcoming events on their website: https://medicine.uky.edu/sites/cure-ky
Share this Article
Latest stories.
May 29, 2024
May 28, 2024
May 23, 2024
- U.S. Department of Health & Human Services

- Virtual Tour
- Staff Directory
- En Español
You are here
News releases.
Media Advisory
Thursday, May 30, 2024
Novel vaccine concept generates immune responses that could produce multiple types of HIV broadly neutralizing antibodies
NIH-funded animal model results will inform vaccine development in humans.

Using a combination of cutting-edge immunologic technologies, researchers have successfully stimulated animals’ immune systems to induce rare precursor B cells of a class of HIV broadly neutralizing antibodies (bNAbs). The findings, published today in Nature Immunology , are an encouraging, incremental step in developing a preventive HIV vaccine.
HIV is genetically diverse making the virus difficult to target with a vaccine, but bNAbs may overcome that hurdle because they bind to parts of the virus that remain constant even when it mutates. Germline targeting is an immune system-stimulating approach that guides naïve (precursor) B cells to develop into mature B cells that can produce bNAbs. A class of bNAbs called 10E8 is a priority for HIV vaccine development because it neutralizes a particularly broad range of HIV variants. The 10E8 bNAb binds to a conserved region of the glycoprotein gp41 on HIV’s surface involved in its entry into human immune cells. Designing an immunogen—a molecule used in a vaccine that elicits a specific immune system response—to stimulate production of 10E8 bNAbs has been challenging because that key region of gp41 is hidden in a recessed crevice on HIV’s surface. Prior vaccine immunogens have not generated bNAbs with the physical structure to reach and bind to gp41.
To address this challenge, the researchers engineered immunogens on nanoparticles that mimic the appearance of a specific part of gp41. They vaccinated rhesus macaque monkeys and mice with those immunogens and elicited specific responses from the 10E8 B cell precursors and induced antibodies that showed signs of maturing into bNAbs that could reach the hidden gp41 region. They observed similar responses when they used mRNA-encoded nanoparticles in mice. The researchers also found that the same immunogens produced B cells that could mature to produce an additional type of gp41-directed bNAb called LN01. Finally, their laboratory analysis of human blood samples found that 10E8-class bNAb precursors occurred naturally in people without HIV, and that their immunogens bound to and isolated naïve human B cells with 10E8-like features. Together these observations suggest that the promising immunization data from mice and macaques has the potential for translation to humans.
The research was conducted by the Scripps Consortium for HIV/AIDS Vaccine Development, one of two consortia supported by the National Institutes of Health’s (NIH) National Institute of Allergy and Infectious Diseases (NIAID). The research also was supported by collaborating partners including the Bill & Melinda Gates Foundation and other NIH Institutes and Offices. According to the authors, these findings support the development of the immunogens as the first part of a multi-step vaccine regimen for humans. Their work further supports research in developing a germline-targeting strategy for priming the immune system to elicit a bNAb called VRC01. This bNAb was discovered by NIAID researchers almost 15 years ago. The goal of this line of research is to develop an HIV vaccine that generates multiple classes of bNAbs to prevent HIV.
Schiffner et al . Vaccination induces broadly neutralizing antibody precursors to HIV gp41. Nature Immunology DOI: 10.1038/s41590-024-01833-w (2024).
Angela Malaspina, program officer in NIAID’s Division of AIDS, is available to discuss this study.
NIAID conducts and supports research—at NIH, throughout the United States, and worldwide—to study the causes of infectious and immune-mediated diseases, and to develop better means of preventing, diagnosing and treating these illnesses. News releases, fact sheets and other NIAID-related materials are available on the NIAID website .
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit www.nih.gov .
NIH…Turning Discovery Into Health ®
Connect with Us
- More Social Media from NIH
Recent advances in treatment and prevention of infectious disease
Review 11 November 2022 Review of preclinical data of PF-07304814 and its active metabolite derivatives against SARS-CoV-2 infection Wujun Chen , 4 more and Dongming Xing 2,528 views 4 citations
Loading... Review 08 September 2022 Remdesivir in treating hospitalized patients with COVID-19: A renewed review of clinical trials Zhenchao Wu , 2 more and Ning Shen 4,771 views 11 citations
Review 29 August 2022 Nucleic acid strategies for infectious disease treatments: The nanoparticle-based oral delivery route Fengqian Chen , 2 more and Li Xu 2,251 views 1 citations
Original Research 24 August 2022 Using an untargeted metabolomics approach to analyze serum metabolites in COVID-19 patients with nucleic acid turning negative Wenyu Chen , 10 more and Ziqing Kong 3,297 views 8 citations
Original Research 12 July 2022 Reduning Attenuates LPS-Induced Human Unmilical Vein Endothelial Cells (HUVECs) Apoptosis Through PI3K-AKT Signaling Pathway Ziyi Wang , 5 more and Zhong Wang 3,107 views 4 citations
Original Research 06 June 2022 Predicted Vancomycin Dosage Requirement in Patients With Hematological Malignancies and Dosage Dynamic Adjustment Xiangqing Song and Yi Wu 1,809 views 2 citations
Loading... Original Research 27 April 2022 Discovery of METTL3 Small Molecule Inhibitors by Virtual Screening of Natural Products Yue Du , 5 more and Xin Tian 6,149 views 32 citations
Original Research 26 April 2022 A Potent Antibiotic Combination of Linezolid and Polymycin B Nonapeptide Against Klebsiella pneumoniae Infection In Vitro and In Vivo Ting Huang , 5 more and Yiwen Chu 2,027 views 5 citations

- Infectious Diseases
Explore the latest in infectious diseases, including community-acquired and nosocomial disease, antibiotic use and stewardship, and more.
Publication
Article type.
This cohort study examines whether systemic fluoroquinolone use is associated with increased risk of acute uveitis or retinal detachment.
This cohort study examines whether outcomes of primary prevention strategies for gastric cancer, including Helicobacter pylori treatment, differ by genetic risk profile among adults in China.
A 43-year-old man presented with a destructive mass on the nose, with multiple areas of erosion, ulcers, and exudate covering the mass. Histological examination showed polymorphous lymphocytic infiltration in the full-thickness dermis. What is your diagnosis?
This case report describes a woman with Creutzfeldt-Jakob disease with findings largely confined to the right precentral gyrus on initial imaging.
- Monoclonal Antibody Protected Majority of Children From Malaria JAMA News May 24, 2024 Global Health Pediatrics Full Text | pdf link PDF free
- Lower Dose of Mpox Vaccine Was Safe, Effective JAMA News May 24, 2024 Mpox (Monkeypox) Vaccination Full Text | pdf link PDF free
- New Drug for UTIs Gets the Greenlight JAMA News May 24, 2024 Urology Reproductive Health Full Text | pdf link PDF free
This Viewpoint discusses the importance of proactively discussing preexposure prophylaxis (PrEP) with Black cisgender women.
This cohort study uses data from 14 national or regional total knee arthroplasty (TKA) registries to compare the rates of revision owing to periprosthetic joint infection among patients who received antibiotic-loaded bone cement or plain bone cement.
This secondary cross-protocol analysis investigates the viral load at COVID-19 diagnosis among previously uninfected and unvaccinated individuals by demographic and clinical characteristics, viral variant, and trial, as well as the ability of viral load to predict severe disease.
This cluster randomized trial examines the effectiveness of a school- and primary care–based multicomponent intervention to increase human papillomavirus vaccination coverage among children aged 11 to 14 years.
This case report describes nonblanching, confluent, purpuric macules and patches on the palmoplantar and dorsal finger surfaces and red to grayish papulovesicles on the dorsal hands and feet, limbs, trunk, and face.
This JAMA Insights provides recommendations for Chagas disease screening, diagnosis, and management in the US
US public health preparedness and response to highly pathogenic avian influenza A(H5N1) viruses are assessed in this survey study conducted by the CDC.
This randomized trial assesses whether acetaminophen would increase the number of days alive and free of organ support to day 28 among critically ill patients with sepsis and respiratory or circulatory organ dysfunction.
This Medical News story discusses the development of experimental combination vaccines for adults.
- WHO Updates Terminology for Pathogens That Spread Through the Air JAMA News May 17, 2024 Global Health Full Text | pdf link PDF free
This cohort study examines long-term clinical outcomes, including mortality and antibiotic-resistant infection with methicillin-resistant Staphylococcus aureus , Clostridium difficile , or vancomycin-resistant Enterococcus , for adults with β-lactam allergies living in western Pennsylvania.
This cross-sectional study identifies the common diagnoses and physician encounter types associated with clotrimazole-betamethasone dipropionate prescriptions among Medicare enrollees in 2021.
- Resistant Trichophyton indotineae Dermatophytosis—An Emerging Pandemic, Now in the US JAMA Dermatology Opinion May 15, 2024 Fungal Infections Skin Infections Dermatology Full Text | pdf link PDF
Select Your Interests
Customize your JAMA Network experience by selecting one or more topics from the list below.
- Academic Medicine
- Acid Base, Electrolytes, Fluids
- Allergy and Clinical Immunology
- American Indian or Alaska Natives
- Anesthesiology
- Anticoagulation
- Art and Images in Psychiatry
- Artificial Intelligence
- Assisted Reproduction
- Bleeding and Transfusion
- Caring for the Critically Ill Patient
- Challenges in Clinical Electrocardiography
- Climate and Health
- Climate Change
- Clinical Challenge
- Clinical Decision Support
- Clinical Implications of Basic Neuroscience
- Clinical Pharmacy and Pharmacology
- Complementary and Alternative Medicine
- Consensus Statements
- Coronavirus (COVID-19)
- Critical Care Medicine
- Cultural Competency
- Dental Medicine
- Dermatology
- Diabetes and Endocrinology
- Diagnostic Test Interpretation
- Drug Development
- Electronic Health Records
- Emergency Medicine
- End of Life, Hospice, Palliative Care
- Environmental Health
- Equity, Diversity, and Inclusion
- Facial Plastic Surgery
- Gastroenterology and Hepatology
- Genetics and Genomics
- Genomics and Precision Health
- Global Health
- Guide to Statistics and Methods
- Hair Disorders
- Health Care Delivery Models
- Health Care Economics, Insurance, Payment
- Health Care Quality
- Health Care Reform
- Health Care Safety
- Health Care Workforce
- Health Disparities
- Health Inequities
- Health Policy
- Health Systems Science
- History of Medicine
- Hypertension
- Images in Neurology
- Implementation Science
- Innovations in Health Care Delivery
- JAMA Infographic
- Law and Medicine
- Leading Change
- Less is More
- LGBTQIA Medicine
- Lifestyle Behaviors
- Medical Coding
- Medical Devices and Equipment
- Medical Education
- Medical Education and Training
- Medical Journals and Publishing
- Mobile Health and Telemedicine
- Narrative Medicine
- Neuroscience and Psychiatry
- Notable Notes
- Nutrition, Obesity, Exercise
- Obstetrics and Gynecology
- Occupational Health
- Ophthalmology
- Orthopedics
- Otolaryngology
- Pain Medicine
- Palliative Care
- Pathology and Laboratory Medicine
- Patient Care
- Patient Information
- Performance Improvement
- Performance Measures
- Perioperative Care and Consultation
- Pharmacoeconomics
- Pharmacoepidemiology
- Pharmacogenetics
- Pharmacy and Clinical Pharmacology
- Physical Medicine and Rehabilitation
- Physical Therapy
- Physician Leadership
- Population Health
- Primary Care
- Professional Well-being
- Professionalism
- Psychiatry and Behavioral Health
- Public Health
- Pulmonary Medicine
- Regulatory Agencies
- Reproductive Health
- Research, Methods, Statistics
- Resuscitation
- Rheumatology
- Risk Management
- Scientific Discovery and the Future of Medicine
- Shared Decision Making and Communication
- Sleep Medicine
- Sports Medicine
- Stem Cell Transplantation
- Substance Use and Addiction Medicine
- Surgical Innovation
- Surgical Pearls
- Teachable Moment
- Technology and Finance
- The Art of JAMA
- The Arts and Medicine
- The Rational Clinical Examination
- Tobacco and e-Cigarettes
- Translational Medicine
- Trauma and Injury
- Treatment Adherence
- Ultrasonography
- Users' Guide to the Medical Literature
- Vaccination
- Venous Thromboembolism
- Veterans Health
- Women's Health
- Workflow and Process
- Wound Care, Infection, Healing
- Register for email alerts with links to free full-text articles
- Access PDFs of free articles
- Manage your interests
- Save searches and receive search alerts
Suggestions or feedback?
MIT News | Massachusetts Institute of Technology
- Machine learning
- Social justice
- Black holes
- Classes and programs
Departments
- Aeronautics and Astronautics
- Brain and Cognitive Sciences
- Architecture
- Political Science
- Mechanical Engineering
Centers, Labs, & Programs
- Abdul Latif Jameel Poverty Action Lab (J-PAL)
- Picower Institute for Learning and Memory
- Lincoln Laboratory
- School of Architecture + Planning
- School of Engineering
- School of Humanities, Arts, and Social Sciences
- Sloan School of Management
- School of Science
- MIT Schwarzman College of Computing
Scientists identify mechanism behind drug resistance in malaria parasite
Press contact :.

Previous image Next image
Share this news article on:
Related links.
- Peter Dedon
- Antimicrobial Resistance Interdisciplinary Research Group
- Singapore-MIT Alliance for Research and Technology (SMART)
- Department of Biological Engineering
Related Topics
- Drug resistance
- Antibiotics
- Biological engineering
- Drug development
- International initiatives
- Collaboration
Related Articles

Satellite-based method measures carbon in peat bogs

Newly discovered bacterial communication system aids antimicrobial resistance

SMART launches research group to advance AI, automation, and the future of work

A novel combination therapy counters antibiotic-resistant Mycobacterium abscessus infections
Previous item Next item
More MIT News

Noubar Afeyan PhD ’87 gives new MIT graduates a special assignment
Read full story →

President Sally Kornbluth’s charge to the Class of 2024

Commencement address by Noubar Afeyan PhD ’87

Microscopic defects in ice influence how massive glaciers flow, study shows

Getting to systemic sustainability

New MIT-LUMA Lab created to address climate challenges in the Mediterranean region
- More news on MIT News homepage →
Massachusetts Institute of Technology 77 Massachusetts Avenue, Cambridge, MA, USA
- Map (opens in new window)
- Events (opens in new window)
- People (opens in new window)
- Careers (opens in new window)
- Accessibility
- Social Media Hub
- MIT on Facebook
- MIT on YouTube
- MIT on Instagram
- Research article
- Open access
- Published: 17 November 2021
The impact of behavioural risk factors on communicable diseases: a systematic review of reviews
- Sara Wood 1 ,
- Sophie E. Harrison 2 , 3 ,
- Natasha Judd 1 , 2 ,
- Mark A. Bellis ORCID: orcid.org/0000-0001-6980-1963 1 , 2 ,
- Karen Hughes 1 , 2 &
- Andrew Jones 4
BMC Public Health volume 21 , Article number: 2110 ( 2021 ) Cite this article
11k Accesses
13 Citations
25 Altmetric
Metrics details
The coronavirus (COVID-19) pandemic has highlighted that individuals with behavioural risk factors commonly associated with non-communicable diseases (NCDs), such as smoking, harmful alcohol use, obesity, and physical inactivity, are more likely to experience severe symptoms from COVID-19. These risk factors have been shown to increase the risk of NCDs, but less is known about their broader influence on communicable diseases. Taking a wide focus on a range of common communicable diseases, this review aimed to synthesise research examining the impact of behavioural risk factors commonly associated with NCDs on risks of contracting, or having more severe outcomes from, communicable diseases.
Literature searches identified systematic reviews and meta-analyses that examined the association between behavioural risk factors (alcohol, smoking, illicit drug use, physical inactivity, obesity and poor diet) and the contraction/severity of common communicable diseases, including infection or associated pathogens. An a priori, prospectively registered protocol was followed (PROSPERO; registration number CRD42020223890).
Fifty-three systematic reviews were included, of which 36 were also meta-analyses. Reviews focused on: tuberculosis, human immunodeficiency virus, hepatitis C virus, hepatitis B virus, invasive bacterial diseases, pneumonia, influenza, and COVID-19. Twenty-one reviews examined the association between behavioural risk factors and communicable disease contraction and 35 examined their association with communicable disease outcomes (three examined their association with both contraction and outcomes). Fifty out of 53 reviews (94%) concluded that at least one of the behavioural risk factors studied increased the risk of contracting or experiencing worse health outcomes from a communicable disease. Across all reviews, effect sizes, where calculated, ranged from 0.83 to 8.22.
Conclusions
Behavioural risk factors play a significant role in the risk of contracting and experiencing more severe outcomes from communicable diseases. Prevention of communicable diseases is likely to be most successful if it involves the prevention of behavioural risk factors commonly associated with NCDs. These findings are important for understanding risks associated with communicable disease, and timely, given the COVID-19 pandemic and the need for improvements in future pandemic preparedness. Addressing behavioural risk factors should be an important part of work to build resilience against any emerging and future epidemics and pandemics.
Peer Review reports
The recent coronavirus (COVID-19) pandemic has highlighted that individuals with potentially modifiable behavioural risk factors that are commonly associated with non-communicable diseases (NCDs), such as smoking, harmful alcohol use, obesity and physical inactivity, are more likely to experience severe symptoms from COVID-19 infection [ 1 ], resulting in greater risk of hospitalisation [ 2 ]. With these behavioural risk factors often having higher prevalence in the poorest communities, COVID-19 has disproportionately impacted those already suffering the greatest risks of ill health, thereby widening health and social inequalities [ 3 ]. Indeed, due to its associations with existing health and social risk factors, COVID-19 has been referred to as a syndemic; one in which existing health and social challenges increase an individual’s susceptibility to disease [ 4 ]. However, whilst addressing behavioural risk factors is routinely considered in the prevention of NCDs, their role in the contraction of communicable disease, and severity of symptoms in those who are infected, has had a lower public health prominence.
Many modifiable behavioural risk factors are highly prevalent among adults and adolescents in both higher (HICs) and lower and middle income countries (LMICs) [ 5 , 6 ], with levels increasing in many LMICs (e.g. obesity, alcohol) [ 7 , 8 ]. As a result, NCDs, such as cancer, respiratory disease and cardiovascular disease are the highest cause of mortality and morbidity in HICs and account for a rapidly increasing proportion of both in LMICs [ 9 ]. Across countries globally, the burden of NCDs has been found to correlate with levels of COVID-19 cases and deaths [ 10 ].
With both international commerce and tourism connecting populations globally, it is highly likely that COVID-19 is only one in a series of existing and emerging infectious diseases likely to impact, to different extents, health and well-being on a global scale [ 11 ]. Although the exact nature or source of any future epidemic or pandemic threat is speculative, behavioural risk factors have also been found to increase the risk of infection and subsequent poorer outcomes across a range of other communicable diseases [ 12 , 13 , 14 ]. Understanding which factors may increase or reduce risk of contraction and severity of disease can provide important intelligence, both in increasing a population’s resilience to infectious disease, and in identifying which communities and individuals may be most at risk from the spread of different types of disease. Although previous research has explored links between behavioural risk factors and individual communicable diseases, few studies have synthesised information across a wider range of communicable diseases and their relationships with behavioural risks. Indeed, such relationships may elucidate how future pandemics threats will exploit behavioural risk factors.
Intending to explore whether communicable diseases and NCDs share a common set of behavioural risk factors, the aim of this review was to provide a synthesis of existing research examining the impact of behavioural risk factors commonly associated with NCDs on the risk of people (adults or children) contracting, or experiencing more severe outcomes from, common communicable diseases. With the breadth of communicable diseases requiring limitation, the focus of this review was on diseases common to high income countries. With an intentionally wide focus on a range of communicable diseases, the review focused specifically on systematic reviews and meta-analyses, clarifying existing knowledge and highlighting gaps in evidence to inform priority areas for future research.
This review was carried out in adherence to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. An a priori protocol was followed and prospectively registered at the National Institute for Health Research international prospective register of systematic reviews (PROSPERO) (registration number CRD42020223890). The focus of this review was limited to behavioural risk factors and communicable diseases common in HICs, regardless of the geographical location of the review. Those more specific to LMICs or certain regions of the world (e.g. tropical diseases) were considered best examined in a separate study.
Search strategy
A systematic search was performed across multiple databases through ProQuest covering the 10-year period 28th October 2010 to 28th October 2020. Preliminary scans of the literature reviews and discussion between members of the research team were used to aid selection of common behavioural risk factors and communicable diseases. Thus, alcohol use, smoking, physical inactivity, obesity, illicit drug use and poor diet were chosen as behavioural risk factors, covering some of the most common behavioural contributors to NCDs [ 15 ]. The same process identified: Tuberculosis (TB), acquired immune deficiency syndrome (AIDS), human immunodeficiency virus (HIV), viral hepatitis, COVID-19, severe acute respiratory syndrome (SARS), middle-east respiratory syndrome (MERS), pneumonia, influenza, and meningitis as communicable diseases (including infections and pathogens) feasible for review. These communicable diseases were broadly consistent with some of the most prevalent disease/infection categories reported in the global burden of disease study for HICs (excluding those categories predominantly affecting specific groups (e.g. maternal, neonatal) or where a component may be associated with non-infectious causes (e.g. diarrheal) [ 9 ]) and with previous outbreaks or epidemics involving HICs [ 16 ]. In addition, it was intended to include diseases that arose from both bacterial and viral pathogens, with a range of transmission types, e.g. airborne, droplet, fomite, blood-borne and contact. Combinations of search terms were developed based on these key risk factors and diseases. Search results were restricted to English language and peer reviewed systematic reviews and meta-analyses. Whilst this strategy restricted literature to that which qualified for inclusion in systematic reviews, it allowed for the inclusion of multiple behavioural risk factors and communicable diseases at the same time. The search was restricted to a 10-year period, allowing coverage of a broad range of behavioural risk factors and communicable diseases, yet limiting the literature to a manageable volume. The full search strategy is available in Supplementary file 1 . Searches also included poor housing conditions as a risk factor given the impact of housing conditions on respiratory disease [ 17 ], but this study focuses specifically on behavioural risk factors.
Study selection and eligibility criteria
To identify eligible studies, the titles and abstracts of studies retrieved were screened by two reviewers, with a sample of 15% screened independently by both reviewers and achieving 98.5% agreement (SH, NJ). Discrepancies were resolved between reviewers. For full text screening, ten reviews (7%) were initially screened by three reviewers (SH, NJ, SW), with results later discussed and discrepancies resolved. Following this, the full text screening was divided across reviewers and any reviews that were not a clear exclude/include (20%) were discussed and agreed between reviewers. This meant that, across all reviews screened by full text, 27% were discussed and agreed by more than one reviewer. Where full texts could not be accessed, authors were contacted to request the full text.
Systematic reviews and meta-analyses of observational studies (including cohort and case-control studies) that examined the association between an identified behavioural risk factor and the contraction or outcomes of an identified communicable disease (including infection or related pathogens) were included in the review. Since there are no single definitions of the selected risk factors across the literature, all reviews that focused on an identified risk factor were included regardless of the definition used in the review (definitions are provided in Tables 1 and 2 and in the results section). An aim of the review was to explore risks associated with illicit drug use in general. However, it was recognised that injection drug use can be a mechanism of transmission for some pathogens relating to included communicable diseases (e.g. HIV, HCV). Studies that focused specifically on injection drug use were therefore included alongside those focusing on drug use more generally. Studies were excluded if they: were not a systematic review or meta-analysis; did not examine the association between an selected behavioural risk factor and the contraction or outcomes of an selected communicable disease; included only selected specialist sub-populations (e.g. sex workers, prisoners), or included sub-populations relating to a risk factor (e.g. people who inject drugs) without a general population comparison; or included behavioural risk factors or communicable diseases not relevant to HICs (see above). No restrictions were made for the age of participants included. A flow chart demonstrating the selection process is presented in Fig. 1 .
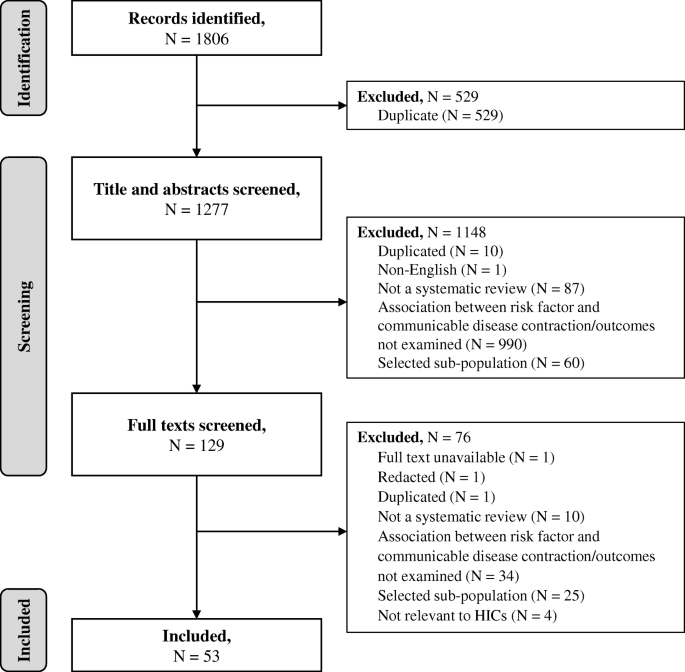
PRISMA flow diagram of study identification, inclusion and exclusion
Data extraction and synthesis
Data were extracted by three reviewers (SH, NJ, SW) into a standardised, pre-piloted form. Each extraction was duplicated across reviewers and discrepancies resolved through discussion. Information extracted from the studies included: title, authors, abstract, behavioural risk factor(s) studied, communicable disease(s) studied (including infection or related pathogens), research question, geographical restrictions, population characteristics, number of reviews included in the systematic review or meta-analysis, main findings (including odds ratios (OR), relative risks (RR) or rate ratios where available), proposed mechanisms of association and conclusions related to identified behavioural risk factor(s) and communicable disease(s). Where information on the number of reviews included for each risk factor was not reported, the corresponding author of the paper was contacted for additional information.
Due to the variety of different communicable diseases, risk factor definitions, outcome measures, and methods of reporting in the included studies, as well as the challenges of conducting meta-analysis for observational studies [ 71 ], findings were not combined statistically through meta-analysis. Instead, a narrative synthesis of the findings was constructed [ 72 ], and effect size ranges reported for each behavioural risk factor. Key information that would have enabled calculation of a common effect size was often not available. To calculate these effect size ranges, it was assumed that ORs, RRs and rate ratios were approximately equivalent, a method suggested for umbrella reviews in these circumstances [ 73 ]. In addition, where studies reported reduced risk of a communicable disease with a health behaviour (e.g. physical activity, no alcohol drinking), an inverse OR (1/OR) for the corresponding risk behaviour was reported. Findings were structured according to the identified behavioural risk factors and their association with a) contraction of the identified communicable diseases, and b) experiencing more severe outcomes from these communicable diseases. Some study conclusions were amended for readability, to aid understanding. Further, where study conclusions were not relevant to the current research question, information was extracted from results sections and amended for readability (see Supplementary file 2 ).
Methodological quality of studies
The methodological quality of included studies was assessed using the Overview Quality Assessment Questionnaire (OQAQ); a frequently used, validated tool for assessing the methodological quality of systematic reviews [ 74 ]. Methodological quality assessment was carried out by three researchers (NJ, SH, SW), with any discrepancies resolved through discussion. Assessment ratings are available in Supplementary file 2 .
The database search yielded 1806 citations, of which 53 were included (Fig. 1 ). Research relating to the following communicable diseases (including infection and pathogens) was identified: TB, HIV, hepatitis C virus (HCV), hepatitis B virus (HBV), invasive bacterial disease (IBD), pneumonia, influenza, and COVID-19. No studies relating to SARS, MERS or meningitis were identified. Thirty-six of the identified systematic reviews also conducted meta-analyses. Reviews used a range of definitions of behavioural risk factors (e.g. current or former smoker, any alcohol use or heavy alcohol use). All definitions were included in the synthesis, and are presented for clarity in each section of the results and in the results tables (Tables 1 and 2 ). Eighteen reviews examined the association between behavioural risk factors and the contraction of a communicable disease only, 32 reviews examined the association between behavioural risk factors and the outcomes from communicable diseases only, and three reviews examined associations with both contraction of and outcomes from communicable diseases. Characteristics of all included reviews and their conclusions can be found in Supplementary file 2 . No systematic review had extensive or major flaws, with most reviews having only minimal or minor flaws (Supplementary file 2 ). Consequently, no reviews were excluded based on methodological quality. A breakdown of reviews by disease and risk factors is provided in Supplementary file 3 .
Behavioural risk factors for communicable diseases
Overall, 50 out of 53 reviews (94%) concluded that at least one of the behavioural risk factors studied increased the risk of contracting or having more severe outcomes of a communicable disease. Across all reviews, effect sizes, where calculated, ranged from 0.83 to 8.22 (Figs. 2 and 3 ; Tables 1 and 2 ). Nineteen out of 21 reviews (90%) concluded that at least one of the behavioural risk factors studied increased the risk of contracting a communicable disease (Table 1 ). Across all contraction reviews, effect sizes, where calculated, ranged from 1.03 to 8.22 (Fig. 2 ). Thirty-two out of 35 reviews (91%) concluded that at least one of the behavioural risk factors studied increased the likelihood of having more severe outcomes from a communicable disease (Table 2 ). Across all outcome reviews, effect sizes, where calculated, ranged from 0.83 to 3.96 (Fig. 3 ).
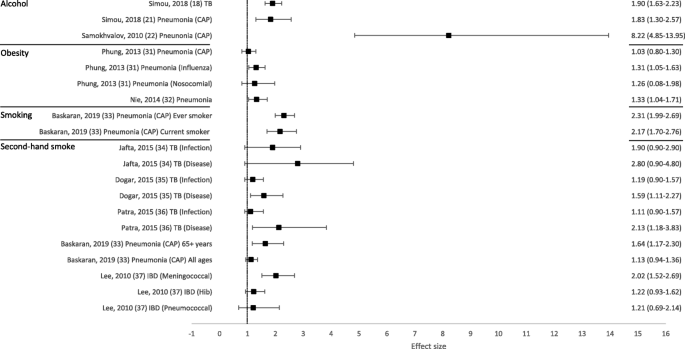
Forest plot of meta-analysis effect sizes: contraction of a communicable disease. Effect sizes refer to odds ratios and relative risks, see Table 1 for more information. CAP = community acquired pneumonia; TB = tuberculosis; IBD = invasive bacterial disease
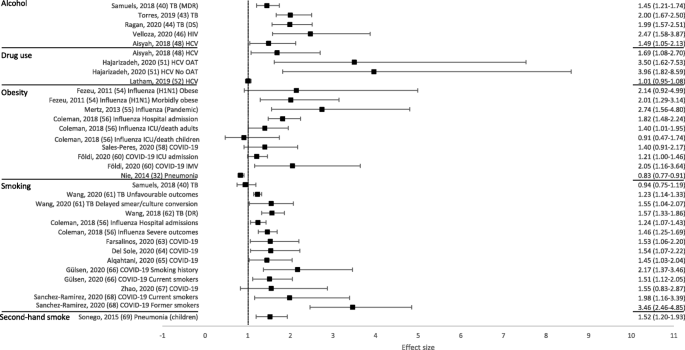
Forest plot of meta-analysis effect sizes: more severe communicable disease outcomes. Effect sizes refer to odds ratios, relative risks and rate ratios, see Table 2 for more information. MDR = multi-drug resistant; TB = tuberculosis; DS = drug-susceptible; HIV = human immunodeficiency virus; HCV = hepatitis C virus; ICU = intensive care unit; IMV = invasive mechanical ventilation; OAT = opioid agonist therapy
Alcohol as a risk factor
Seventeen reviews included alcohol as a risk factor for a communicable disease, with a range of definitions used: any alcohol consumption [ 19 , 23 , 41 , 43 , 44 , 46 , 47 , 49 ]; any alcohol use or higher amounts [ 18 , 21 ]; binge drinking or alcohol misuse [ 20 ]; alcohol misuse [ 40 ]; alcohol use disorder (AUD) [ 22 , 45 ]; alcoholism [ 39 , 42 ]; or current/history of excess use [ 48 ]. The majority of reviews reported an increased risk of contraction (5/6 reviews; Table 1 ) and more severe outcomes (10/12 reviews; Table 2 ). Across all alcohol reviews, effect sizes, where calculated, ranged from 1.83–8.22 for contraction (Fig. 2 ) and 1.45–2.47 for severe outcomes (Fig. 3 ). Alcohol use (any use, higher amounts, binge drinking or AUD) was reported to increase the risk of contracting TB [ 18 ], HIV [ 20 ], pneumonia [ 21 , 22 ] and invasive pneumococcal diseases (IPD) [ 23 ]. One review did not draw a conclusion, but reported mixed findings for the association between alcohol consumption and contraction of TB [ 19 ]. Alcohol use (any use, misuse, current/history of excess use, alcoholism or AUD) was reported to increase the risk of having more severe outcomes from TB [ 39 , 40 , 41 , 42 , 43 , 44 ], HIV [ 45 , 46 ], and HCV [ 48 , 49 ]. One review reported mixed findings and made no clear conclusion about the association of alcohol consumption and TB outcomes [ 19 ], and one review reported variable results among studies examining the association between alcohol consumption and the progression of HIV [ 47 ].
Illicit drug use as a risk factor
Fifteen reviews examined the association between illicit drug use and communicable disease contraction or outcomes, with a wide range of definitions used: drug abuse [ 19 , 41 ]; illicit drug use [ 25 ]; regular/problem cocaine use [ 50 ]; recent drug use [ 51 ]; and injection drug use [ 20 , 24 , 26 , 27 , 28 , 29 , 39 , 48 , 52 , 53 ]. The majority of reviews reported an increased risk of contraction (8/9 reviews; Table 1 ) and more severe outcomes (6/8 reviews; Table 2 ). Across all drug use reviews, effect sizes, where calculated (for more severe outcomes only), ranged from 1.01–3.96 (Fig. 3 ). Both injection drug use and illicit drug use were reported to increase the risk of contracting TB [ 24 ] and HIV [ 20 , 24 , 25 ], whilst the prevalence of HCV was found to be higher among people who inject drugs (PWID) compared to general population or community groups [ 26 , 27 , 28 , 29 ]. One review did not draw a conclusion but reported mixed findings for the association between drug abuse and TB contraction [ 19 ]. Both injecting drug use and drug use/abuse were reported to increase the risk of having more severe outcomes from TB [ 39 , 41 ], HIV [ 50 ] and HCV [ 48 , 51 ]. Furthermore, among those with HBV, the prevalence of hepatitis D (HDV; co-infection with HDV is considered a more severe form of viral hepatitis) was substantially higher for PWID compared to a mixed population with no risk factors [ 53 ]. One review did not draw a conclusion but reported mixed findings for the association between drug abuse and TB outcomes [ 19 ], and one review concluded that treatment outcomes for HCV were similar between people who currently were and were not injecting drugs [ 52 ].
Physical inactivity as a risk factor
One systematic review was identified examining the association between physical activity and communicable disease contraction or outcomes. This study reported an association between increased prolonged, moderate aerobic exercise and reduced influenza-related mortality, and improved immunocompetence [ 38 ].
Obesity as a risk factor
Ten reviews focused on the relationship between obesity [ 30 , 31 , 54 , 55 , 56 , 57 , 58 , 60 ], or overweight and obesity [ 32 , 59 ], and communicable disease risk. The majority of reviews reported an increased risk of contraction (3/3 reviews; Table 1 ) and more severe outcomes (8/9 reviews; Table 2 ). Across all obesity reviews, effect sizes, where calculated, ranged from 1.03–1.33 for contraction (Fig. 2 ) and 0.83–2.74 for severe outcomes (Fig. 3 ). Obesity was reported to increase the risk of contracting influenza [ 30 ] and pneumonia [ 31 , 32 ]. Obesity was reported to increase the risk of having more severe outcomes from influenza [ 30 , 54 , 55 , 56 ] and COVID-19 [ 58 , 59 , 60 ]. One review concluded that most studies showed some degree of association between higher body mass index (BMI) and a worse clinical presentation of COVID-19 and the need for hospitalisation. This review suggested that obesity seemed to predict poor clinical evolution in patients with COVID-19, but that studies in the review had limited methodological quality [ 57 ]. However, one review, which concluded that obesity increased the risk of contracting pneumonia, also found that obese individuals had a lower mortality risk from pneumonia [ 32 ].
Smoking as a risk factor
Eighteen reviews examined the association between smoking (current, past or both) and communicable disease contraction or outcomes. The majority of reviews reported some evidence of an increased risk of contraction (3/3 reviews; Table 1 ) and more severe outcomes (13/15 reviews; Table 2 ). Across all smoking reviews, effect sizes, where calculated, ranged from 2.17–2.31 for contraction (Fig. 2 ) and 0.94–3.46 for severe outcomes (Fig. 3 ). Smoking was reported to increase the risk of contracting HIV [ 20 ], pneumonia [ 33 ] and invasive pneumococcal disease (IPD) [ 23 ]. Further, smoking was reported to increase the risk of having more severe outcomes from TB [ 19 , 41 , 42 , 43 , 61 , 62 ], influenza [ 56 ], and COVID-19 [ 63 , 64 , 65 , 66 , 68 ]. Two reviews reported no associations between smoking and more severe outcomes from communicable diseases, including death from TB [ 39 ] and TB treatment outcomes [ 40 ]. One review reported that active smoking may increase the risk of severe COVID-19, but found the result was heavily influenced by one study [ 67 ].
Second-hand smoke as a risk factor
Six reviews focused on second-hand smoke as a risk factor for a communicable disease. The majority of reviews reported some evidence of an increased risk of contraction (4/5 reviews; Table 1 ) and more severe outcomes (1/1 review; Table 2 ). Across all second-hand smoking reviews, effect sizes, where calculated, ranged from 1.11 to 2.80 for contraction (Fig. 2 ), and the one effect size calculated for severe outcomes was 1.52 (Fig. 3 ). One review suggested that second-hand smoke exposure increased the risk of TB infection and disease [ 34 ]. The remaining four reviews reported at least some evidence of second-hand smoke exposure increasing the risk of contracting a communicable disease, including TB [ 35 , 36 ], pneumonia (among those aged 65+ only) [ 33 ], and IBD (invasive meningococcal disease [IMD] only) [ 37 ]. Second-hand smoke exposure was reported to increase the risk of severe outcomes from acute lower respiratory infections (ALRIs), including pneumonia [ 69 ].
Poor diet as a risk factor
Only two reviews were identified that examined the association between poor diet and communicable disease outcomes, and no reviews examining the association between poor diet and communicable disease contraction were identified. One review found that vitamin D status may influence the course of HIV disease [ 70 ]. The second review reported that a high intake of polyunsaturated fatty acids was associated with non-response to HCV antiviral therapy [ 49 ].
The key finding of this systematic review is that behavioural risk factors play a significant role in the risk of contracting, and having more severe outcomes from, common communicable diseases. To the authors’ knowledge, this is the first time that a review has brought together studies exploring the impact of behavioural risk factors on a range of communicable diseases. Whilst the focus on selected communicable diseases and use of systematic reviews has led to inevitable gaps, the findings nevertheless provide strong evidence that both NCDs and communicable diseases share a common set of behavioural risk factors. This work indicates that the prevention of communicable disease is likely to be most successful if it involves the prevention of behavioural risk factors. These findings are timely, in light of the COVID-19 pandemic, and highlight potential additional benefits of addressing behavioural risk factors ahead of any future epidemics or pandemics. While the specific diseases that may be involved can only be speculated, they are likely to share at least some characteristics with diseases in this review.
Although this review has not examined the mechanisms connecting behavioural risk factors and communicable diseases, there are likely to be multiple mechanisms. Behavioural factors, such as alcohol use, smoking, obesity, and illicit drug use, are well documented to impair the immune system. For instance, smoking is known to influence both innate and adaptive immunity [ 75 ]. Impairments to the immune system can make individuals more susceptible to communicable diseases and less able to control or recover from infection, leading to worse outcomes [ 76 , 77 , 78 , 79 , 80 , 81 ]. Use of alcohol/drugs may also reduce the efficacy of treatment for communicable diseases [ 82 ]. The presence of comorbidities, such as diabetes and cardiovascular disease, in individuals with behavioural risk factors has also been implicated in the increased risk of communicable diseases [ 83 , 84 ]. However, behavioural risk factors, such as obesity, are also reported to independently influence communicable diseases, after adjusting for comorbidities [ 55 ]. Behavioural mechanisms may also be important, particularly for alcohol and drug use, which may reduce risk perception [ 78 ], interfere with the uptake of services, or lead to poorer treatment adherence [ 85 ]. Additionally, behavioural risk factors may be likely to appear in combination, for example combined alcohol use and smoking [ 86 ], to further increase influences on communicable diseases. Furthermore, behaviours associated with drug use, such as injecting drugs, have a high efficiency of transmission of communicable diseases and reinfection with communicable diseases [ 87 ]. Having a communicable disease could also lead to the presence of behavioural risk factors (e.g. alcohol may be used as a way of coping with the emotional distress of diseases such as HIV and HCV [ 88 ]). Finally, there may be social mechanisms, such as the social marginalisation of heavy drinkers that affects health service use or treatment [ 89 ], or social issues such as homelessness, incarceration and poverty, which may increase the risk of both behavioural risk factors and communicable diseases [ 90 , 91 ]. It is likely that there are multiple ways in which these different physiological, behavioural and social factors come together to affect the likelihood of transmission and severity of communicable disease, which require further investigation.
With behavioural risk factors influencing the contraction and severity of communicable diseases, their prevention is likely to play a role in addressing future communicable disease burden, potentially through improvements in the immune system, bodily functioning and risk behaviours. As the recent COVID-19 pandemic has highlighted, their prevention is also likely to impact on communicable disease burden through the potential reduction of NCDs commonly associated with behavioural risk factors, which can also alter immune system function [ 92 ] and increase the risk of communicable disease complications and death [ 10 , 93 ]. The review findings are important in understanding communicable disease risk, and timely, in light of COVID-19. They suggest that improvements in the prevention of behavioural risk factors may serve to reduce the negative impacts of future epidemics or pandemics, building resilience and helping to address the pressing need for greater investment in pandemic preparedness [ 94 ]. Indeed, COVID-19 should not only be a reminder that good communicable disease control is necessary, but that the more successful we are in addressing behavioural risk factors, the better we will also be at reducing the burden of communicable disease, including future epidemics or pandemics. The finding that both communicable diseases and NCDs share a common set of behavioural risk factors also lends support for a more holistic understanding of these two disease categories. For instance, research suggests that NCDs and communicable diseases can interact; whilst NCDs can increase the risk and severity of communicable diseases (e.g. individuals with diabetes, hypertension and respiratory illnesses are more likely to affected by COVID-19 [ 95 ]), at least some diseases previously considered NCDs are now known to have an infectious origin (e.g. HBV is a cause of heptatocellular carcinoma [ 96 ]).
Although the focus of this review is on HICs, findings will be of importance to LMICs, which often experience a much higher burden of communicable disease [ 9 ] and where, for many countries, the prevalence of behavioural risk factors is increasing [ 7 , 8 ]. Due to ageing populations, the negative impacts of globalisation, and ill-equipped health systems, these countries are also facing a rapidly growing burden of NCDs [ 97 , 98 , 99 ], which may reduce resistance to infection, increase communicable disease complications, or interfere with its treatment [ 100 , 101 ]. In the current global society, any negative effects of rising behavioural risk factors and related NCDs on communicable disease transmission have the potential to affect not only LMICs, but health and well-being globally.
Across both HICs and LMICs, behavioural risk factors and related NCDs are known to cluster in disadvantaged populations [ 97 , 102 , 103 , 104 ], with poverty contributing to behavioural risk factors and NCDs, and vice versa [ 97 ]. Disadvantaged communities are more likely, therefore, to experience dual burdens of NCDs and communicable disease, contributing to social and economic health inequalities. In the UK for instance, people living in the most disadvantaged communities have been over twice as likely to die from COVID-19 as those in the least disadvantaged areas [ 105 ]. Preventing behavioural risk factors, particularly among disadvantaged populations, is likely to play an important role in reducing future global and national health inequalities, as well as the unequal burden of future pandemics.
There are some limitations to this work. The wide-ranging nature of the research allowed for a broad view of the links between behavioural risk factors and communicable diseases. However, this did not allow for the exploration of causal pathways of specific associations. Further research exploring these pathways would aid understanding and inform prevention. The use of systematic reviews to achieve a broader range of information also meant that newer empirical research may have been missed, only more widely researched topics for which there is enough information to conduct a systematic review would have been included, and more in-depth information such as potential interactions between risk factors could not be included. With no single definitions of behavioural risk factors agreed across the literature, it was not possible to standardise the definitions of risk factors in this review, meaning that there was often variation in the definitions included in each risk factor category, hampering discussion of relationships. Low socio-economic status (SES) and other factors associated with low SES, such as poor housing, are likely to be an important element in the link between behavioural risk factors and communicable diseases, although little is currently known about the influence of low SES and associated factors. It was not possible to explore the role of low SES within the review since many of the reviews included did not explore low SES in their analyses. Many of the included studies are global syntheses, however, the relationships between behavioural risk factors and communicable diseases may vary between countries. Only papers written in English were included, meaning that research in other languages may have been missed. Finally, conclusions should be considered with publication bias in mind; papers are more likely to be published if they reveal significant effects rather than null findings [ 106 ], so those reporting that behavioural risk factors are associated with communicable diseases are more likely to be identified.
This work identified several gaps in the current systematic review literature relating to specific behavioural risk factors and common communicable diseases, including studies examining the association of physical inactivity and poor dietary habits with communicable diseases, which warrant urgent further exploration. For instance, recently published literature has indeed highlighted the important role of physical inactivity in severe COVID-19 risk [ 2 , 107 ]. Due to the study being limited to systematic reviews only, a comprehensive comparison of behavioural risk factors across different disease types could not be provided; although future reviews could provide such comparisons. However, it was noted that reviews examining the association of communicable diseases with alcohol and illicit drug use largely focused on TB, HIV and hepatitis, whereas reviews examining association with obesity largely focused on pneumonia, influenza and COVID-19 (see Supplementary file 3 ). Further research understanding the more intricate ways in which individual behavioural risk factors are linked to specific types of disease, and the mechanisms by which they are linked, would provide a valuable framework for understanding how current and future communicable diseases may affect different population groups. Finally, findings highlight an opportunity for future research to examine the efficacy of behavioural risk factor prevention efforts in reducing communicable disease burden.
Behavioural risk factors play a significant role in the risk of contracting, and having more severe outcomes from, common communicable diseases. These risk factors are largely modifiable or preventable. Prevention of communicable diseases is likely to be most successful if it involves the prevention of behavioural risk factors that are commonly associated with NCDs, particularly among disadvantaged populations. These findings are important for understanding risks associated with communicable disease, and timely, given the current COVID-19 pandemic and need for improvements in future pandemic preparedness. Addressing behavioural risk factors should be an important part of work to build resilience against any emerging and future epidemics and pandemics. Furthermore, the pandemic can offer a timely, teachable moment for the public on how improvements to general health, through addressing risk behaviours commonly associated with NCDs, may help protect them from infections like COVID-19 in the future.
Availability of data and materials
Not applicable.
Abbreviations
Acquired immune deficiency syndrome
Acute lower respiratory infections
Body mass index
Hepatitis C virus
Hepatitis D virus
High-income country
Human immunodeficiency virus
Invasive bacterial disease
Invasive pneumococcal disease
Invasive meningococcal disease
Lower-middle income country
Middle east respiratory syndrome
Non-communicable disease
Overview quality assessment questionnaire
Preferred reporting items for systematic reviews and meta-analyses
International prospective register of systematic reviews
Relative risk
Severe acute respiratory syndrome
Tuberculosis
World Health Organization and the United Nations Development Programme. Responding to non-communicable diseases during and beyond the COVID-19 pandemic. 2020. Available from: https://www.who.int/publications/i/item/WHO-2019-nCoV-Non-communicable_diseases-Evidence-2020.1 . Accessed 24 June 2021.
Google Scholar
Hamer M, Kivimäki M, Gale CR, Batty GD. Lifestyle risk factors, inflammatory mechanisms, and COVID-19 hospitalization: a community-based cohort study of 387,109 adults in UK. Brain Behav Immun. 2020;87:184–7. https://doi.org/10.1016/j.bbi.2020.05.059 .
Article CAS PubMed PubMed Central Google Scholar
Bambra C, Riordan R, Ford J, Matthews F. The COVID-19 pandemic and health inequalities. J Epidemiol Community Health. 2020;74(11):964–8. https://doi.org/10.1136/jech-2020-214401 .
Article PubMed Google Scholar
Horton R. Offline: COVID-19 is not a pandemic. Lancet. 2020;396(10255):874. https://doi.org/10.1016/S0140-6736(20)32000-6 .
World Health Organization. The Global Health Observatory. Available from: https://www.who.int/data/gho . Accessed 8 Nov 2021.
Global Burden of Disease Collaborative Network. GBD Results Tool. 2021. Available from: http://ghdx.healthdata.org/gbd-results-tool . Accessed 24 June 2021.
Uddin R, Lee E-Y, Khan SR, Tremblay MS, Khan A. Clustering of lifestyle risk factors for non-communicable diseases in 304,779 adolescents from 89 countries: a global perspective. Prev Med An Int J Devoted to Pract Theory. 2020;131:8. https://doi.org/10.1016/j.ypmed.2019.105955 .
Article Google Scholar
Ford ND, Patel SA, Narayan KMV. Obesity in low- and middle-income countries: burden, drivers, and emerging challenges. Annu Rev Public Health. 2017;38(1):145–64. https://doi.org/10.1146/annurev-publhealth-031816-044604 .
World Health Organization. Global status report on alcohol and health 2018. Geneva: World Health Organization; 2018.
Azarpazhooh MR, Morovatdar N, Avan A, Phan TG, Divani AA, Yassi N, et al. COVID-19 pandemic and burden of non-communicable diseases: an ecological study on data of 185 countries. J Stroke Cerebrovasc Dis. 2020;29(9):1–9. https://doi.org/10.1016/j.jstrokecerebrovasdis.2020.105089 .
Bloom DE, Cadarette D. Infectious disease threats in the twenty-first century: strengthening the global response. Front Immunol. 2019;10:549. https://doi.org/10.3389/fimmu.2019.00549 .
Article PubMed PubMed Central Google Scholar
Hamer M, O’Donovan G, Stamatakis E. Lifestyle risk factors, obesity and infectious disease mortality in the general population: linkage study of 97,844 adults from England and Scotland. Prev Med (Baltim). 2019;123:65–70. https://doi.org/10.1016/j.ypmed.2019.03.002 .
Baik I, Curhan GC, Rimm EB, Bendich A, Willett WC, Fawzi WW. A prospective study of age and lifestyle factors in relation to community-acquired pneumonia in US men and women. Arch Intern Med. 2000;160(20):3082–8. https://doi.org/10.1001/archinte.160.20.3082 .
Article CAS PubMed Google Scholar
Creswell J, Raviglione M, Ottmani S, Migliori GB, Uplekar M, Blanc L, et al. Tuberculosis and noncommunicable diseases: neglected links and missed opportunities. Eur Respir J. 2011;37(5):1269–82. https://doi.org/10.1183/09031936.00084310 .
Majid E, Elio R. Behavioral and dietary risk factors for noncommunicable diseases. N Engl J Med. 2013;369(10):954–64. https://doi.org/10.1056/NEJMra1203528 .
Article CAS Google Scholar
World Health Organization. Disease outbreaks. 2021. Available from: https://www.who.int/emergencies/diseases/en/ . Accessed 24 June 2021.
Wimalasena NN, Chang-Richards A, Wang KI-K, Dirks KN. Housing risk factors associated with respiratory disease: a systematic review. Int J Environ Res Public Health. 2021;18(6):2815. https://doi.org/10.3390/ijerph18062815 .
Simou E, Britton J, Leonardi-Bee J. Alcohol consumption and risk of tuberculosis: a systematic review and meta-analysis. Int J Tuberc Lung Dis. 2018;22(11):1277–85. https://doi.org/10.5588/ijtld.18.0092 .
Mohidem NA, Hashim Z, Osman M, Shaharudin R, Farrah MM, Makeswaran P. Demographic, socio-economic and behavior as risk factors of tuberculosis in Malaysia: a systematic review of the literature. Rev Environ Health. 2018;33(4):407–21. https://doi.org/10.1515/reveh-2018-0026 .
Rumbwere Dube BN, Marshall TP, Ryan RP, Omonijo M. Predictors of human immunodeficiency virus (HIV) infection in primary care among adults living in developed countries: a systematic review. Syst Rev. 2018;7(1):82. https://doi.org/10.1186/s13643-018-0744-3 .
Simou E, Britton J, Leonardi-Bee J. Alcohol and the risk of pneumonia: a systematic review and meta-analysis. BMJ Open. 2018;8(8):e022344. https://doi.org/10.1136/bmjopen-2018-022344 .
Samokhvalov AV, Irving HM, Rehm J. Alcohol consumption as a risk factor for pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2010;138(12):1789–95. https://doi.org/10.1017/S0950268810000774 .
Cruickshank HC, Jefferies JM, Clarke SC. Lifestyle risk factors for invasive pneumococcal disease: a systematic review. BMJ Open. 2014;4(6):e005224. https://doi.org/10.1136/bmjopen-2014-005224 .
Pimpin L, Drumright LN, Kruijshaar ME, Abubakar I, Rice B, Delpech V, et al. Tuberculosis and HIV co-infection in European Union and European economic area countries. Eur Respir J. 2011;38(6):1382–92. https://doi.org/10.1183/09031936.00198410 .
Zhao Y, Luo T, Tucker JD, Wong WCW. Risk factors of HIV and other sexually transmitted infections in China: a systematic review of reviews. PLoS One. 2015;10(10):e0140426. https://doi.org/10.1371/journal.pone.0140426 .
Goel A, Seguy N, Aggarwal R. Burden of hepatitis C virus infection in India: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2019;34(2):321–9. https://doi.org/10.1111/jgh.14466 .
Mahmud S, Akbarzadeh V, Abu-Raddad LJ. The epidemiology of hepatitis C virus in Iran: systematic review and meta-analyses. Sci Rep. 2018;8(1):1. https://doi.org/10.1038/s41598-017-18296-9 .
Al Kanaani Z, Mahmud S, Kouyoumjian SP, Abu-Raddad LJ. The epidemiology of hepatitis C virus in Pakistan: systematic review and meta-analyses. R Soc Open Sci. 2018;5(4):180257. https://doi.org/10.1098/rsos.180257 .
Fadlalla FA, Mohamoud YA, Mumtaz GR, Abu-Raddad LJ. The epidemiology of hepatitis C virus in the Maghreb region: systematic review and meta-analyses. PLoS One. 2015;10(3):e0121873. https://doi.org/10.1371/journal.pone.0121873 .
Falagas M, Koletsi PK, Baskouta E, Rafailidis PI, Dimopoulos G, Karageorgopoulos DE. Pandemic a(H1N1) 2009 influenza: review of the southern hemisphere experience. Epidemiol Infect. 2011;139(1):27–40. https://doi.org/10.1017/S0950268810002037 .
Phung DT, Wang Z, Rutherford S, Huang C, Chu C. Body mass index and risk of pneumonia: a systematic review and meta-analysis. Obes Rev. 2013;14(10):839–57. https://doi.org/10.1111/obr.12055 .
Nie W, Zhang Y, Jee SH, Jung KJ, Li B, Xiu Q. Obesity survival paradox in pneumonia: a meta-analysis. BMC Med. 2014;12(1):61. https://doi.org/10.1186/1741-7015-12-61 .
Baskaran V, Murray RL, Hunter A, Lim WS, McKeever TM. Effect of tobacco smoking on the risk of developing community acquired pneumonia: a systematic review and meta-analysis. PLoS One. 2019;14(7):e0220204. https://doi.org/10.1371/journal.pone.0220204 .
Jafta N, Jeena PM, Barregard L, Naidoo RN. Childhood tuberculosis and exposure to indoor air pollution: a systematic review and meta-analysis. Int J Tuberc Lung Dis. 2015;19(5):596–602. https://doi.org/10.5588/ijtld.14.0686 .
Dogar OF, Pillai N, Safdar N, Shah SK, Zahid R, Siddiqi K. Second-hand smoke and the risk of tuberculosis: a systematic review and a meta-analysis. Epidemiol Infect. 2015;143(15):3158–72. https://doi.org/10.1017/S0950268815001235 .
Patra J, Bhatia M, Suraweera W, Morris SK, Patra C, Gupta PC, et al. Exposure to second-hand smoke and the risk of tuberculosis in children and adults: a systematic review and meta-analysis of 18 observational studies. PLoS Med. 2015;12(6):e1001835. https://doi.org/10.1371/journal.pmed.1001835 .
Lee C-C, Middaugh NA, Howie SRC, Ezzati M. Association of secondhand smoke exposure with pediatric invasive bacterial disease and bacterial carriage: a systematic review and meta-analysis. PLoS Med. 2010;7(12):e1000374. https://doi.org/10.1371/journal.pmed.1000374 .
Song Y, Ren F, Sun D, Wang M, Baker JS, István B, et al. Benefits of exercise on influenza or pneumonia in older adults: a systematic review. Int J Environ Res Public Health. 2020;17(8):2655. https://doi.org/10.3390/ijerph17082655 .
Article CAS PubMed Central Google Scholar
Waitt CJ, Squire SB. A systematic review of risk factors for death in adults during and after tuberculosis treatment. Int J Tuberc Lung Dis. 2011;15(7):871–85. https://doi.org/10.5588/ijtld.10.0352 .
Samuels JP, Sood A, Campbell JR, Khan FA, Johnston JC. Comorbidities and treatment outcomes in multidrug resistant tuberculosis: a systematic review and meta-analysis. Sci Rep. 2018;8(1):1–13. https://doi.org/10.1038/s41598-018-23344-z .
Rajendran M, Zaki RA, Aghamohammadi N. Contributing risk factors towards the prevalence of multidrug-resistant tuberculosis in Malaysia: a systematic review. Tuberculosis. 2020;122:1. https://doi.org/10.1016/j.tube.2020.101925 .
Azhar G. DOTS for TB relapse in India: a systematic review. Lung India. 2012;29(2):147–53. https://doi.org/10.4103/0970-2113.95320 .
Torres NMC, Rodríguez JJQ, Andrade PSP, Arriaga MB, Netto EM. Factors predictive of the success of tuberculosis treatment: a systematic review with meta-analysis. PLoS One. 2019;14(12):e0226507. https://doi.org/10.1371/journal.pone.0226507 .
Ragan EJ, Kleinman MB, Sweigart B, Gnatienko N, Parry CD, Horsburgh CR, et al. The impact of alcohol use on tuberculosis treatment outcomes: a systematic review and meta-analysis. Int J Tuberc Lung Dis. 2020;24(1):73–82. https://doi.org/10.5588/ijtld.19.0080 .
Azar MM, Springer SA, Meyer JP, Altice FL. A systematic review of the impact of alcohol use disorders on HIV treatment outcomes, adherence to antiretroviral therapy and health care utilization. Drug Alcohol Depend. 2010;112(3):178–93. https://doi.org/10.1016/j.drugalcdep.2010.06.014 .
Velloza J, Kemp CG, Aunon FM, Ramaiya MK, Creegan E, Simoni JM. Alcohol use and antiretroviral therapy non-adherence among adults living with HIV/AIDS in sub-saharan Africa: a systematic review and meta-analysis. AIDS Behav. 2020;24(6):1727–42. https://doi.org/10.1007/s10461-019-02716-0 .
Ge S, Sanchez M, Nolan M, Liu T, Savage CL. Is alcohol use associated with increased risk of developing adverse health outcomes among adults living with human immunodeficiency virus: a systematic review. J Addict Nurs. 2018;29(2):96–118. https://doi.org/10.1097/JAN.0000000000000220 .
Aisyah DN, Shallcross L, Hully AJ, O’Brien A, Hayward A. Assessing hepatitis C spontaneous clearance and understanding associated factors—a systematic review and meta-analysis. J Viral Hepat. 2018;25(6):680–98. https://doi.org/10.1111/jvh.12866 .
Sublette VA, Douglas MW, McCaffery K, George J, Perry KN. Psychological, lifestyle and social predictors of hepatitis C treatment response: a systematic review. Liver Int. 2013;33(6):894–903. https://doi.org/10.1111/liv.12138 .
Peacock A, Tran LT, Larney S, Stockings E, Santo T, Jones H, et al. All-cause and cause-specific mortality among people with regular or problematic cocaine use: a systematic review and meta-analysis. Addiction. 2021;116(4):725–42. https://doi.org/10.1111/add.15239 .
Hajarizadeh B, Cunningham EB, Valerio H, Martinello M, Law M, Janjua NZ, et al. Hepatitis C reinfection after successful antiviral treatment among people who inject drugs: a meta-analysis. J Hepatol. 2020;72(4):643–57. https://doi.org/10.1016/j.jhep.2019.11.012 .
Latham NH, Doyle JS, Palmer AY, Vanhommerig JW, Agius P, Goutzamanis S, et al. Staying hepatitis C negative: a systematic review and meta-analysis of cure and reinfection in people who inject drugs. Liver Int. 2019;39(12):2244–60. https://doi.org/10.1111/liv.14152 .
Chen HY, Shen DT, Ji DZ, Han PC, Zhang WM, Ma JF, et al. Prevalence and burden of hepatitis D virus infection in the global population: a systematic review and meta-analysis. Gut. 2019;68(3):512–21. https://doi.org/10.1136/gutjnl-2018-316601 .
Fezeu L, Julia C, Henegar A, Bitu J, Hu FB, Grobbee DE, et al. Obesity is associated with higher risk of intensive care unit admission and death in influenza a (H1N1) patients: a systematic review and meta-analysis. Obes Rev. 2011;12(8):653–9. https://doi.org/10.1111/j.1467-789X.2011.00864.x .
Mertz D, Kim TH, Johnstone J, Lam P-P, Science M, Kuster SP, et al. Populations at risk for severe or complicated influenza illness: systematic review and meta-analysis. BMJ. 2013;347(aug23 1):f5061. https://doi.org/10.1136/bmj.f5061 .
Coleman BL, Fadel SA, Fitzpatrick T, Thomas S. Risk factors for serious outcomes associated with influenza illness in high- versus low- and middle-income countries: systematic literature review and meta-analysis. Influenza Other Respir Viruses. 2018;12(1):22–9. https://doi.org/10.1111/irv.12504 .
Peres KC, Riera R, Martimbianco ALC, Ward LS, Cunha LL. Body Mass Index and Prognosis of COVID-19 Infection. A Systematic Review. Front Endocrinol (Lausanne). 2020;11:562.
de Carvalho Sales-Peres SH, de Azevedo-Silva LJ, RCS B, de Carvalho Sales-Peres M, da Silvia Pinto AC, Santiago Junior JF. Coronavirus (SARS-CoV-2) and the risk of obesity for critically illness and ICU admitted: Meta-analysis of the epidemiological evidence. Obes Res Clin Pract. 2020;14(5):389–97. https://doi.org/10.1016/j.orcp.2020.07.007 .
de Siqueira JVV, Almeida LG, Zica BO, Brum IB, Barceló A, de Siqueira Galil AG. Impact of obesity on hospitalizations and mortality, due to COVID-19: a systematic review. Obes Res Clin Pract. 2020;14(5):398–403. https://doi.org/10.1016/j.orcp.2020.07.005 .
Földi M, Farkas N, Kiss S, Zádori N, Váncsa S, Szakó L, et al. Obesity is a risk factor for developing critical condition in COVID-19 patients: a systematic review and meta-analysis. Obes Rev. 2020;21(10):e13095. https://doi.org/10.1111/obr.13095 .
Wang EY, Arrazola RA, Mathema B, Ahluwalia IB, Mase SR. The impact of smoking on tuberculosis treatment outcomes: a meta-analysis. Int J Tuberc Lung Dis. 2020;24(2):170–5. https://doi.org/10.5588/ijtld.19.0002 .
Wang M-G, Huang W-W, Wang Y, Zhang Y-X, Zhang M-M, Wu S-Q, et al. Association between tobacco smoking and drug-resistant tuberculosis. Infect Drug Resist. 2018;11:873–87. https://doi.org/10.2147/IDR.S164596 .
Farsalinos K, Barbouni A, Poulas K, Polosa R, Caponnetto P, Niaura R, et al. Current smoking, former smoking, and adverse outcome among hospitalized COVID-19 patients: a systematic review and meta-analysis. Ther Adv Chronic Dis. 2020;11:1–14. https://doi.org/10.1177/2040622320935765 .
Del Sole F, Farcomeni A, Loffredo L, Carnevale R, Menichelli D, Vicario T, et al. Features of severe COVID-19: a systematic review and meta-analysis. Eur J Clin Investig. 2020;50(10):1–7. https://doi.org/10.1111/eci.13378 .
Alqahtani JS, Oyelade T, Aldhahir AM, Alghamdi SM, Almehmadi M, Alqahtani AS, et al. Prevalence, severity and mortality associated with COPD and smoking in patients with COVID-19: a rapid systematic review and meta-analysis. PLoS One. 2020;15(5):e0233147. https://doi.org/10.1371/journal.pone.0233147 .
Gülsen A, Yigitbas BA, Uslu B, Drömann D, Kilinc O. The effect of smoking on COVID-19 symptom severity: systematic review and meta-analysis. Pulm Med. 2020;2020:7590207–11. https://doi.org/10.1155/2020/7590207 .
Zhao Q, Meng M, Kumar R, Wu Y, Huang J, Lian N, et al. The impact of COPD and smoking history on the severity of COVID-19: a systemic review and meta-analysis. J Med Virol. 2020;92(10):1915–21. https://doi.org/10.1002/jmv.25889 .
Sanchez-Ramirez DC, Mackey D. Underlying respiratory diseases, specifically COPD, and smoking are associated with severe COVID-19 outcomes: a systematic review and meta-analysis. Respir Med. 2020;171:106096. https://doi.org/10.1016/j.rmed.2020.106096 .
Sonego M, Pellegrin MC, Becker G, Lazzerini M. Risk factors for mortality from acute lower respiratory infections (ALRI) in children under five years of age in low and middle-income countries: a systematic review and meta-analysis of observational studies. PLoS One. 2015;10(1):17. https://doi.org/10.1371/journal.pone.0116380 .
Giusti A, Penco G, Pioli G. Vitamin D deficiency in HIV-infected patients: a systematic review. Nutr Diet Suppl. 2011;3:101–11. https://doi.org/10.2147/NDS.S6921 .
Metelli S, Chaimani A. Challenges in meta-analyses with observational studies. Evid Based Ment Heal. 2020;23(2):83–7 Available from: http://ebmh.bmj.com/content/23/2/83.abstract .
Popay J, Roberts H, Sowden A, Petticrew M, Arai L, Rodgers M, et al. Guidance on the conduct of narrative synthesis in systematic Reviews. A Product from the ESRC Methods Programme. Version 1. 2006. Available from: https://www.lancaster.ac.uk/media/lancaster-university/content-assets/documents/fhm/dhr/chir/NSsynthesisguidanceVersion1-April2006.pdf . Accessed 24 June 2021.
Fusar-Poli P, Radua J. Ten simple rules for conducting umbrella reviews. Evid Based Ment Health. 2018;21(3):95–100. https://doi.org/10.1136/ebmental-2018-300014 .
Pussegoda K, Turner L, Garritty C, Mayhew A, Skidmore B, Stevens A, et al. Identifying approaches for assessing methodological and reporting quality of systematic reviews: a descriptive study. Syst Rev. 2017;6(1):117. https://doi.org/10.1186/s13643-017-0507-6 .
Qiu F, Liang C-L, Liu H, Zeng Y-Q, Hou S, Huang S, et al. Impacts of cigarette smoking on immune responsiveness: up and down or upside down? Oncotarget. 2017;8(1):268–84. https://doi.org/10.18632/oncotarget.13613 .
Díaz LE, Montero A, González-Gross M, Vallejo AI, Romeo J, Marcos A. Influence of alcohol consumption on immunological status: a review. Eur J Clin Nutr. 2002;56(S3):S50–3. https://doi.org/10.1038/sj.ejcn.1601486 .
Sattar N, McInnes IB, McMurray JJV. Obesity is a risk factor for severe COVID-19 infection: multiple potential mechanisms. Circulation. 2020;142(1):4–6. https://doi.org/10.1161/CIRCULATIONAHA.120.047659 .
Andreasson S, Chikritzhs T, Dangardt F, Holder H, Naimi T, Sherk A, et al. Alcohol and society 2021: alcohol and the coronavirus pandemic: individual, societal and policy perspectives. Stockholm: Medicine, SIGHT, Movendi International & IOGT-NTO; 2021.
Trivedi GY, Saboo B. The risk factors for immune system impairment and the need for lifestyle changes. J Soc Heal Diabetes. 2020;8(01):025–8. https://doi.org/10.1055/s-0040-1715778 .
Pacifici R, Zuccaro P, Pichini S, Roset PN, Poudevida S, Farré M, et al. Modulation of the immune system in cannabis users. JAMA. 2003;289(15):1929–31. https://doi.org/10.1001/jama.289.15.1929-a .
Friedman H, Eisenstein TK. Neurological basis of drug dependence and its effects on the immune system. J Neuroimmunol. 2004;147(1-2):106–8. https://doi.org/10.1016/j.jneuroim.2003.10.022 .
Pandrea I, Happel KI, Amedee AM, Bagby GJ, Nelson S. Alcohol’s role in HIV transmission and disease progression. Alcohol Res Heal. 2010;33:203–18.
Vas P, Hopkins D, Feher M, Rubino F, Whyte M. Diabetes, obesity and COVID −19: a complex interplay. Diabetes Obes Metab. 2020;22(10):1892–6. https://doi.org/10.1111/dom.14134 .
Poirier P, Alpert MA, Fleisher LA, Thompson PD, Sugerman HJ, Burke LE, et al. Cardiovascular evaluation and management of severely obese patients undergoing surgery. Circulation. 2009;120(1):86–95. https://doi.org/10.1161/CIRCULATIONAHA.109.192575 .
Gonzalez A, Barinas J, O’Cleirigh C. Substance use: impact on adherence and HIV medical treatment. Curr HIV/AIDS Rep. 2011;8(4):223–34. https://doi.org/10.1007/s11904-011-0093-5 .
Poortinga W. The prevalence and clustering of four major lifestyle risk factors in an English adult population. Prev Med (Baltim). 2007;44(2):124–8. https://doi.org/10.1016/j.ypmed.2006.10.006 .
Maher L, Jalaludin B, Chant KG, Jayasuriya R, Sladden T, Kaldor JM, et al. Incidence and risk factors for hepatitis C seroconversion in injecting drug users in Australia. Addiction. 2006;101(10):1499–508. https://doi.org/10.1111/j.1360-0443.2006.01543.x .
Moitra E, Anderson BJ, Herman DS, Hayaki J, Pinkston MM, Kim HN, et al. Examination of using alcohol to cope, depressive symptoms, and perceived social support in persons with HIV and Hepatitis C. AIDS Care. 2020 Oct;32(10):1238–45. https://doi.org/10.1080/09540121.2020.1734177 .
Room R. Stigma, social inequality and alcohol and drug use. Drug Alcohol Rev. 2005;24(2):143–55. https://doi.org/10.1080/09595230500102434 .
Fazel S, Bains P, Doll H. Substance abuse and dependence in prisoners: a systematic review. Addiction. 2006;101(2):181–91. https://doi.org/10.1111/j.1360-0443.2006.01316.x .
Story A, Murad S, Roberts W, Verheyen M, Hayward AC. Tuberculosis in London: the importance of homelessness. Problem drug use and prison. Thorax. 2007;62(8):667–71. https://doi.org/10.1136/thx.2006.065409 .
Berbudi A, Rahmadika N, Tjahjadi AI, Ruslami R. Type 2 diabetes and its impact on the immune system. Curr Diabetes Rev. 2020;16(5):442–9. https://doi.org/10.2174/1573399815666191024085838 .
Clark A, Jit M, Warren-Gash C, Guthrie B, Wang HHX, Mercer SW, et al. Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Glob Heal. 2020;8(8):e1003–17. https://doi.org/10.1016/S2214-109X(20)30264-3 .
Fan VY, Jamison DT, Summers LH. Pandemic risk: how large are the expected losses? Bull World Health Organ. 2018;96(2):129–34. https://doi.org/10.2471/BLT.17.199588 .
Nikoloski Z, Alqunaibet AM, Alfawaz RA, Almudarra SS, Herbst CH, El-Saharty S, et al. Covid-19 and non-communicable diseases: evidence from a systematic literature review. BMC Public Health. 2021;21(1):1068. Available from. https://doi.org/10.1186/s12889-021-11116-w .
Xie Y, Hepatitis B. Virus-associated hepatocellular carcinoma. Adv Exp Med Biol. 2017;1018:11–21. https://doi.org/10.1007/978-981-10-5765-6_2 .
World Health Organization. Global status report on noncommunicable diseases 2010. Geneva: World Health Organization; 2011.
Islam SMS, Purnat TD, Phuong NTA, Mwingira U, Schacht K, Fröschl G. Non-Communicable Diseases (NCDs) in developing countries: a symposium report. Global Health. 2014;10:81.
Kostova D, Chaloupka FJ, Frieden TR, Henning K, Paul J, Osewe PL, et al. Noncommunicable disease risk factors in developing countries: policy perspectives. Prev Med (Baltim). 2017;105:S1–3. https://doi.org/10.1016/j.ypmed.2017.09.027 .
Kostova D, Husain M, Sugerman D, Hong Y, Saraiya M, Keltz J, et al. Synergies between communicable and noncommunicable disease programs to enhance global health security. Emerg Infect Dis J. 2017;23(13):S40–467. https://doi.org/10.3201/eid2313.170581 .
Remais JV, Zeng G, Li G, Tian L, Engelgau MM. Convergence of non-communicable and infectious diseases in low- and middle-income countries. Int J Epidemiol. 2013;42(1):221–7. https://doi.org/10.1093/ije/dys135 .
Marmot M, Bell R. Social determinants and non-communicable diseases: time for integrated action. BMJ. 2019;364:10–2. https://doi.org/10.1136/bmj.l251 .
Hosseinpoor AR, Bergen N, Kunst A, Harper S, Guthold R, Rekve D, et al. Socioeconomic inequalities in risk factors for non communicable diseases in low-income and middle-income countries: results from the world health survey. BMC Public Health. 2012;12(1):912. https://doi.org/10.1186/1471-2458-12-912 .
Allen L, Williams J, Townsend N, Mikkelsen B, Roberts N, Foster C, et al. Socioeconomic status and non-communicable disease behavioural risk factors in low-income and lower-middle-income countries: a systematic review. Lancet Glob Heal. 2017;5(3):e277–89. https://doi.org/10.1016/S2214-109X(17)30058-X .
Caul S. Deaths involving COVID-19 by local area and socioeconomic deprivation : deaths occurring between 1 March and 31 July 2020. 2020. Available from: https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/deaths/bulletins/deathsinvolvingcovid19bylocalareasanddeprivation/deathsoccurringbetween1marchand31july2020 . Accessed 24 June 2021.
Knobloch K, Yoon U, Vogt PM. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. J Cranio-Maxillofacial Surg. 2011;39(2):91–2. https://doi.org/10.1016/j.jcms.2010.11.001 .
Sallis R, Young DR, Tartof SY, Sallis JF, Sall J, Li Q, et al. Physical inactivity is associated with a higher risk for severe COVID-19 outcomes: a study in 48 440 adult patients. Br J Sports Med. 2021;0:1–8.
Download references
Acknowledgements
We would like to thank Rebecca Hill and Nel Griffith, Public Health Wales, for their assistance in reviewing this manuscript.
This project was funded by Public Health Wales.
Author information
Authors and affiliations.
Policy and International Health, World Health Organization Collaborating Centre on Investment for Health and Well-being, Public Health Wales, Wrexham, UK
Sara Wood, Natasha Judd, Mark A. Bellis & Karen Hughes
Public Health Collaborating Unit, School of Medical and Health Sciences, Bangor University, Wrexham, UK
Sophie E. Harrison, Natasha Judd, Mark A. Bellis & Karen Hughes
Institute for Applied Human Physiology, School of Human and Behavioural Sciences, Bangor University, Bangor, UK
Sophie E. Harrison
Health Protection and Screening Services, Public Health Wales, Cardiff, UK
Andrew Jones
You can also search for this author in PubMed Google Scholar
Contributions
MB and KH contributed to the conception, design and direction of the project. SW managed the project with SH and NJ. NJ and SH undertook the literature searches and conducted the initial data screening. NJ, SH and SW conducted the full text screening, data extraction and quality assessment. SH synthesised the findings. AJ and all other authors contributed to the manuscript write up / revisions. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Mark A. Bellis .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1., additional file 2., additional file 3., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Wood, S., Harrison, S.E., Judd, N. et al. The impact of behavioural risk factors on communicable diseases: a systematic review of reviews. BMC Public Health 21 , 2110 (2021). https://doi.org/10.1186/s12889-021-12148-y
Download citation
Received : 07 October 2021
Accepted : 22 October 2021
Published : 17 November 2021
DOI : https://doi.org/10.1186/s12889-021-12148-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Infectious disease
- Risk factors
BMC Public Health
ISSN: 1471-2458
- General enquiries: [email protected]
- Campus News
- Student News
- UK HealthCare
- UK Happenings
- Arts & Culture
- Professional News
New consortium to advance research on infectious disease prevention, treatment

LEXINGTON, Ky. (May 29, 2024) — A new partnership between researchers at the University of Kentucky will combat the threat of infectious diseases through research directed at prevention and treatment.
The Consortium for Understanding and Reducing Infectious Diseases in Kentucky (CURE-KY) will foster multidisciplinary collaborations to address the burden of infectious diseases in the Commonwealth and beyond.
“I am thrilled by the boundless possibilities our collaborative efforts promise at the University of Kentucky. With a steadfast commitment to advancing knowledge and fostering transdisciplinary alliances, we aim to propel infectious disease research to new heights, creating a healthier Kentucky and nurturing future scientific leaders,” said Ilhem Messaoudi, Ph.D., CURE-KY founder and chair of the Department of Microbiology, Immunology, and Molecular Genetics in the College of Medicine.
CURE-KY grew out of the Emerging Themes for Research Program, which is part of the Research Leadership Academy (RLA) supported by the Office of the Vice President for Research and was built on the heels of UK’s COVID-19 Unified Research Experts (CURE) Alliance that was quickly assembled to support a full range of COVID-related research.
“A key facet of the Research Leadership Academy is to empower our researchers to tackle research questions that are critical to Kentucky and do so with a collaborative and innovative approach,” said Lisa Cassis, UK’s vice president for research. “CURE-KY is an exemplary model of this strategy. I am excited to see the range of this team’s scientific expertise and their dedication to advancing the health of Kentucky through vital research.”
CURE-KY brings together a diverse group of experts from the College of Medicine, College of Public Health, College of Pharmacy, Martin-Gatton College of Agriculture, Food and Environment and Markey Cancer Center.
CURE-KY will focus on research in five areas: microbial pathogenesis; vaccines, therapeutics and antimicrobial resistance; public health and community engagement; animal health; and engineering and systems.
The consortium will also work to address vaccine hesitancy and misinformation to foster trust between Kentuckians and the scientific community.
“CURE-KY leverages the expertise of some of the best and brightest physicians and scientists specializing in infectious disease treatment and prevention. With the power of this unified team, the University of Kentucky will equip the Commonwealth with the information, resources and health care solutions necessary to combat dangerous diseases,” said Charles “Chipper” Griffith III, M.D., dean of the College of Medicine .
“The College of Public Health is dedicated to bettering public health, which is tightly integrated with the mission of CURE-KY. From studying prevention, control and understanding transmission dynamics to assessing risk, promoting health equity and engaging communities in disease control efforts, this work is critical toward our goal of creating a healthier Kentucky,” said Heather Bush, Ph.D., dean of the College of Public Health .
"We are proud to be a part of the CURE-KY consortium, which represents a pivotal step forward in our ongoing commitment to improve public health in Kentucky,” said R. Kip Guy, Ph.D., dean of the College of Pharmacy . “The collective expertise on our campus in drug discovery, vaccine development and infectious disease research is essential for addressing the complex challenges presented in Kentucky and beyond.”
“From Dr. Li’s groundbreaking research on infectious diseases in animals to the breadth of research in plant sciences, engineering and biotechnology, our college is a powerhouse of expertise that propels transdisciplinary projects like CURE-KY, fostering innovation and collaboration at every turn," said Nancy Cox, vice president for land-grant engagement and dean of the UK Martin-Gatton College of Agriculture, Food and Environment .
“Kentucky is home to the highest cancer rates in the country, including several cancers caused by infectious diseases such as HPV and hepatitis C. Cancer prevention is a key component of Markey’s mission to significantly reduce Kentucky’s cancer burden. By better understanding and combating diseases that can lead to cancer, CURE-KY will contribute to this mission and make progress toward our ultimate goal of conquering cancer in the Commonwealth,” said B. Mark Evers, M.D., director of the Markey Cancer Center .
“We have an excellent team of scientists here at UK and on our leadership team. Together we can tackle infectious diseases from an array of perspectives to make a difference in the health of our community,” said Messaoudi.
You can find more information about CURE-KY and upcoming events on their website: https://medicine.uky.edu/sites/cure-ky
As the state’s flagship, land-grant institution, the University of Kentucky exists to advance the Commonwealth. We do that by preparing the next generation of leaders — placing students at the heart of everything we do — and transforming the lives of Kentuckians through education, research and creative work, service and health care. We pride ourselves on being a catalyst for breakthroughs and a force for healing, a place where ingenuity unfolds. It's all made possible by our people — visionaries, disruptors and pioneers — who make up 200 academic programs, a $476.5 million research and development enterprise and a world-class medical center, all on one campus.
In 2022, UK was ranked by Forbes as one of the “Best Employers for New Grads” and named a “Diversity Champion” by INSIGHT into Diversity, a testament to our commitment to advance Kentucky and create a community of belonging for everyone. While our mission looks different in many ways than it did in 1865, the vision of service to our Commonwealth and the world remains the same. We are the University for Kentucky.
Latest Stories
Uk researcher named an inspiring woman in plant biology, uk’s #icanendthetrend youth advisory board members garner national award, ned crankshaw named dean of uk college of design, ‘great teacher’ mark fillmore emphasizes importance of student research, uk celebrates opening of engineering lab designed to advance us manufacturing.
Volume 4, Number 3—September 1998 THEME ISSUE ICEID 1998
About emerging infectious diseases, new and reemerging diseases: the importance of biomedical research.
Cite This Article
Figure 1 . Examples of new and reemerging diseases.
A generation ago, it was suggested that the threat of infectious diseases would soon become an artifact of history. Today, as we approach the new millennium, the folly of this position is increasingly clear. My 87-year-old father recently reminded me of this. In the course of his lifetime, spent almost entirely in New York City, he has witnessed two pandemics of extraordinary impact: the global influenza pandemic of 1918-1919, which killed more than 20 million people worldwide, and the HIV/AIDS pandemic, which began to accelerate in the early 1980s and continues unabated in some parts of the world. In addition, at least 30 other new and reemerging diseases and syndromes have been recognized since the 1970s, including liver disease due to hepatitis C virus, Lyme disease, foodborne illness caused by Escherichia coli O157:H7 and Cyclospora , waterborne disease due to Cryptosporidium , hantavirus pulmonary syndrome, and human disease caused by the avian H5N1 influenza virus ( Figure 1 ). Clearly, we remain vulnerable to new and reemerging diseases.
New diseases are superimposed on endemic diseases such as diarrheal diseases, malaria, tuberculosis (TB), and measles, which continue to exact a huge toll. Indeed, malaria and TB, among others, are reemerging in a drug-resistant form. Today, infectious diseases remain the leading cause of death worldwide and the third leading cause of death in the United States. Many pathogens are becoming increasingly resistant to standard antimicrobial drugs, making treatment difficult and in some cases impossible. Moreover, chronic conditions generally considered noninfectious actually have been found to have a microbial etiology.
Awareness of Emerging Infections
The challenges posed by infectious diseases are recognized by the public and the media, as well as by political leaders and policy makers at the highest levels of government. There is a growing awareness that we live a global community, that diseases do not recognize borders, and that the U.S. public health community has an important role to play in fostering global health.
The Importance of Research
The infectious diseases community faces a difficult challenge: coping with ongoing problems such as malaria and TB while preparing for the inevitable emergence of diseases that are unknown or are recognized but will reemerge in a more threatening form. Available resources must be maximized by sustaining and increasing collaboration between federal agencies, academia, industry, and nongovernmental agencies, all of which play important roles in the fight against infectious diseases.
Figure 2 . Emerging infectious diseases: a research approach.
Within the federal government, the Centers for Disease Control and Prevention's (CDC) work in detecting and tracking pathogens is critical, especially with regard to diseases that have recently emerged or have the potential for emergence. Equally important, and complementary to CDC's efforts, is basic and clinical research supported by the National Institutes of Health (NIH) and other agencies. Historically, basic research has led to important, often serendipitous, advances that have illuminated the etiology of sometimes mysterious diseases and facilitated the development of diagnostics, therapies, and vaccines ( Figure 2 ).

Figure 3 . Emerging diseases funding (National Institute of Allergy and Infectious Diseases).
At the National Institute of Allergy and Infectious Diseases (NIAID) at NIH, we have increased funding for emerging diseases from $39.3 million in fiscal year 1993 to an estimated (president's budget) $85.0 million in fiscal 1999 ( Figure 3 ). Approximately 21% of the NIAID non-AIDS infectious diseases budget is devoted to emerging infectious diseases.
With the help of our advisory committees, we have defined five priorities in emerging and reemerging diseases research: 1) supporting the application of relevant scientific knowledge and new technologies to the detection, identification, and interdiction of emerging diseases, by expanding research on ecologic and environmental factors influencing disease emergence and transmission; 2) supporting the application of recent discoveries and new biomedical technologies to the identification, management, and control of emerging diseases, by expanding research on microbial changes and adaptations that influence disease emergence; 3) providing fundamental information for developing prevention and treatment strategies that can be employed to ameliorate disease impact, by expanding research on host susceptibility to emerging or reemerging pathogens; 4) supporting the development and validation of vaccines, therapeutics, and other control strategies for specific diseases with the potential to emerge or reemerge; and 5) strengthening the current U.S. research and training infrastructure for detecting and responding to outbreaks of infectious diseases.
Among many studies domestically and internationally, NIAID sponsors five international programs in tropical infectious diseases, most of which have components both in the United States and in the countries where the incidence of these diseases is greatest. It is essential to engage scientists in host countries and work with them collaboratively, both to tap their expertise as well as to help them build research infrastructure on their home soil.
Successful Partnerships
The public and private sectors, including government, academia, and industry, bring complementary skills and perspectives to the research endeavor. Cross-sector collaboration can yield extraordinary dividends. A cogent example is the development of protease inhibitors for the treatment of HIV disease.
After HIV was identified in 1983, researchers funded by NIH and others began to intensively study the structural and regulatory genes of HIV and the role these genes and their products play in the replication cycle of the virus. This work led to an understanding of the importance of the HIV protease enzyme and methods to express, purify, and crystallize the enzyme. Building on these findings, researchers in the private sector designed and produced specific inhibitors of HIV protease and worked closely with the Food and Drug Administration, NIH, and others to assess protease inhibitors in clinical trials.
The first of four licensed protease inhibitors reached the market in December 1995. Given in combination with at least two other antiretroviral drugs, protease inhibitors dramatically reduce levels of plasma viremia in a substantial proportion of patients. Both controlled and observational studies show that these potent regimens can provide a substantial clinical benefit.
Although drug combinations that include protease inhibitors have helped many patients, it is far too soon to become complacent or declare victory. Many patients have not benefited from the new drugs or cannot tolerate their side effects, and drug resistance will inevitably become more widespread. The development of the next generation of antiretroviral agents is crucial and will require the skills of investigators in both the public and private sectors. However, the cost of antiretroviral drugs will probably keep them beyond the reach of much of the developing world; therefore, the development of an HIV vaccine is of paramount importance.
Malaria Initiatives at NIH
Until relatively recently, AIDS was virtually the only emerging disease with global impact that was widely discussed in the United States; however, other diseases such as malaria and TB have actually caused more illnesses and deaths over the past 2 decades.
Malaria kills up to three million persons each year, most of them children in sub-Saharan Africa. In the past year, NIH has worked with research organizations and donor agencies from around the world to form a coalition called the Multilateral Initiative on Malaria. This unprecedented initiative will enhance international collaborations, encourage the involvement in malaria research of scientists from malaria-endemic countries, and identify additional malaria research resources. In addition, NIH has bolstered its long-term commitment to malaria research. NIH-supported malaria projects—many in collaboration with other government and international agencies—include 1) a new repository of materials available to researchers worldwide; 2) basic, field-based, and clinical research on all phases of malaria research; and 3) projects to determine the genetic sequences of important malaria species.
Responding to Avian H5N1 Influenza

Figure 4 . Response to H5N1 avian influenza outbreak in Hong Kong.
An outbreak of avian H5N1 influenza in Hong Kong recently alarmed the medical community and the world. The multinational response to this outbreak has involved the close collaboration of many organizations ( Figure 4 ). As part of NIH's long-standing research into respiratory viruses, we had in our reagent repository the specific antisera needed to quickly develop test kits that were used effectively by CDC and others for detecting and tracking the virus. We also have supported the rapid production of a recombinant vaccine against avian influenza virus for use in laboratory and health-care personnel at risk. Without a strong research base, the rapid response to this emergency would not have been possible.
Vaccine Development
With avian flu, malaria, AIDS, and other new and reemerging diseases, an important goal of NIH is the development of vaccines. If just four recently developed vaccines (hepatitis B, rotavirus, Haemophilus influenzae type b, and acellular pertussis) were universally administered, more than three million deaths could be prevented each year.
Historically, scientific advances in microbiology and related disciplines have driven the development of new vaccines. For example, the identification of microbial toxins, as well as methods to inactivate them, allowed the development of some of our earliest vaccines, including those for diphtheria and tetanus. In the 1950s, new tissue culture techniques ushered in a new generation of vaccines, including measles, mumps, and rubella. In recent years we have seen rapid advances in our understanding of the immune system and host-pathogen interactions, as well as technical advances such as recombinant DNA technology, peptide synthesis, and gene sequencing. Each of these has facilitated the development of new vaccines and vaccine candidates for important pathogens.
Sequence information can be used in many ways and promises to be useful in identifying antigens to incorporate into vaccines, as well as determining the factors that influence the antigenicity or virulence of a microbe. The complete genetic sequences of more than 13 microorganisms have now been published. More than 60 other sequencing projects for medically important pathogens, such as Plasmodium spp., Mycobacterium spp., Chlamydia trachomatis , Vibrio cholerae, and Neisseria gonorrhoeae, are under way.

Figure 5 . Benefits of emerging diseases research.
The importance of basic research to the control of emerging and reemerging diseases cannot be overemphasized. Emerging diseases research encompasses many disciplines, and research advances that fall under the rubric of emerging diseases will be relevant not only to specific diseases being studied but to a broad range of disciplines such as vaccinology, immunology, and drug development ( Figure 5 ). In turn, research in these areas is critical to advances in emerging and reemerging diseases. With a sustained commitment to basic research and cross-sector collaboration, important scientific findings and technological advances can be translated into improved global health and reduced susceptibility to new microbial threats.
Acknowledgment
The author thanks Greg Folkers for helpful discussion related to the preparation of this manuscript.
- Institute of Medicine, Board on International Health. America's vital interest in global health. Washington: National Academy Press; 1997 .
- Centers for Disease Control and Prevention . Staphylococcus aureus with reduced susceptibility to vancomycin–United States, 1997. MMWR Morb Mortal Wkly Rep . 1997 ; 46 : 765 – 6 . PubMed Google Scholar
- Centers for Disease Control and Prevention . Update: isolation of avian influenza A (H5N1) viruses from humans–Hong Kong, 1997-1998. MMWR Morb Mortal Wkly Rep . 1998 ; 26 : 1245 – 7 .
- The CVI strategic plan: managing opportunity and change: a vision of vaccination for the 21st century. Geneva: Children's Vaccine Initiative, 1997 . Sponsored by UNICEF, United Nations Development Program, World Health Organization, World Bank, Rockefeller Foundation.
- Two cheers for the multilateral malaria initiative. [editorial] . Nature . 1997 ; 388 : 211 . DOI PubMed Google Scholar
- Fauci AS . Biomedical research in an era of unlimited aspi-rations and limited resources. Lancet . 1996 ; 348 : 1002 – 3 . PubMed Google Scholar
- The Institute for Genomic Research . TIGR Microbial Database [database online] [cited 1998 Apr 1]. Available from: URL: http://www.tigr.org .
- World Health Organization . World Health Report 1997–conquering suffering, enriching humanity. Geneva: The Organization; 1997 .
- Figure 1 . Examples of new and reemerging diseases.
- Figure 2 . Emerging infectious diseases: a research approach.
- Figure 3 . Emerging diseases funding (National Institute of Allergy and Infectious Diseases).
- Figure 4 . Response to H5N1 avian influenza outbreak in Hong Kong.
- Figure 5 . Benefits of emerging diseases research.
DOI: 10.3201/eid0403.980308
Table of Contents – Volume 4, Number 3—September 1998
Exit Notification / Disclaimer Policy
- The Centers for Disease Control and Prevention (CDC) cannot attest to the accuracy of a non-federal website.
- Linking to a non-federal website does not constitute an endorsement by CDC or any of its employees of the sponsors or the information and products presented on the website.
- You will be subject to the destination website's privacy policy when you follow the link.
- CDC is not responsible for Section 508 compliance (accessibility) on other federal or private website.
Article Citations
Highlight and copy the desired format.
Metric Details
Volume 4, Number 3—September 1998
Article Views: 10296
Data is collected weekly and does not include downloads and attachments. View data is from .
Citations: 26
What is the altmetric attention score.
The Altmetric Attention Score for a research output provides an indicator of the amount of attention that it has received. The score is derived from an automated algorithm, and represents a weighted count of the amount of attention Altmetric picked up for a research output.
May 22, 2024
Innovative Thinking Could Make New Sickle Cell Treatments More Accessible
The cost of new gene-based sickle cell treatments isn’t the only barrier to access. Coming up with new ways to treat the whole disease—and person—could make treatment more equitable
By Shobita Parthasarathy
Pixelimage/Getty Images
Last fall, to great fanfare, US regulators approved two gene therapies for sickle cell disease , and the European Union and UK soon followed. Many people hope that these treatments will provide a “ functional cure ” for the genetic condition, which causes rigid, misshapen red blood cells that lead to anemia, episodes of extreme pain , blood vessel and organ damage, stroke risk and lower life expectancy. These sickle cell therapies also bring us closer to an age of “ CRISPR medicine ” in which new gene-editing tools could be used to fix a range of debilitating genetic diseases, including Duchenne muscular dystrophy and cancer.
For the eight million people across the world, including the 100,000 in the U.S., who have sickle cell disease, these gene therapies could be life-changers. Yet immediately after the approvals, attention turned to the prohibitive cost of the treatments. One company sells its therapy for $2.2 million, and the other does so for $3.1 million. Other gene-based treatments have similar price tags .
Reducing costs for these therapies is important. It is unlikely to make life much better for most people with sickle cell and those with other genetic diseases, however. Many will simply be unable or unwilling to take these treatments, which require multiple hospital visits (and often overnight stays) over the course of many months. Indeed, we already know that many people aren’t getting what treatments already exist in the U.S. for sickle cell for a variety of reasons. Of the people with the disease who have multiple pain episodes related to blood-flow blockage per year, only 35 percent receive the most common and cheapest medication, hydroxyurea, and the majority of them receive this medication by itself. Only 13.2 percent of people with multiple pain episodes receive one or more of any of the newer medications approved since 2017, either in combination with hydroxyurea or alone.
On supporting science journalism
If you're enjoying this article, consider supporting our award-winning journalism by subscribing . By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
People already struggle to get treatment for sickle cell disease, so reducing the cost of gene therapies will not make treatment any more accessible.
Understanding this, what if we were to treat barriers to care other than cost as innovation problems? What if the best way for technology to serve society, and particularly to advance equity, is for governments and innovators to focus on more than creating potentially lifesaving treatments? Some might argue that social and health challenges are simply not the responsibility of scientists or biotechnology companies. But if the goal is for people to be healthy rather than to simply put inaccessible products on the market, these challenges must be.
Currently, many scientists, physicians and people with the disease view the treatments as an important step toward racial equity after generations of discrimination and mistreatment—sickle cell disease mostly affects people of African, South Asian and Middle Eastern descent.
Yet many people with sickle cell disease have limited incomes and are unlikely to pay for the therapies out-of-pocket; 50 percent of people with the disease are on Medicaid. In response, both private insurers and the Centers for Medicare & Medicaid Services (CMS) are developing “ outcomes-based ” reimbursement plans for cell and gene therapies in general. Under these plans, drug companies would reimburse payers if a treatment is less successful than expected over a prespecified number of years.
But this is only part of the equation: many people with sickle cell simply do not have the care infrastructure—including a primary care physician or access to a specialist hematologist—needed to participate in the full course of the gene-therapy treatment. This absence of care infrastructure may be the result of precarities, including a lack of employment, housing, or health care or wariness of the scientific and medical institutions where people have experienced racism for decades. People working service jobs and receiving hourly wages cannot leave work easily or frequently. People who want to have children may not want to participate either because the new treatments involve chemotherapies that can affect fertility.
Beyond the idea of a functional cure, for most people with sickle cell, their immediate priority is to alleviate the extreme pain they experience. In fact, as described by Ghanian sickle cell expert Felix Israel Domeno Konotey-Ahulu in a 1975 article, the names for the disease in several West African languages— chwechweechwe (Ga), nwiiwii (Fante), nuidudui (Ewe) and ahotutuo (Twi)—are onomatopoetic representations of the relentless and repetitive gnawing pain people with sickle cell experience.
Because sickle cell disease is considered a blood disorder, however, scientists and industry have focused their attention on correcting the blood cell dysfunction. Little scientific effort is focused on developing new pain treatments for this population or studying the efficacy of existing therapies for managing pain. Instead, when people with this disease visit emergency departments seeking pain relief, they are often assumed to have a substance use disorder.
So how might we trigger innovation that responds to public needs and takes people’s lived experiences seriously? First, governments must change who sets research agendas and how. Technical experts, including high-level advisory committees, currently set priorities for most funding agencies. This leads to prioritizing scientific and industrial priorities, such as attention to novelty and efficiency, over the needs of the individuals affected by a disease.
People with sickle cell disease, for example, and other experts who understand the impacts of disease in society rarely play a role; of the members of the U.S. National Health, Lung and Blood Institute’s Sickle Cell Disease Advisory Committee, only two are grassroots advocates for people with the disease. This is the case for many underserved illnesses in society.
What if people who understand sickle cell disease historically, psychologically and socially sat on this committee? They might steer more attention toward pain and addressing racism in science, technology and medicine. They might also encourage collaborations across the clinical, social and basic research arms of the National Institutes of Health rather than defining research projects according to scientific or medical specialty.
Second, governments can steer funding toward interdisciplinary research projects that focus on more immediate benefits. They could encourage true collaborations among biomedical scientists, social scientists and people with the disease so that knowledge about the technology’s likely effects inform its design and development. (The National Science Foundation is trying to pioneer this approach through its new Responsible Design, Development and Deployment of Technologies program.) People with sickle cell disease and social scientists with relevant expertise could also sit on grant review committees to ensure that the portfolio of funded projects fits with their knowledge and concerns and doesn’t just focus on the molecular determinants of the disease.
Third, the private sector must also take responsibility. One way to create an incentive is through indexes, such as those that exist for environmental sustainability, that publicly assess the social and equity impacts of a company. These indexes would evaluate the real social consequences of a company’s products, including whether and how it included people with a disease in its innovation processes, rather than force people to simply trust claims about a technology’s transformative potential.
To develop technologies that successfully solve serious health and social problems and to ensure that people benefit from innovation, we need the public and private sectors to invest in high-impact interdisciplinary projects early in the innovation process and—above all—to value the expertise of individuals with a disease. Otherwise treatments that cost billions to create and cost millions to administer will continue to be out of reach for those who need them the most.
This is an opinion and analysis article, and the views expressed by the author or authors are not necessarily those of Scientific American.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Review Article
- Published: 22 March 2021
Nanotechnology approaches for global infectious diseases
- Ameya R. Kirtane ORCID: orcid.org/0000-0002-3779-0363 1 , 2 , 3 na1 ,
- Malvika Verma ORCID: orcid.org/0000-0001-7043-5120 1 , 4 , 5 na1 ,
- Paramesh Karandikar ORCID: orcid.org/0000-0002-3529-2037 1 , 2 ,
- Jennifer Furin 6 ,
- Robert Langer ORCID: orcid.org/0000-0003-4255-0492 1 , 2 , 5 , 7 , 8 &
- Giovanni Traverso ORCID: orcid.org/0000-0001-7851-4077 3 , 5 , 9
Nature Nanotechnology volume 16 , pages 369–384 ( 2021 ) Cite this article
37k Accesses
243 Citations
71 Altmetric
Metrics details
- Biomaterials
- Nanobiotechnology
Infectious diseases are a major driver of morbidity and mortality globally. Treatment of malaria, tuberculosis and human immunodeficiency virus infection are particularly challenging, as indicated by the ongoing transmission and high mortality associated with these diseases. The formulation of new and existing drugs in nano-sized carriers promises to overcome several challenges associated with the treatment of these diseases, including low on-target bioavailability, sub-therapeutic drug accumulation in microbial sanctuaries and reservoirs, and low patient adherence due to drug-related toxicities and extended therapeutic regimens. Further, nanocarriers can be used for formulating vaccines, which represent a major weapon in our fight against infectious diseases. Here we review the current burden of infectious diseases with a focus on major drivers of morbidity and mortality. We then highlight how nanotechnology could aid in improving existing treatment modalities. We summarize our progress so far and outline potential future directions to maximize the impact of nanotechnology on the global population.
Similar content being viewed by others

A guide to vaccinology: from basic principles to new developments

Lipid nanoparticles for mRNA delivery
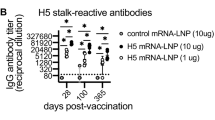

Development of a nucleoside-modified mRNA vaccine against clade 2.3.4.4b H5 highly pathogenic avian influenza virus
Infections are a dominant contributor to the global disease burden. High mortality rates are associated with lower respiratory infections, diarrhoea, tuberculosis (TB), human immunodeficiency virus (HIV) infection and malaria (Fig. 1a ) 1 . Mortality rates are highest in developing countries, where resources such as vaccines and anti-infectives may be limited (Fig. 1b ) 1 . Temporal trends in mortality show that although the overall number of deaths is declining, the gap between deaths in high socio-demographic index (SDI) countries and low SDI countries remains substantial (Fig. 1c ) 1 . Unfortunately, clinical trials for these infectious diseases (IDs) are also lagging compared with conditions such as cancer and cardiovascular diseases (Fig. 1d ) 1 and there is thus an urgent need to identify practical and impactful strategies that enable better treatment of IDs.
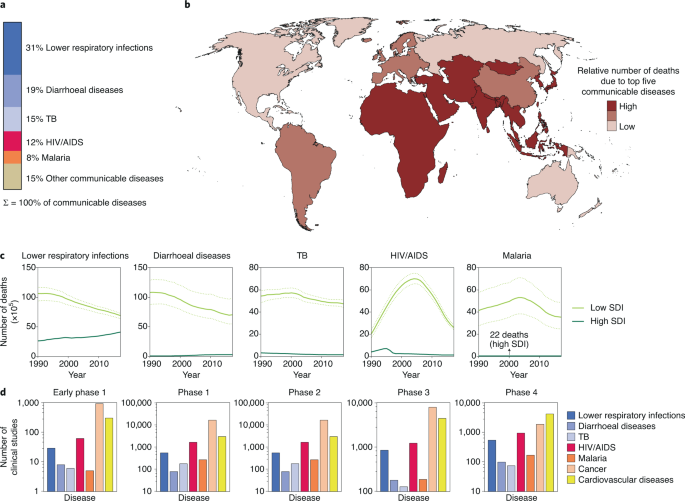
IDs represent a chronic healthcare burden around the world and as such are high priority targets for investigational drugs. a , The communicable diseases with the five highest mortality rates are lower respiratory infections, diarrhoeal diseases, TB, HIV infection and malaria. b , Low SDI countries are disproportionately affected by these top five communicable diseases. c , Temporal trends in mortality rate for the five diseases. Although mortality rates from these communicable diseases is declining, there is considerable disparity between low and high SDI countries. The solid lines represent the mean values and the dotted lines show the uncertainty interval. d , The number of clinical studies listed in clinicaltrials.gov for various diseases. Data in a – c are from ref. 1 . Credit: Ohaiyoo/Adobe.
A host of challenges must be overcome to effectively manage IDs. The lack of safe and effective drugs is central to our inability to treat IDs 2 . In some cases, a lack of drug efficacy can be attributed to pathogen resistance, which then requires a more expensive drug regimen for cure 3 . Challenges to treating IDs are often compounded in low SDI countries. Poor patient adherence to therapies and the need for sustained patient monitoring are major obstacles to effective treatment 4 . Poor procurement practices, the inability to pay for drugs and poor stability of drug products at high temperature and humidity prevent access to effective treatments 5 . A concerted effort on scientific and policy levels is necessary to overcome these challenges.
Nanotechnology has the potential to transform both detection and treatment of a wide range of diseases. These technologies, involving systems with a diameter of about one-thousandth of the thickness of a hair, stand to substantially impact the globe’s main sources of morbidity and mortality. Nanosystems have been the subject of intensive research over the past few decades, resulting in Food and Drug Administration (FDA)-approved chemotherapeutics, anaesthetics, imaging agents, nutritional supplements and others 6 . Not surprisingly, nanotechnology has been extensively evaluated to improve the treatment of IDs. In this Review, we examine the opportunities presented by nanotechnology for the treatment of IDs and discuss preclinical and clinical progress in this area with a focus on HIV, TB and malaria. Additionally, we recognize the ongoing impact nanotechnology is having in the form of successful mRNA and protein vaccines for SARS-CoV-2. Finally, we discuss challenges to translating these technologies from the laboratory to the clinic. We do not include polymicrobial IDs, other monomicrobial IDs, metal nanoparticles 7 and vaccination 8 , 9 —all of which are areas of intensive research.
Nanotechnology in treating IDs
The availability and correct use of safe and efficacious medications are imperative for treating IDs. Nanotechnology-based approaches have been the topic of intensive preclinical evaluation to improve the therapeutic index of ID drugs and simplify their use. We highlight promising preclinical studies and discuss challenges to their clinical translation.
Sustained systemic delivery of anti-infectives
The management of several IDs involves chronic treatment, which places a major burden on the patient and the health care system. HIV-infected individuals require lifelong treatment to ensure control of the viral burden 10 . To treat TB, a combination of pills must be consumed over a period of several months to years—often multiple times a day 11 . Prolonged and complicated dosing regimens with high pill burden result in lower patient adherence and, ultimately, failure of treatment. At times, providers administer medications only once a day for programmatic ease even though this may not achieve optimal drug levels. Hence there is interest in developing systems that reduce dosing frequency and ease the dosing regimen. Injectable nanocarriers that deliver drugs for sustained periods have been actively pursued. Broadly, these systems can be classified into two categories—systems that control drug release with an excipient (such as a polymer or lipid) and those that rely on the slow dissolution of poorly soluble drug crystals in interstitial fluid (Fig. 2 ).
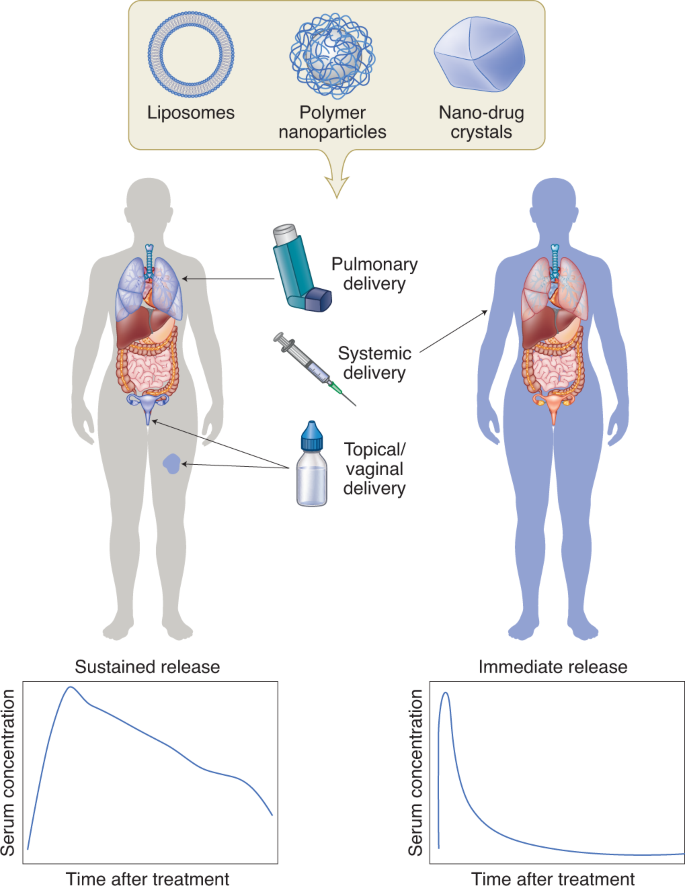
The encapsulation of drugs in nanocarriers such as liposomes, polymer nanoparticles, or prepared drug nanocrystals can enable sustained drug delivery in a localized manner (left). These systems have been evaluated for treating local infections of the female reproductive tract, lungs and skin. Injectable nanocarriers are also being explored for systemic drug delivery (right).
Considerable research has focused on polyester-based drug delivery systems that degrade in the presence of physiological esterases (for example poly(lactide-co-glycolide) (PLGA) and poly(caprolactone)). Drug release occurs via bulk degradation of the polymer and drug diffusion 12 and can be modified by altering the hydrophobicity of the monomer, polymer chain length and particle size 13 . Other polymers used for sustained release include poly(anhydrides), poly(orthoesters), poly(cyanoacrylates) and poly(amides) 12 .
Sustained release can also be obtained by encapsulating drugs in liposomes, which are lipid vesicles with the capacity to load both hydrophobic and hydrophilic drugs. NeXstar Pharmaceuticals developed MiKasomes—a liposomal formulation of amikacin 14 , a drug requiring frequent dosing and continuous therapeutic monitoring. In a rat model, intravenously administered Mikasomes distributed to several tissues, where they released drug gradually resulting in an eightfold increase in drug half-life. Despite early positive results in clinical trials for urinary tract infections and Mycobacterium infections 15 , development of the formulation was discontinued in 2000.
A drawback of some polymeric and liposomal formulations is that they require large quantities of excipients to control drug release. This increases the size of the injection and limits the amount of drug that can be administered. To circumvent this, aqueous dispersions of nano-milled drug crystals are being developed 16 , 17 . Following subcutaneous or intramuscular injection, drug absorption into systemic circulation is dictated by the rate of dissolution of the drug crystals in the interstitial fluid 16 . Hence, physicochemical properties of the drug and size of the drug crystals play an important role in determining drug release kinetics. A common technique to prolong drug absorption is to convert the drug into salt forms of lower solubility or into prodrugs that have greater hydrophobicity 18 .
An important consideration with sustained release systems is the potential for prolonged exposure to sub-therapeutic concentrations, which may lead to drug resistance 19 . Dwindling amounts of drug released during the terminal phase of the release period may lead to a ‘long pharmacologic tail’ 20 , a period of slowly declining drug levels. Pulsatile release formulations, instead of sustained release formulations, may help address this issue. Alternatively, minimally invasive means of removing the depot at the end of the therapeutic period may be attractive.
Local delivery
As epithelial surfaces are common sites of entry and residence for pathogens, regional drug delivery to these surfaces holds great promise. Compared with systemic administration, local delivery to the site of infection is likely to enhance on-target (and minimize off-target) drug exposure. Of course, its utility is limited for systemic infections. Preclinical studies, predominantly in rodents, have identified several advantages of using nanoformulation for local therapies. Encapsulation of drugs in nanocarriers allows for sustained drug delivery, thereby reducing dosing frequency. Nanocarriers can be engineered to release drugs in the presence of certain triggers and provide temporal control on drug exposure. Nanocarriers may enhance drug uptake in cells and improve efficacy against intracellular pathogens. Finally, encapsulation in nanocarriers may protect labile drugs (such as nucleic acids) in harsh physiological conditions such as low pH or the presence of enzymes.
Vaginal drug delivery
HIV and herpes simplex virus (HSV) can enter the body via the vaginal tract. Prophylactic local administration of microbicides can reduce the chance of infection. Ensuring maximal coverage throughout the vaginal tract at the time of infection is likely to minimize the chances of infection. Several barriers impede complete coverage of the vaginal tract and prolonged residence of the drug delivery system 21 . First, formulations can leak out of the vaginal tract following application of a drug delivery system. Second, the vaginal mucosa is covered by a layer of mucous (a viscous biopolymer made of glycoproteins called mucins), which acts as a diffusional barrier trapping the drug particulate matter. Third, the mucous layer is shed frequently, and particles trapped within it can be lost 22 . Fourth, the vaginal surface has several folds or rugae reducing drug accessibility. Finally, the low pH and enzymes present in the vaginal fluid may degrade susceptible drugs 21 .
To enhance vaginal residence, a common theme has been to develop systems that are rapidly transported across the mucous into the mucosa. In a study from the Saltzman group 23 , the authors compared the vaginal uptake of dye-loaded PLGA nanoparticles that were surface coated with either avidin or poly(ethylene glycol) (PEG) or left uncoated. Leakage out of the vaginal tract was highest for uncoated nanoparticles (~13% of the dose within 0.5 h). Vaginal lavage, which presumably enabled collection of the mucous layer, was rich in the mucoadhesive avidin-coated nanoparticles. The highest tissue uptake was achieved with PEG-coated nanoparticles; however, tissue uptake was limited to the lower reproductive tract. Improved tissue uptake of PEGylated nanoparticles may be attributed to their higher diffusivity through the mucous layer—an observation shown using multiple particle tracking experiments by the Hanes lab 24 . An alternative strategy to enhance vaginal retention is to administer nanoparticles embedded in films. Although nanoparticle dispersions can rapidly leak out of the vagina, films may hold the nanoparticles in place for some duration. Cunha-Reis et al. demonstrated that nanoparticles loaded with an anti-HIV drug (efavirenz) and embedded in a water-soluble film showed lower leakage and enabled higher tissue concentrations than nanoparticle dispersions 25 . However, the improved tissue concentrations were observed only within the first hour of administration, and not at later times. These studies highlight two challenges with nanoparticle-based vaginal drug delivery systems. First, there is minimal nanoparticle uptake in the upper reproductive tract, bringing into question the coverage offered by these systems. Second, in rodent models, drug concentrations become undetectable at about 24 h post-dose, suggesting that sustained delivery may be possible for only a relatively short time. The issue of residence may be overcome by loading nanocarriers in macro-drug delivery systems such as vaginal rings, which have been clinically shown to have extended residence times.
Despite their limitations, vaginal delivery with nanosystems has shown therapeutic potential in murine models. Ensign and colleagues showed that PEGylated PLGA nanoparticles dosed in a hypotonic solution were rapidly taken up into vaginal folds via advection 26 . Acyclovir monophosphate nanoparticles protected ~54% of the mice that were infected with HSV-2 30 min post-treatment. In contrast, the soluble drug protected only 16% of the mice from viral challenge. In a mouse model of lethal HSV-2 infection, intravaginal administration of oligofectamine-complexed small interfering RNA (siRNA) targeting genes encoding viral envelope and DNA binding proteins resulted in improved survival compared with untreated controls 27 . Interestingly, PLGA nanoparticles containing cationic spermidine have been used for siRNA-based knockdown of the same target, and have shown better protection than lipofectamine lipoplexes 28 . The authors found that the PLGA nanoparticles had a similar knockdown efficiency to lipoplexes, but elicited lower inflammation (as observed by measuring neutrophil invasion using immunohistochemistry). They attributed the improved protective activity of PLGA nanoparticles to their ability to cause minimal inflammation. It must be noted that in all studies described here, exposure to the pathogen was within a few minutes to hours of treatment. Such treatment schedules are difficult to reduce to practice in resource-limited settings 10 , underscoring the need for long-acting local systems. Even so, the recent regulatory success of Starpharma’s VivaGel, a dendrimer antimicrobial with demonstrated efficacy against HIV, HSV and bacterial vaginosis and formulated both as gels and as a surface coating for condoms, highlight the vast potential for growth and the versatility of nanotechnological systems in this area.
Pulmonary delivery
Pulmonary delivery is an attractive route for the treatment of respiratory infections as it may enable greater on-target drug exposure. However, formulations that cannot penetrate mucous layers and biofilms have low pulmonary bioavailability due to rapid enzymatic deactivation, sequestration and elimination by coughing 22 , 29 . In the lungs, mucous is secreted by goblet cells within the epithelium of mucous membranes. Biofilms are produced when microbes aggregate on a surface and produce an extracellular matrix comprising high-molecular-weight polysaccharides (such as alginate and N -acetyl glucosamine), DNA and proteins 30 .
Greater local bioavailability of therapeutics has been observed following pulmonary administration compared with oral or intravenous administration. In mouse studies, pulmonary delivery of amorphous itraconazole nanoparticles resulted in a tenfold higher drug concentration in the lungs compared with oral administration of its commercial formulation (Sporanox) and the unformulated drug 31 . In a murine Aspergillus fumigatus infection model, the same group observed an increased median survival in the pulmonary nanoparticle treated group (7.5 days) compared with oral Sporanox treated group (5 days) 32 . These benefits cannot be attributed completely to nanoformulation as changing the route of administration from oral to pulmonary is likely to contribute heavily to this improvement. However, the poor aqueous solubility of itraconzole may have precluded the pulmonary administration of the unformulated drug. In a study with ciprofloxacin (a drug with higher water solubility), Wong et al. compared the activity of liposome-encapsulated and free drug upon pulmonary administration 33 . The half-life of the liposome-encapsulated drug in mouse lungs was about double that of the free drug (~3 h versus ~1.5 h). Importantly, in a Francisella tularensis infection model, survival of mice in the untreated and free-drug-treated groups was similar, in that all mice succumbed to the infection within 14 days. In contrast, there were no deaths observed in mice treated with pulmonary liposomes. Interestingly, the authors found the liposomes to be effective even when administered via the intravenous route. Direct comparison of the pharmacokinetics of intravenous and pulmonary liposomes would have yielded important information regarding on-target and off-target exposure.
Bacteria regulate their cooperative growth and form biofilms by secreting and sensing molecules that signal for virulence. Inhibitors of this quorum sensing phenomenon have the potential to disrupt biofilms. However, the mucous covering, especially in patients suffering from cystic fibrosis, retards drug delivery to the bacterial biofilms. To overcome mucous resistance in the respiratory tract, Nafee and colleagues encapsulated the quorum sensing inhibitor in lipid nanoparticles surface coated with polysorbate 80—a co-polymer surfactant containing PEG 34 . In comparison with the free drug, the nanoparticle-encapsulated quorum sensing inhibitor showed better in vitro control of virulence as measured by quantifying levels of a bacterial product (pyocyanin). Confocal microscopy also demonstrated that polysorbate 80-coated nanoparticles diffused across porcine mucous. However, transport of the quorum sensing inhibitor in its free-drug form and nanoparticulate form was not assessed. Although these studies introduce an interesting concept, in vivo evaluation in a model of cystic fibrosis will be critical.
Another biofilm-disrupting approach involves the use of nitrous oxide-releasing polymeric nanoparticles. To enable sustained release of nitric oxide, which has a short half-life, Duong et al. 35 conjugated nitric oxide with star-shaped polymers. Using a bacterial reporter strain, the authors confirmed sustained release of nitric oxide from the star-shaped polymer nanoparticles in vitro. Furthermore, they found that nanoparticles were able to decrease Pseudomonas aeruginosa biofilm formation by >70% compared with untreated controls 35 . However, in vivo validation was not performed.
Mucolytic agents, such as N -acetyl cysteine, can be used to improve the penetration of nanocarriers by reducing disulphide bonds in mucous. N -acetyl cysteine enhanced the diffusivity of DNA-loaded poly(lysine) polyplexes in the sputum of cystic fibrosis patients by more than fivefold 36 . To validate the benefit of N -acetyl cysteine in vivo, the authors treated mice with inflammatory lipopolysaccharide from P. aeruginosa , which led to enhanced mucous secretion in the lungs. Compared with healthy mice, the transfection efficiency of the polyplex in mice treated with lipopolysaccharide was reduced by about tenfold. Impressively, pre-treatment with N -acetyl cysteine restored transfection efficiency in the inflammation model. This study demonstrates the inhibitory effects of mucous on drug delivery to lungs and the benefits of combining mucolytics with nanotherapeutics.
Topical delivery
Chronic and non-healing wounds are a source of increasing healthcare cost and mortality 37 . These wounds occur due to breakdowns in the wound repair pathway and chronic infection caused by opportunistic pathogens. Unlike acute infections, which occur due to the presence of a large burden of individual (planktonic) pathogens and have a more serious symptomatology, chronic infections are localized and recur periodically. Furthermore, while acute infections may be managed effectively by medications, chronic infections do not respond to conventional antibiotic therapy. Interestingly, a study found that 60% of clinical samples from chronic infection sites contained biofilms compared with 6% found in acute infections 38 . These results suggest that the presence of biofilms plays a role in chronic infections and non-healing wounds, and strategies that overcome biofilm resistance may help treat these infections.
Topical drug delivery shares both challenges and therapeutic targets with pulmonary delivery—biofilm-forming opportunistic pathogens such as Staphylococcus aureus and P. aeruginosa have been implicated in both lung and skin infections. As such, many principles of engineering biofilm-penetrating nanosystems for pulmonary delivery can be leveraged for topical delivery. Sustained release of nitric oxide was achieved by fabricating nanoparticles containing reducing sugars and sodium nitrite. These nanoparticles were capable of releasing nitric oxide for over a month. In murine skin infection models of S. aureus and methicillin-resistant S. aureus biofilms, nitric oxide-releasing nanoparticles decreased overall wound bacterial burden compared with both unloaded nanoparticles and the control group 39 .
One strategy for treating wound infections is the application of silver solutions using a cotton gauge. However, due to the short retention and sudden exposure to high concentrations, the treatment needs to be applied very frequently and it can be toxic to host cells 40 . One proposed solution is to encapsulate porous silver microparticles in a poly(lactide) nanofibre scaffold. It was hypothesized that the slower release of silver from the scaffold may minimize its toxicity to human cells while maintaining its anti-infective properties. The scaffold did enable the slow release of silver; however, toxicity to host cells was not reduced 40 . Further work in this field could be valuable.
Targeted delivery to sites of infection
There is considerable interest in designing systems that enable targeted drug delivery to sites of infection. This is motivated by cases of low drug penetration into infected tissues (for example, low penetration of antiretrovirals into the brain and lymph nodes 10 and anti-TB drugs into cavitary lesions 41 ), drug distribution into sites of toxicity (such as aminoglycosides in the ear 42 ) and killing of commensal microbiota 43 . Encapsulation in nanocarriers offers the possibility of achieving drug targeting (Fig. 3 ), but there is considerable room for improvement. Improved on-target accumulation occurs because of enhanced nanocarrier permeability at sites of infection compared with uninfected tissue. Nanocarrier surfaces can also be functionalized with ligands that bind to infected tissues or microbes. This latter strategy has been termed active targeting or ligand-mediated targeting. Strategies that do not rely on specific ligands have thus been dubbed passive targeting.
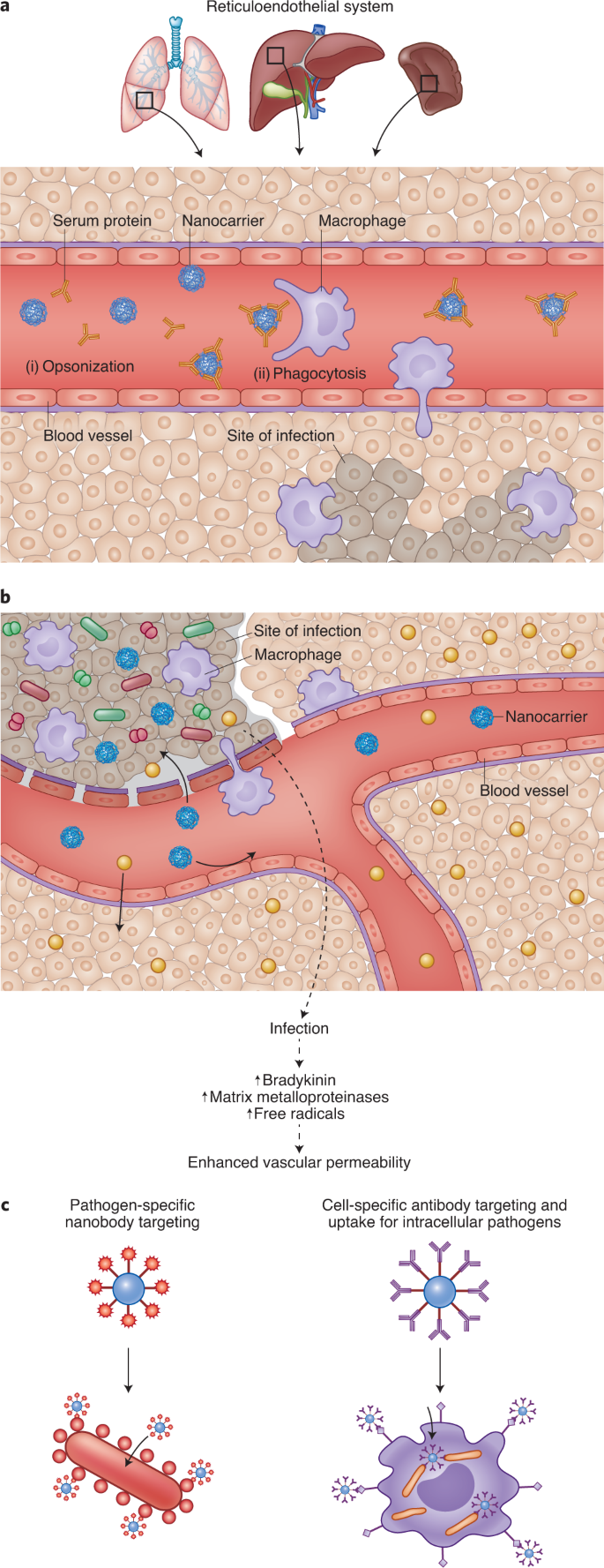
Targeting delivery of drugs to sites of infection can improve efficacy and reduce toxicity. a , On systemic injection, serum proteins are adsorbed on the surface of nanocarriers (opsonization; i) leading to phagocytosis by macrophages (ii). Hence, nanocarriers have been used for targeting drugs to macrophages, which are common sites of infection. b , Vascular permeability at sites of infection is higher than that observed in non-infected tissue. Increased vascular permeability is due to pathogens (shown as red and green shapes), which activate the kallikrein-kinin pathway, and due to inflammation, which results in enhanced reactive oxygen species. While entry of free drug (yellow circles) is possible at both uninfected and infected sites, entry of nanocarriers (blue circles) at uninfected sites is limited. This phenomenon has been exploited for targeting anti-infectives to sites of infection. c , Coating nanocarriers with ligands that bind receptors on the surface of infected host cells (right) or pathogens (left) have also been used for increasing drug targeting.
Passive targeting
Nanoencapsulation of anti-infectives has been pursued for two divergent goals—targeting drugs to macrophages and to infected tissues. The mechanisms by which nanocarriers deposit drugs at these two sites, and hence formulation strategies, are distinct.
Macrophages are common targets for bacteria (such as Mycobacterium tuberculosis ), fungi (for example Aspergillus species) and viruses (such as HIV). As nanocarriers are predominantly cleared by these cells (Fig. 3a ), they have been widely used for targeting drugs to macrophages. Administering the antiretroviral azidothymidine in poly(hexacyanoacrylate) nanoparticles improved its accumulation in reticuloendothelial system (RES) organs such as the liver, lungs and spleen 44 . In rats, 60% of the drug dose was found in the RES organs 8 h after treatment with nanoparticles. In contrast, after treatment with the soluble drug alone, only 12% of the drug was recovered in the RES tissues.
Nanocarriers have been used to enhance drug accumulation at sites of infection other than the RES organs. This is based on the observation that blood vessels at infection sites are leaky, promoting greater nanocarrier entry (Fig. 3b ). To study vascular permeability at infection sites, Evan’s blue was injected intravenously into guinea pigs following intradermal injection of microbial peptidases. As the dye binds to albumin in the blood, accumulation of this dye in tissues has been used as a marker of vascular permeability. Although there was no accumulation of the dye at sites of saline injection, dye accumulation was elevated at sites of peptidase injection. These effects were observed with enzymes isolated from Candida albicans 45 , Pseudomonas aeruginosa , Aspergillus meleus and so on 46 . There have also been several studies that show higher accumulation of nanocarriers at sites of infection compared with non-infected sites (reviewed in ref. 47 ). In these studies, following a unilateral bacterial infection in the calf muscle of rats, the animals were treated intravenously with radiolabelled liposomes. In one study 48 , there was an ~40-fold greater liposome concentration (per cent injected dose per gram of tissue) in the abscess compared with the muscle. The change in vascular permeability and drug targeting depended, in part, on the infectious agent.
Enhanced circulation time can lead to more passes across the target and non-target tissues. Modelling studies suggest that if permeability into the target tissue is greater, longer circulation times can improve targeting efficiency 49 . Longer circulation times can be achieved by reducing the uptake of nanocarriers by macrophages. Hence, strategies orthogonal to those used to target drugs to macrophages are required. One approach involves modifying the surface of the nanocarriers with hydrophilic polymers such as PEG, which is thought to provide steric hindrance to opsonization 50 . The effect of circulation half-life on infection site accumulation was studied in a rat model of unilateral lung Klebsiella pneumoniae infection 51 , 52 . Gallium-67-labelled liposomes with different half-lives were administered to infected rats. Liposomes, regardless of their circulation half-life showed higher accumulation in the infected lung compared with the uninfected lung, validating the enhanced permeability discussed above. Furthermore, the accumulation of liposomes was proportional to the intensity of infection. Importantly, in rats that had the most severe lung infection, PEG-coated liposomes (half-life = 27 h) resulted in approximately fourfold higher concentrations compared with uncoated liposomes (half-life = 19 h) 51 . The benefit of PEG coating was reduced at low infection severity.
Ligand-mediated (active) and trigger-mediated targeted drug delivery
The expression of certain targets/receptors in diseased areas may be elevated. Conjugating ligands that bind these targets to the surface of nanocarriers can increase their accumulation in diseased sites (Fig. 3c ). Ligand-conjugated nanocarriers can also increase their cell uptake, aiding in the treatment of intracellular pathogens.
The mannose receptor is expressed on tissue-resident macrophages and dendritic cells, and serves to clear pathogens expressing mannosylated glycoproteins 53 . Nanocarriers functionalized with sugar moieties can highjack this uptake mechanism and enhance uptake into tissue-resident macrophages. Ciprofloxacin-loaded mannose-functionalized liposomes were administered as a pulmonary spray to rats, and drug exposure in alveolar macrophages was studied 54 . Mannose-functionalized liposomes showed a 1.5-fold higher exposure in alveolar macrophages compared with unmodified liposomes. In comparison with the free drug, the targeted liposomes showed a 23-fold higher bioavailability in the alveolar macrophages. However, these studies were carried out in an uninfected model, and therapeutic efficacy was not studied.
Nanocarriers targeting epitopes displayed on the surface of the pathogen can improve drug delivery into infection sites. Arias et al. encapsulated pentamidine in PLGA nanoparticles for treating Trypanosoma brucei , the causative agent of African sleeping disease 55 . The nanoparticles were functionalized with a fragment of an antibody called a nanobody. These fragments were 7.5% of the mass of the original antibody, which enabled them to penetrate the glycoprotein coat on the microbe and access its epitope. Nanoparticle-based treatment enabled a tenfold dose reduction in vivo.
In vivo phage display screening has identified cyclic peptides that bind the surface of S. aureus 56 . The distribution of vancomycin-loaded nanoparticles functionalized with this fluorescein-tagged peptide was measured by confocal microscopy and indicated preferential accumulation in S. aureus -infected lungs. In a 20 day efficacy study, 100% of the mice administered the peptide-functionalized nanoparticles survived, compared with roughly 40% survival in the non-functionalized-nanoparticle and free-vancomycin cohorts. These data suggest that targeted nanoparticles enable greater therapeutic efficacy and may allow reduction of antibiotic doses.
The use of antibody-based targeting has also been employed in hyperthermia-based treatment of S. aureus infection. Kim et al. 57 decorated streptavidin-coated magnetic nanoparticles with biotinylated anti-protein A monoclonal antibodies. Binding of the antibody-functionalized nanoparticles to cultures of S. aureus was quantified by flow cytometry and demonstrated a 2.5-fold increase in binding compared with the IgG-coated nanoparticles. Local injection of anti-protein A-functionalized nanoparticles was observed to be nearly twice as effective at killing S. aureus in murine cutaneous infection models compared with the control 57 . Similar antibody-targeted nanoparticles have been described in the literature 58 , 59 but did not include determination of in vivo efficacy. Considerable inquiry is being conducted on similar ligand-based targeting in the context of drug delivery to cancers and specific tissues, but further development of this technology in the space of ID is required.
Drug molecules released while nanocarriers are in circulation are subject to non-selective distribution. Hence, there is value in developing systems that are binary, whereby no drug is released in systemic circulation and then all of the content is released upon encountering the pathogen. Nanocarriers that disintegrate rapidly in the presence of bacterial phosphatase 60 (made from diblock copolymers of poly(phosphoesters) and PEG) and lipase 61 (made from triblock copolymers of poly(phosphoester), poly(caprolactone) and PEG) have been used for pathogen-triggered release of vancomycin. In a zebrafish model of methicillin-resistant S. aureus infection, the phosphatase-sensitive nanoparticles enhanced survival by 20 percentage points over free-drug controls. Stimuli-sensitive nanoparticles have also been used for treatment of sepsis, where the goal was to reduce bacterial load and inflammation in the lungs 62 . Nanoparticles were made with a cationic polyester (poly(β-amino ester)) to enable their degradation in the presence of bacterial enzymes and reduced pH, commonly found at sites of infection. The nanoparticles were surface functionalized with an antibody that binds intercellular adhesion molecule-1, a protein upregulated on the surface of endothelial cells of infected tissues. The nanoparticles were dually loaded with an antibiotic and an anti-inflammatory agent. In a mouse model of pulmonary P. aeruginosa infection, treatment with targeted nanoparticles improved survival over the free-drug control by nearly 50 percentage points. A common limitation of these studies is that comparison of the stimuli-sensitive nanoparticles with non-degradable nanoparticles was not performed. Polymers such as poly(β-amino esters) also degrade rapidly even in the absence of enzymes, further underscoring the need for including the non-degradable nanoparticle controls.
Although nanoencapsulation may enhance drug accumulation at sites of infection, there are certain limitations when translating from rodent models to humans. It is not clear if enhanced permeability is limited to rodent models or if it is observed in humans as well. Furthermore, some studies show that the degree of targeting is directly correlated with the severity of infection 51 , 52 . Hence, whether targeting can be achieved to treat low titres of persistent infection is unclear. It may also be interesting to compare the numbers of bacteria/viruses used in disease models in rats to those found in humans. If the numbers used in rodents are inordinately high, the data obtained thus far may need reinterpretation. Although drug accumulation at sites of infection is increased in rodent models, a substantial fraction of the drug is at off-target locations. Drugs with toxicity at these non-target sites are not good candidates for nanoencapsulation. It must also be determined whether the increase in on-target accumulation is high enough to justify reformulation. Metrics such as the drug targeting index (which compares the ratio of nanocarrier bioavailability at sites of efficacy and toxicity to those of the drug) 49 may prove valuable in this regard. For triggered-release systems, some drug leakage is generally observed even in the absence of triggers. The specificity of release is unclear as even host enzymes may act as triggers. Moreover, due to the non-specific biodistribution of nanocarriers that typically precedes triggering, drug release at non-target sites cannot be avoided. Most importantly, it must be noted that the intravenous route of administration may not be practical in resource-limited settings or in mass drug administration campaigns. Hence, it will be critical to understand whether these targeting strategies are applicable if the nanocarriers are administered via clinically relevant routes of administration.
Overcoming resistance to anti-infectious agents
Pathogen resistance is a critical and ever-growing obstacle to treatment of IDs. Resistance results from the secretion of drug-inactivating enzymes and/or the pathogens switching to a state of reduced metabolic activity, forming biofilms, adopting an intracellular life cycle or being obligate intracellular pathogens 3 .
Poor drug diffusion across the host cell membrane and its active efflux via drug transporters can reduce activity against intracellular pathogens 63 . Another complexity is that both drug molecules and pathogens are unevenly distributed within host cells, and in the absence of colocalization, efficacy is compromised (Fig. 4a ). For example, aminoglycosides largely accumulate in the lysosomes and are likely to be ineffective against cytosolic pathogens 64 . Hence, strategies that enhance intracellular accumulation of drugs and targeting to subcellular locations may improve efficacy.
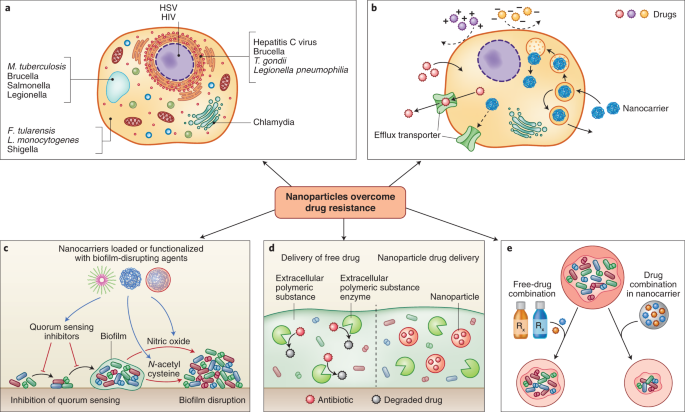
Nanocarrier systems can be designed to evade physical and chemical mechanisms for microbial drug resistance. a , Pathogens can escape the action of anti-infectives by residing in intracellular foci that are poorly accessible to drugs 147 . b , Highly polar drugs can have low uptake into cells, and drug internalized in the cell can subsequently be removed by efflux transporters. Both factors can protect intracellular pathogens. Nanocarriers have been used to improve cellular uptake of drugs and improve their activity. c , Nanocarriers can be loaded or surface functionalized with moieties that disrupt quorum sensing or the biofilm matrix. d , Nanocarriers can protect drug-degrading enzymes present in the biofilm. e , Co-encapsulation of drug combinations in nanocarriers has been shown to produce improved efficacy over co-administration of the free-drug combination.
Encapsulation in nanocarriers offers opportunities to increase cell uptake of anti-infectives and alter their intracellular disposition (Fig. 4b ). Lutwyche et al. attempted to improve the delivery of gentamicin to cytoplasmic strains such as Salmonella typhirium and Listeria monocytogenes 65 . Gentamycin was encapsulated in fusogenic liposomes that are stable under neutral conditions but disintegrate under the acidic conditions of the endolysosomes. Upon disintegration, the lipids fuse with the endosomal membrane, destabilizing it and releasing the drug in the cytoplasm. To validate the acid-mediated breakdown of the liposomes, the authors mixed two liposomal formulations—one formulation containing a fluorescence resonance energy transfer pair, and one formulation without. Owing to the proximity of the fluorescent molecules, this mixture had low baseline fluorescence. However, upon acidification of the mixture, the liposomes disintegrated and the lipids mixed, resulting in dequenching of the fluorescence. In an in vitro model of recombinant S. typhirium infection, gentamicin-loaded pH-sensitive liposomes showed threefold higher activity than the free drug. Despite higher cell uptake of pH-insensitive liposomes, they showed inferior activity to the pH-sensitive formulation. To understand the mechanism, authors loaded pH-sensitive and insensitive liposomes with a marker that fluoresced upon release and metabolism by cellular enzymes. Confocal microscopy showed that only pH-sensitive liposomes released the marker intracellularly in murine macrophages. Marker release was abrogated by bafilomycin, an inhibitor of endosome acidification. This study showed excellent mechanistic details of the role of each formulation component, and the need for intracellular targeting.
Clemens et al. developed mesoporous silica nanoparticles loaded with isoniazid for treating intracellular M. tuberculosis 66 . The pores of the nanoparticles were plugged with β-cyclodextrin, which prevents the drug from leaking out at a neutral pH. However, at an acidic pH, the plug is lost, resulting in drug release. Sémiramoth and colleagues conjugated penicillin G to hydrophobic squalene using a pH-sensitive ester bond 67 . Nanoparticles formed from this conjugate were rapidly taken up by cells and released drug in the endolysosomes. This resulted in improved activity over the free drug.
Reverse engineering of bacterial cell uptake mechanisms on the surface of nanocarriers has been used to improve intracellular delivery. Invasin, a Yersinia pseudotuberculosis protein, binds integrins on host cells, causes reorganization of its cytoskeleton and enables bacterial uptake into membrane enclosed structures. Labouta and colleagues 68 reverse engineered this mechanism on the surface of liposomes by conjugating them to the C-terminal fragment of the invasin—InvA497. In vitro confocal microscopy studies in Hep-2 cells showed that InvA497-functionalized liposomes had increased cell binding (>30-fold) compared with albumin-functionalized liposomes, and this was abrogated in competition experiments. InvA497-funtionalized gentamycin-loaded liposomes caused a 30% reduction in bacterial load in an in vitro model of Y. pseudotuberculosis infection, whereas albumin-functionalized liposomes had no effect 68 . This group has shown that surface functionalization with InvA497 is beneficial for enhancing both in vitro cell uptake and antimicrobial efficacy of polymeric nanoparticles 69 . This is a highly innovative concept that may benefit from in vivo evaluation. Furthermore, in the event that these systems are applied orally, it may be important to understand the stability of the peptides in the enzyme-rich gastrointestinal environment. If the nanocarriers are administered systemically, it will be important to understand immune response to these systems as they are functionalized with peptides found in bacteria, and its effect on their pharmacokinetics and tolerability.
Intracellular delivery is of particular importance with molecules such as messenger RNA, which have gained much attention due their application in vaccination. Most polymer-based nucleic acid and protein delivery systems are inspired by initial work done by Langer and Folkman 70 , which showed that macromolecules such as proteins could be encapsulated in small polymer-based carriers—a system deemed unlikely to work due to the use of organic solvents during synthesis and the perceived imperviousness of polymers to macromolecules 71 . Following this initial proof-of-concept with polymers, encapsulation of RNA in lipid carriers 72 , and their utility in vaccination 73 , was demonstrated. A key feature of nanocarriers is that they aid in the transport of mRNA across the cell membrane, enable endolysosomal escape and release the nucleic acid in the cytoplasm 74 . In one example 75 , self-replicating mRNA encoding an antigen protein was complexed with a cationic dendrimer-based polymer. Animals treated with mRNA nanoparticles produced antibody-based and CD8 + T-cell immune responses, and were completely protected from lethal challenges of Ebola virus, H1N1 influenza and Toxoplasma gondii . mRNA vaccines based on lipid nanocarriers have also been tested in the clinic by companies such as Moderna, Acuitas Therapeutics, Pfizer, BioNTech, GlaxoSmithKline, Sanofi, Translate Bio and others. Initial results with lipid-nanoparticle-formulated mRNA vaccines against H10N8 and H7N9 influenza viruses and SARS-CoV-2 indicate that these vaccines are well tolerated and that they produce a robust humoral response 76 , 77 . Specifically, mRNA-based vaccines have shown >90% efficacy against SARS-CoV-2. The stability of mRNA-based vaccines during long-term storage and transport could also be major obstacles to their application in resource-limited settings. Peptide-based antigen assembled into nanoparticles have also been developed for vaccination against SARS-CoV-2 78 .
As bacteria and nanocarriers can accumulate in intracellular vesicles, nanoencapsulation has been used for intracellular targeting of antibiotics. Using confocal and transmission electron microscopy, Couvreur and colleagues showed that poly(cyanoacrylate) nanoparticles colocalized with S. typhirium in phagosomes of macrophages, resulting in enhanced intracellular targeting of ampicillin 79 . In another study, Toti et al. found that PLGA nanoparticles loaded with a fluorescent dye accumulated with chlamydial inclusion bodies in infected human lung epithelial cells 80 . In vitro studies revealed that antibiotics (azithromycin and rifampicin) were effective if applied immediately after infection, but not if applied after 24 or 48 h. However, encapsulation in nanoparticles partly restored their activities even when applied at 24 and 48 h after infection.
Nanoparticles can prevent, disrupt and inhibit or disperse bacteria from biofilm infections 30 (Fig. 4c ). By forming a diffusion barrier, the extracellular polymeric matrix in the biofilm protects bacteria from high antibiotic concentrations, often leading to chronic infections. Teirlinck et al. showed that laser irradiation of gold nanoparticles resulted in the formation of nanobubbles that disrupted this diffusional barrier 81 . Local disruption of the biofilms increased the permeability and therefore susceptibility to the antibiotic tobramycin. Other examples of nanocarrier-mediated biofilm disruption are discussed earlier in this Review in the context of pulmonary drug delivery.
The evolution of antibiotic-degrading enzymes is a major mechanism causing bacterial resistance. These enzymes, such as β-lactamases and aminoglycoside-modifying enzymes, are readily transferred through horizontal gene transfer and have been implicated in the pattern of increasing drug resistance 3 . Nanoparticles have been used to protect antibiotic agents from enzymatic degradation and resensitize resistant bacteria to their bactericidal effects (Fig. 4d ). Shaaban et al. developed poly(caprolactone) nanoparticles loaded with imipenem 82 and found that drug-loaded nanoparticles conferred 250- and 62.5-fold decreases in minimal inhibitory concentrations compared with free imipenem in in vitro cultures of imipenem-resistant K. pneumoniae and P. aeruginosa , respectively. Furthermore, to verify the ability of the nanocarriers to protect the drug from carbapenem-degrading enzymes, the imipenem formulations were tested with and without the addition of exogenous carbapenemase in Escherischia coli . The results demonstrated that while free imipenem had negligible effect when co-administered with the enzyme, the nanoparticles had comparable bactericidal effects regardless of whether carbapenemase had been co-administered. Liposomes have also been used for protecting drugs against bacterial enzymes 83 . When co-administered with exogenous β-lactamase, piperacillin-loaded liposomes provided twofold growth inhibition compared with the free drug co-administered with β-lactamase. The authors posited that the lactamases were either unable to penetrate the liposome or were sterically hindered by the liposomal surface, but were unable to confirm these hypotheses experimentally. Further testing in animal biofilm models and mechanistic confirmation must be done to reliably demonstrate the ability of nanoparticles to protect therapeutic payloads from enzymatic degradation.
Multiple antimicrobial agents can be packaged within the same nanoparticle to increase the likelihood of overcoming existing drug resistance mechanisms instead of using one drug alone (Fig. 4e ). Schiffelers and colleagues evaluated the efficacy of gentamicin and ceftazidime as single agents and in combination when formulated as free-drug solutions and as liposomes 84 . Testing was performed in a rat unilateral lung infection model with drug-sensitive and drug-resistant K. pneumoniae . In the drug-sensitive model, ten doses of the soluble drug combination treatment resulted in complete survival. Impressively, only a single dose of the liposomal combination produced the same effect. In the drug-resistant model, the dose of the drug combination was increased compared with that used in the drug-sensitive model—the dose of ceftazidime was 4-fold higher, and that of gentamicin was 66-fold higher. Ten treatments at these elevated doses produced complete survival in the drug-resistant model. However, only two treatments with substantially lower doses of the liposomal combination also produced 100% survival. This system is attractive for two reasons—it enabled dose reduction and the treatment of drug-resistant infection; and it dramatically reduced the number of administration events required to produce efficacy. This latter feature is of great value in resource-limited settings.
Advanced preclinical and clinical nanotechnologies
In this section, we highlight the most advanced nanotechnologies in major IDs—HIV infection, TB and malaria. The reader will glean that nanotechnology has been most actively studied in the clinic and in large animals for the treatment and prevention of HIV infection (Table 1 ). In contrast, for malaria and TB, nanotechnology has been pursued with less gusto. For these IDs, work has mainly been limited to preclinical trials in rodents. We also hope to convey that the most advanced nanotechnologies are non-complex, yet highly impactful. Hence, perhaps for nanotechnology to benefit the patient, it needs to satisfy certain tenets such as simplicity in design, clinical need and financial enthusiasm.
HIV infection
HIV infection is one of the major IDs contributing to high rates of morbidity and mortality globally 85 . HIV primarily targets CD4 + T cells (Fig. 5 ) and weakens the patient’s immune system making the individual susceptible to opportunistic infections. Although access to antiretroviral therapy has continued to improve for people living with the disease around the world—reducing morbidity, mortality and ongoing transmission—more than 900,000 people continue to die of acquired immunodeficiency syndrome each year. In 2017, there were 1.8 million people who became newly infected with HIV. Moreover, new HIV infections are increasing at an alarming rate in some populations, including people of colour, men who have sex with men and younger women, as well as in certain geographic regions including Russia and other eastern European countries 86 , 87 .
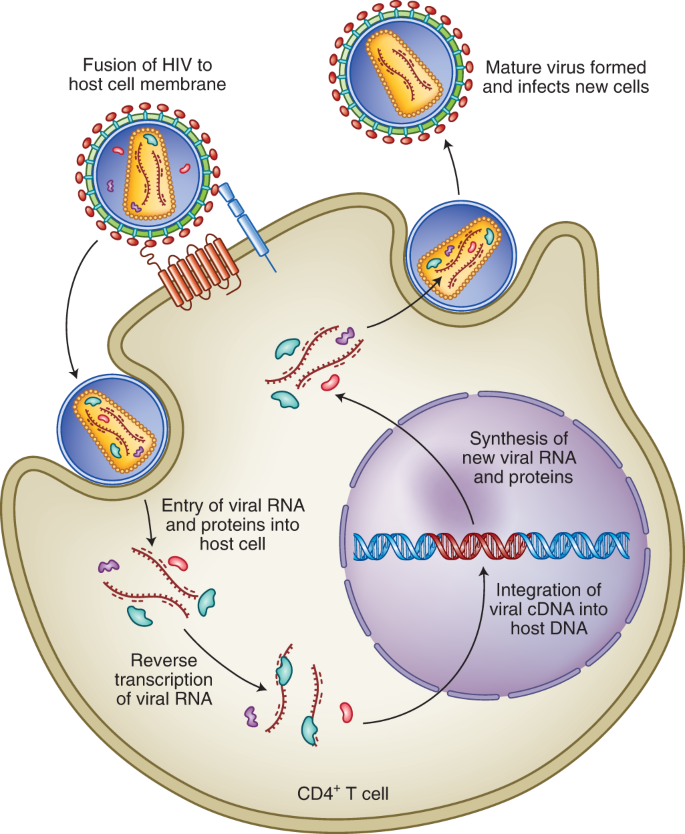
Fusion of the viral membrane with the host cell membrane enables entry of viral contents into the host cells. Viral RNA is reverse transcribed in complementary DNA (cDNA), which can be integrated into the host genome. This is followed by the formation of new copies of the viral RNA and proteins, which assemble into new viral particles that can infect new cells. Antiretrovirals work by inhibiting one or more steps of this pathway 10 .
Long-acting injectable antiretrovirals are the most clinically advanced nanotechnology in HIV treatment. Long-acting injectable nanoparticles reduce dosing frequency, which stands to improve patient adherence. They also allow drugs to be administered to dysphagic patients, for example individuals infected with HIV and suffering from opportunistic oesophageal infections 88 . Moreover, there has been some interest in developing long-acting nanoparticles for prevention—including both pre-exposure prophylaxis (which requires long-term administration of two medications to achieve protection) and post-exposure prophylaxis (where administration of a single injection or long-acting oral medication would be especially valuable) 89 , 90 . Specifically, nanoparticles of two antiretrovirals, cabotegravir and rilpivirine, are being actively pursued in the clinic. Cabotegravir 16 and rilpivirine 17 were chosen owing to their efficacy at low oral doses and low water solubility.
The pharmacokinetics, safety and efficacy of rilpivirine and cabotegravir nanosuspensions (surfactant-stabilized drug nanocrystals with a mean diameter of 200 nm) have been evaluated in clinical studies. Both nanosuspensions were safe and devoid of major adverse effects 88 . Estimated half-lives of rilpivirine and cabotegravir in nanosuspensions were 44–61 days (ref. 91 ) and 25–54 days (ref. 91 ), respectively, enabling administration every 1–3 months. In contrast, half-lives of rilpivirine and cabotegravir in tablets have been estimated to be ~1–2 days (refs. 91 , 92 ). In Phase IIb clinical trials, the combination of rilpivirine and cabotegravir nanosuspensions enabled disease suppression equivalent to oral tablets of cabotegravir and abcavir-lamivudine 93 .
Two other injectable nanoparticles have shown promising results in non-human primates. One of these formulations was developed for the prolonged systemic delivery of dolutegravir, which has relatively high water solubility 18 , 94 . A water-insoluble prodrug of dolutegravir was produced by esterification with myristic acid and formulated into nanoparticles. In rhesus macaques, prodrug nanoparticles maintained therapeutic concentrations of dolutegravir for ~40 days and extended the half-life of dolutegravir to ~19.5 days. It is believed that the nanoparticles are engulfed by macrophages, leading to intracellular activation of the prodrug, followed by release of the active moiety from the macrophages 18 , 94 . Lipid-based nanoparticles loaded with three to four antiretrovirals including lopinavir, ritonavir and tenofovir have also been studied in non-human primates 95 , 96 . Nanoencapsulation enhanced circulation times of the antiretrovirals and increased their uptake into the mononuclear cells in peripheral blood and lymph nodes of macaques. This is especially notable for antiretrovirals, as HIV persists in macrophages for prolonged durations, even in patients receiving antiretroviral therapy.
Long-acting injectable antiretrovirals are widely considered breakthrough interventions; however, certain challenges remain. Despite most advanced nanosuspensions containing >90% drug, clinical use of this technology has been limited to drugs that have a daily oral dose of <25–50 mg. Patients treated with injectable nanosuspensions are exposed to the drug for at least a month, and there is no mechanism to withdraw exposure in cases of adverse reactions. This problem has been addressed in clinical trials by providing patients with a lead-in period of oral drug therapy before the injection. An alternative approach was recently tested in mice and involves injecting a solution of the drug and polymer in an organic solvent subcutaneously 97 . A surgically removable drug depot (~0.75 cm diameter in mice) forms as the organic solvent diffuses out of the injection site. Finally, in most clinical trials, the nanosuspensions have been administered by medical practitioners, and it is likely that patients will need to visit their caregiver for each injection. It remains to be seen whether the need for chronic visits to a clinic deters patient adherence. The ability to self-medicate at home with infrequently administered oral dosage forms may help address this issue. For this, we have developed an orally administered gastric retentive dosage form that can reduce dosing frequency from daily to weekly 98 . However, personal preferences may be highly varied and providing multiple options to the patient may aid in maximizing patient adherence.
Oral antiretroviral nanoparticles have been developed for paediatric use with the intent to replace tablets (which are difficult to swallow) and alcohol-containing solutions (which contain harmful excipients). These nanoparticles are produced by a novel emulsion template freeze-drying approach and have been loaded with lopinavir and efavirenz 99 . In rats, the optimized nanoparticle formulation was bioequivalent to lopinavir dissolved in alcohol and propylene glycol 99 . The authors believe that the nanoparticles are probably absorbed via the lymphatic vessels in the gastrointestinal tract. However, further work may be required to validate this observation. Efavirenz and lopinavir nanoparticles made using this strategy are currently awaiting clinical testing (NCT02631473).
Malaria, caused by the single-celled complex protozoan of the Plasmodium genus (Fig. 6 ), is the most prevalent parasitic disease in the world. In 2017, malaria cases rose for the second year in a row, reaching 219 million cases 100 . Around 90% of all malaria cases and deaths are in Africa, where the disease costs the continent’s economy US$12 billion yr −1 in direct losses and reduces GDP growth by 1.3% (ref. 101 ). Despite over 3 billion people being at risk of infection, funding for the global malaria response has plateaued since 2010, reaching only US$3.1 billion in 2017, which is less than half of the US$6.6 billion yr −1 funding target for 2020 100 . As part of the investment, both short-term and long-term solutions to malaria must be considered, such as using nanotechnology for treatment and vaccination. A vital consideration for development of new approaches will be their cost-effectiveness, given the target product profiles of US$1 day −1 for treatment of malaria.
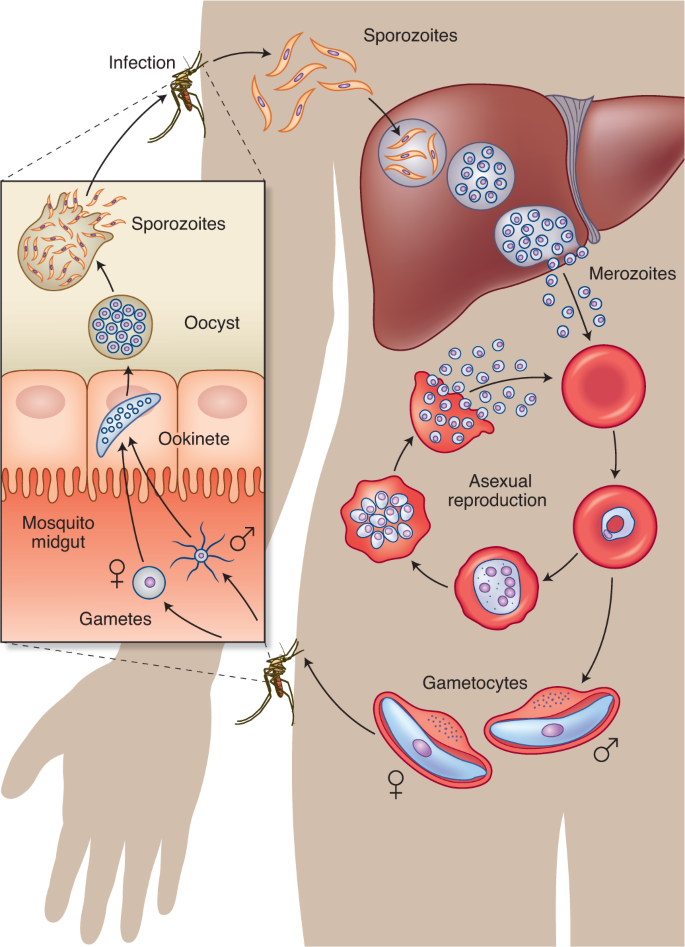
The infection is spread to a human through the bite of a female Anopheles mosquito, which injects the parasite Plasmodium in the form of sporozoites that migrate through the skin into the circulation. Within minutes, the sporozoites reach the liver, where they invade hepatocytes and proliferate to form merozoites. Each infected hepatocyte releases thousands of merozoites into the bloodstream, beginning a cycle of invasion of RBCs. Merozoites replicate asexually through ring, trophozoite and schizont stages within RBC vacuoles to form new daughter cell merozoites that egress and infect other RBCs. Meanwhile, a small fraction of merozoites sexually differentiate into male and female gametocytes, which are ingested into the midgut of a mosquito from peripheral blood when it bites an infected human. The gametocytes further develop into gametes, zygotes, ookinetes, oocysts and finally sporozoites, which can resume the cycle of human infection when they migrate to the mosquito’s salivary glands and are injected into another human during a blood meal 148 .
Malaria is mainly treated using chemotherapy, with artemisinin-based combination therapy being integral to curing the disease 102 . Unfortunately, due to the low uptake of artemisinin in parasite-infected red blood cells (RBCs), these drugs provide only symptomatic relief at low doses 103 . Killing parasites in the RBCs requires prolonged treatment with high doses, which results in toxicities, potential for missed doses and development of resistance 104 . Given that blood stage parasites are the main therapeutic target, researchers are developing various approaches to target antimalarials to infected RBCs 105 , 106 , 107 , thereby maximizing their therapeutic window. Covalent attachment of heparin and monoclonal antibodies to liposomes has been proven to selectively target infected RBCs in vitro 105 , 108 . These functionalized liposomes preferentially adhere to surface proteins decorating infected RBCs and reduce the overall parasitaemia. Further studies of parasite resistance and in vivo testing of these systems will be needed, while potentially prioritizing heparin–liposome formulations due to their cheaper cost. Although liposomal drug delivery is an established platform with proven biocompatibility, the oral route of administration is typically not used due to instability in the gastrointestinal tract 109 . Therefore, researchers must continue to develop other targeting strategies to maximize the benefit to those living in malaria-endemic regions, where oral administration is preferred. These can include designing nanocarriers to selectively target other Plasmodium stages, such as at the parasite stage in the liver or during the transmissible sexual stages.
Nanotechnology can also improve the oral bioavailability of poorly water-soluble antimalarials, such artemether and tafenoquine. NanoAbsorb is a solid microemulsion pre-concentrate formulation that can safely deliver the widely used drug artemether with higher antimalarial efficacy in vivo than the marketed formulation Larither 110 . Another example is the use of self-microemulsifying drug delivery systems, which can be filled in hard gelatine capsules and rapidly transform into microemulsions in gastrointestinal fluids 111 . Long-chain-triglyceride-based self-microemulsifying systems containing β-artemether were demonstrated to be safe and have a notable improvement in the antimalarial activity in vivo compared with Larither. An oral lipid-based nano-emulsion of tafenoquine with sizes <20 nm enhanced drug solubility, bioavailability and efficacy in vivo while also reducing the toxicity 112 . These approaches, using oily vehicles to enhance solubilization in the gastrointestinal tract, improve drug bioavailability and patient adherence.
Nanoformulation of multiple combined drugs is an efficient method for designing innovative therapeutics and can prevent resistance of the malaria parasite. Polymer–drug conjugates were synthesized into nanoparticles with substituted poly(phosphazenes) and the combination of primaquine and dihydroartemisinin. The combination therapy exhibited promising antimalarial efficacy in vivo against a resistant parasite strain 113 . Artemether and lumefantrine are well accepted as a combination therapy for treating uncomplicated malaria, yet the current marketed formulation degrades in the gastrointestinal tract, leading to unpredictable pharmacokinetics. Using co-loaded nanostructured lipid carriers with these drugs, researchers showed that the systems resulted in greater clearance of parasites from infected mice and had a clear benefit over individual drug-loaded carriers in terms of survival period and parasitaemia progression 114 .
There is only one malaria vaccine candidate that has received a positive regulatory assessment: RTS,S/AS01 (RTS,S) (Mosquirix), which is an injectable vaccine that provides partial protection against malaria in young children 115 . The antigen is formulated as virus-like particle and combined with a liposomal adjuvant system. In July 2015, European regulators approved the use of the vaccine, although it is at most 50% effective 115 . Vaccine development is an active area of research, and a recent study used spontaneous nanoliposome antigen particleization to show efficacy of a multistage, multi-antigen vaccine in mice and rabbits 116 . Further research and development with affordable large-scale production is necessary for a successful malaria vaccine, and nanotechnology could play a critical role in providing new approaches to protect all ages.
TB, which claims the lives of over 3,500 people every day, is the world’s leading killer among IDs 117 . According to the World Health Organization (WHO), 10 million people developed TB in 2017 with a global economic burden amounting to US$12 billion annually 117 , 118 . The burden of TB is concentrated in Asia and Africa—only 6% of global cases were in the WHO European Region and WHO Region of the Americas 117 . Furthermore, M. tuberculosis (Fig. 7 ) is the most critical pathogen in the global antimicrobial resistance crisis 119 . Unless radical action is taken, drug-resistant strains of M. tuberculosis will account for 25% of antimicrobial resistance-related deaths and cost the global economy US$16.7 trillion by the year 2050 119 .
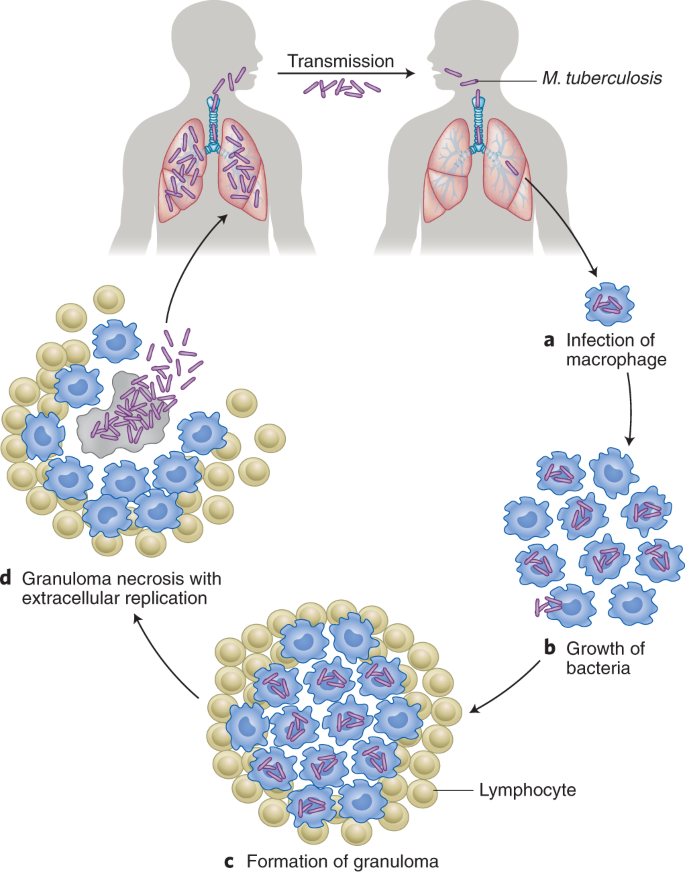
TB is caused by the inhalation of droplet nuclei containing M. tuberculosis that enter the respiratory tract and infect the alveoli of the lungs. During the first stage, alveolar macrophages recognize, engulf and attempt to destroy the bacilli. The second stage of symbiosis occurs when the bacilli grow logarithmically within infected alveolar macrophages that are unable to stop the bacilli from growing. Eventually, the infected areas transform into a granuloma, a hallmark of TB characterized by a wall of lymphocytes and severe chest pain. This third stage of the disease results in a solid caseous centre, where the bacteria can survive for years. Most humans infected with TB do not exhibit progression of the disease, as it remains in a latent state. It is estimated that 1.7 billion people, or 23% of the world’s population, have latent TB. However, some infected individuals progress to the fourth and final stage of the disease, where the caseous centre liquefies and cavitates to fill the lungs with free-floating bacteria and spread in the lungs, causing pulmonary TB. These bacteria can disseminate to more distant tissues and organs via the lymphatic system or bloodstream to result in extrapulmonary TB. The cycle is complete when an infected TB individual coughs, sneezes, speaks or sings to release highly transmissible infectious droplet nuclei into the air for a susceptible individual to inhale 149 .
WHO guidelines recommend treating drug-susceptible TB for at least 6 months with an oral drug regimen of four antibiotics taken daily 11 . Because of the prolonged and frequent dosing, and side effects, patients find it difficult to adhere to these regimens and are at risk of developing drug-resistant strains 4 . Several of the current antibiotics are poorly soluble, unstable in gastric acid and unable to penetrate the alveolar macrophages where the bacilli reside 120 . Given that bedaquiline was the first new drug approved in more than 40 years (ref. 121 ) and the dearth of others in the pipeline, reformulating existing drugs in nanocarriers is an attractive way of having an immediate impact on the treatment of TB.
Several oral nanotechnology-based systems have been designed with existing TB drugs and tested in animal models. Solid lipid nanoparticles have been tested in rodents with the goal of improving the bioavailability of rifampicin and the combination of rifampicin, isoniazid and pyrazinamide 122 , 123 . In the lungs and spleen of infected mice, no tubercle bacilli could be detected after five oral doses of drug-loaded solid lipid nanoparticles administered every tenth day, whereas 46 daily doses of oral free drugs were required to achieve the same therapeutic benefit 123 . Polymer-based nanoparticles (PLGA and sodium alginate) have also been developed to improve the oral bioavailability of rifampicin, isoniazid, ethambutol, pyrazinamide and streptomycin 124 , 125 , 126 , 127 .
Inhalable nanocarriers of TB drugs have been a major focus of research given that pulmonary TB is the most common TB form 128 . Researchers have developed nebulized solid lipid particles 129 and sodium alginate nanoparticles 130 incorporating rifampicin, isoniazid and pyrazinamide to improve targeted bioavailability in the lungs and reduce hepatotoxicity. Interestingly, pulmonary administration of rifampicin-loaded solid lipid nanoparticles in rats enabled delivery to alveolar macrophages 131 . These examples highlight the promise of pulmonary administration of nanocarriers, although they must be designed for delivery with portable and easy-to-operate cost-effective devices such as inhalers.
There is no effective vaccine that comprehensively protects against TB infection. Bacillus Calmette–Guerin (BCG), which was introduced in 1921, is the sole approved TB vaccine, but it offers only limited protection 132 . Therefore, researchers are attempting to improve the existing BCG vaccine and derive new vaccines to prevent TB 133 . When guinea pigs were administered an aerosolized formulation of BCG nanomicroparticles, they generated better immune protection and enhanced resistance to TB infection than when they were immunized with the standard parenteral BCG formulation 134 . The nanomicroparticles are a single-particle form with two nanometre-scale axes and a third of micrometre dimension. These dimensions facilitate better aerosolization compared with particles with similar dimensions in all axes. Pulmonary administration of the common TB antigen 85B conjugated to poly(propylene sulphide) nanoparticles demonstrated improved protection in mice after they were challenged with aerosolized M. tuberculosis (ref. 135 ). Researchers are also exploring chitosan, a biodegradable and biocompatible polycationic polymer, for its ability to bind and protect DNA from nuclease degradation and for its mucoadhesive properties. A nano-chitosan-based recombinant DNA vaccine elevated the immunologic and protective effects against TB in a mouse model 136 , and chitosan has a role as a vaccine adjuvant in the prevention of TB 137 .
It is also recommended that people who are infected with TB but do not have active disease (latent TB) receive treatment to eradicate the mycobacteria and prevent progress to active disease 138 . These preventative regimens can last for 9 months and require multidrug therapy. Given that most people with TB infection do not have symptoms, it can be challenging for them to complete their regimens 139 . Nanotechnology can potentially simplify dosing and minimize adverse events in this population, thereby greatly contributing to the elimination of TB.
Challenges and outlook
The examples above highlight many promising therapeutic strategies for developing nanomedicines to treat and prevent IDs. However, to have the maximum impact, nanotechnology solutions will need to overcome several financial, manufacturing and regulatory challenges. First, reducing the cost will be the major hurdle in developing new drugs and nanotechnology-based systems. For global health solutions, development of new drugs is expensive. Reformulating existing drugs in nanocarriers may help to achieve similar targets of enhanced efficacy and safety at a considerably lower cost. However, one currently approved nano-system (an amphotericin B liposomal formulation) is not cost-effective in developing countries 140 . Excipients used in nanosystems, such as lyoprotectants, can increase the cost of many treatments. Second, the synthesis and storage conditions of some nanoparticles may not be conducive to conditions in low-resource countries 141 . Researchers will need to enable the reproducibility and bulk production while considering the environmental effects of these nanosystems. Nanomedicines are likely to be three-dimensional constructs of multiple components with preferred spatial arrangements. Subtle changes in process or composition can adversely affect the complex composition of nanomedicines 142 , 143 . Third, regional and national differences within and between regulatory authorities, especially when running multi-centric international trials, will be a challenge during the clinical trial stage 144 . Finally, patient acceptability of these nanosystems will need to be addressed. In 2006, the International Centre for Technology Assessment and other consumer groups filed a legal petition against the FDA for perceived lack of initiative in regulating nanomaterial-containing products under their jurisdiction. The FDA created a Nano Task Force in response 145 . To achieve successful translation of nanomedicines, a network of stakeholders will need to converge. This includes academics, investors from industry and the government, and contract research and manufacturing organizations. Nanotechnology is a multi-disciplinary field that also requires intellectual property and commercialization strategies to grow 146 . Nonetheless, the rapid translation of nanotechnologies from the bench to the people, and the impact that these systems have had on the SARS-CoV-2 pandemic provide great promise. This example suggests that similar feats may be possible in treating other infectious diseases as well.
IDs are a major driver of morbidity and mortality globally, and their impact on low SDI countries is particularly grave. Simplifying the use of medicines and making drugs safer and more efficacious can improve patients’ quality of life and reduce disease burden. In this Review, we highlighted how nanotechnology-based approaches can enable oral drug administration, infrequent drug administration and drug targeting to sites of infection, ultimately improving treatment efficacy. Finally, we discussed examples of nanotechnologies that are in clinical trials for the treatment of HIV and in preclinical development for the treatment of malaria and TB. Innovative and cost-effective nanotechnologies that take into consideration the challenges that are encountered low SDI countries are most likely to benefit patients.
Roth, G. A. et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the global burden of disease Study 2017. Lancet 392 , 1736–1788 (2018).
Article Google Scholar
Kinch, M. S., Patridge, E., Plummer, M. & Hoyer, D. An analysis of FDA-approved drugs for infectious disease: antibacterial agents. Drug Discov. Today 19 , 1283–1287 (2014).
Article CAS Google Scholar
Munita, J. M. & Arias, C. A. Mechanisms of antibiotic resistance. Microbiol. Spectrum 4 , VMBF-0016-2015. (2016).
Sabaté, E. Adherence to Long-Term Therapy: Evidence for Action (WHO, 2003).
Pheage, T. Dying from Lack of Medicines (United Nations, 2016); https://www.un.org/africarenewal/magazine/december-2016-march-2017/dying-lack-medicines
Anselmo, A. C. & Mitragotri, S. Nanoparticles in the clinic. Bioeng. Transl. Med. 1 , 10–29 (2016).
Huh, A. J. & Kwon, Y. J. ‘Nanoantibiotics’: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 156 , 128–145 (2011).
Irvine, D. J., Hanson, M. C., Rakhra, K. & Tokatlian, T. Synthetic nanoparticles for vaccines and immunotherapy. Chem. Rev. 115 , 11109–11146 (2015).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 17 , 261–279 (2018).
Kirtane, A. R., Langer, R. & Traverso, G. Past, present, and future drug delivery systems for antiretrovirals. J. Pharm. Sci. 105 , 3471–3482 (2016).
Treatment of Tuberculosis: Guidelines 4th edn (WHO, 2010); https://www.who.int/tb/publications/2010/9789241547833/en/
Langer, R. Drug delivery and targeting. Nature 392 , 5–10 (1998).
CAS Google Scholar
Makadia, H. K. & Siegel, S. J. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers 3 , 1377–1397 (2011).
Fielding, R. M., Lewis, R. O. & Moon-McDermott, L. Altered tissue distribution and elimination of amikacin encapsulated in unilamellar, low-clearance liposomes (MiKasome). Pharm. Res. 15 , 1775–1781 (1998).
Schiffelers, R., Storm, G. & Bakker-Woudenberg, I. Liposome-encapsulated aminoglycosides in pre-clinical and clinical studies. J. Antimicrob. Chemother. 48 , 333–344 (2001).
Trezza, C., Ford, S. L., Spreen, W., Pan, R. & Piscitelli, S. Formulation and pharmacology of long-acting cabotegravir. Curr. Opin. HIV AIDS 10 , 239–245 (2015).
Williams, P. E., Crauwels, H. M. & Basstanie, E. D. Formulation and pharmacology of long-acting rilpivirine. Curr. Opin. HIV AIDS 10 , 233–238 (2015).
Mcmillan, J. et al. Pharmacokinetics of a long-acting nanoformulated dolutegravir prodrug in rhesus macaques. Antimicrob. Agents Chemother. 62 , e01316-17 (2017).
Wistrand-Yuen, E. et al. Evolution of high-level resistance during low-level antibiotic exposure. Nat. Commun. 9 , 1599 (2018).
Landovitz, R. J., Kofron, R. & McCauley, M. The promise and pitfalls of long-acting injectable agents for HIV prevention. Curr. Opin. HIV AIDS 11 , 122–128 (2016).
das Neves, J., Nunes, R., Machado, A. & Sarmento, B. Polymer-based nanocarriers for vaginal drug delivery. Adv. Drug Deliv. Rev. 92 , 53–70 (2015).
Leal, J., Smyth, H. D. C. & Ghosh, D. Physicochemical properties of mucus and their impact on transmucosal drug delivery. Int. J. Pharm. 532 , 555–572 (2017).
Cu, Y., Booth, C. J. & Saltzman, W. M. In vivo distribution of surface-modified PLGA nanoparticles following intravaginal delivery. J. Control. Release 156 , 258–264 (2011).
Lai, S. K. et al. Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus. Proc. Natl Acad. Sci. USA 104 , 1482–1487 (2007).
Cunha-Reis, C. et al. Nanoparticles-in-film for the combined vaginal delivery of anti-HIV microbicide drugs. J. Control. Release 243 , 43–53 (2016).
Ensign, L. M. et al. Mucus-penetrating nanoparticles for vaginal drug delivery protect against herpes simplex virus. Sci. Transl. Med. 4 , 138ra79 (2012).
Palliser, D. et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 439 , 89–94 (2006).
Steinbach, J. M., Weller, C. E., Booth, C. J. & Saltzman, W. M. Polymer nanoparticles encapsulating siRNA for treatment of HSV-2 genital infection. J. Control. Release 162 , 102–110 (2012).
Roy, I. & Vij, N. Nanodelivery in airway diseases: challenges and therapeutic applications. Nanomedicine 6 , 237–244 (2010).
Han, C. et al. Recent developments in the use of nanoparticles for treatment of biofilms. Nanotechnol. Rev. 6 , 383–404 (2017).
Vaughn, J. M. et al. Single dose and multiple dose studies of itraconazole nanoparticles. Eur. J. Pharm. Biopharm. 63 , 95–102 (2006).
Alvarez, C. A. et al. Aerosolized nanostructured itraconazole as prophylaxis against invasive pulmonary aspergillosis. J. Infect. 55 , 68–74 (2007).
Wong, J. P. et al. Liposome delivery of ciprofloxacin against intracellular Francisella tularensis infection. J. Control. Release 92 , 265–273 (2003).
Nafee, N. et al. Antibiotic-free nanotherapeutics: ultra-small, mucus-penetrating solid lipid nanoparticles enhance the pulmonary delivery and anti-virulence efficacy of novel quorum sensing inhibitors. J. Control. Release 192 , 131–140 (2014).
Duong, H. T. T. et al. Nanoparticle (star polymer) delivery of nitric oxide effectively negates Pseudomonas aeruginosa biofilm formation. Biomacromolecules 15 , 2583–2589 (2014).
Suk, J. S. et al. N-acetylcysteine enhances cystic fibrosis sputum penetration and airway gene transfer by highly compacted DNA nanoparticles. Mol. Ther. 19 , 1981–1989 (2011).
Sen, C. K. et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 17 , 763–771 (2009).
James, G. A. et al. Biofilms in chronic wounds. Wound Repair Regen. 16 , 37–44 (2008).
Martinez, L. R. et al. Antimicrobial and healing efficacy of sustained release nitric oxide nanoparticles against Staphylococcus aureus skin infection. J. Invest. Dermatol. 129 , 2463–2469 (2009).
Mohiti-Asli, M., Pourdeyhimi, B. & Loboa, E. G. Skin tissue engineering for the infected wound site: biodegradable PLA nanofibers and a novel approach for silver ion release evaluated in a 3D coculture system of keratinocytes and Staphylococcus aureus . Tissue Eng. Pt C 20 , 790–797 (2014).
Sarathy, J. P. et al. Prediction of drug penetration in tuberculosis lesions. ACS Infect. Dis. 2 , 552–563 (2016).
Huth, M. E., Ricci, A. J. & Cheng, A. G. Mechanisms of aminoglycoside ototoxicity and targets of hair cell protection. Int. J. Otolaryngol. 2011 , 937861 (2011).
Tedijanto, C., Olesen, S. W., Grad, Y. H. & Lipsitch, M. Estimating the proportion of bystander selection for antibiotic resistance among potentially pathogenic bacterial flora. Proc. Natl Acad. Sci. USA 115 , E11988–E11995 (2018).
Löbenberg, R., Araujo, L., von Briesen, H., Rodgers, E. & Kreuter, J. Body distribution of azidothymidine bound to hexyl-cyanoacrylate nanoparticles after i.v. injection to rats. J. Control. Release 50 , 21–30 (1998).
Kaminishi, H., Tanaka, M., Cho, T., Maeda, H. & Hagihara, Y. Activation of the plasma kallikrein-kinin system by Candida albicans proteinase. Infect. Immun. 58 , 2139–2143 (1990).
Molla, A., Yamamoto, T., Akaike, T., Miyoshi, S. & Maeda, H. Activation of hageman factor and prekallikrein and generation of kinin by various microbial proteinases. J. Biol. Chem. 264 , 10589–10594 (1989).
Laverman et al. Liposomes for scintigraphic detection of infection and inflammation. Adv. Drug Deliv. Rev. 37 , 225–235 (1999).
Laverman, P. et al. Microscopic localization of PEG-liposomes in a rat model of focal infection. J. Control. Release 75 , 347–355 (2001).
Siegel, R. A., Kirtane, A. R. & Panyam, J. Assessing the benefits of drug delivery by nanocarriers: a partico/pharmacokinetic framework. IEEE Trans. Biomed. Eng. 64 , 2176–2185 (2017).
Gref, R. et al. Biodegradable long-circulating polymeric nanospheres. Science 263 , 1600–1603 (1994).
Bakker-Woudenberg, I. A. et al. Liposomes with prolonged blood circulation and selective localization in Klebsiella pneumoniae -infected lung tissue. J. Infect. Dis. 168 , 164–171 (1993).
Bakker-Woudenberg, I. A., Lokerse, A. F., ten Kate, M. T. & Storm, G. Enhanced localization of liposomes with prolonged blood circulation time in infected lung tissue. Biochim. Biophys. Acta 1138 , 318–326 (1992).
Azad, A. K., Rajaram, M. V. S. & Schlesinger, L. S. Exploitation of the macrophage mannose receptor (CD206) in infectious disease diagnostics and therapeutics. J. Cytol. Mol. Biol. 1 , 1000003 (2014).
Google Scholar
Chono, S., Tanino, T., Seki, T. & Morimoto, K. Efficient drug targeting to rat alveolar macrophages by pulmonary administration of ciprofloxacin incorporated into mannosylated liposomes for treatment of respiratory intracellular parasitic infections. J. Control. Release 127 , 50–58 (2008).
Arias, J. L. et al. Nanobody conjugated PLGA nanoparticles for active targeting of African trypanosomiasis. J. Control. Release 197 , 190–198 (2015).
Hussain, S. et al. Antibiotic-loaded nanoparticles targeted to the site of infection enhance antibacterial efficacy. Nat. Biomed. Eng. 2 , 95–103 (2018).
Kim, M.-H. et al. Magnetic nanoparticle targeted hyperthermia of cutaneous Staphylococcus aureus infection. Ann. Biomed. Eng. 41 , 598–609 (2013).
Luo, Y. et al. Targeted nanoparticles for enhanced X-ray radiation killing of multidrug-resistant bacteria. Nanoscale 5 , 687–694 (2013).
Millenbaugh, N., Baskin, J., DeSilva, M., Elliott, W. R. & Glickman, R. Photothermal killing of Staphylococcus aureus using antibody-targeted gold nanoparticles. Int. J. Nanomed. https://doi.org/10.2147/IJN.S76150 (2015).
Xiong, M.-H. et al. Bacteria-responsive multifunctional nanogel for targeted antibiotic delivery. Adv. Mater. 24 , 6175–6180 (2012).
Xiong, M.-H. et al. Lipase-sensitive polymeric triple-layered nanogel for ‘on-demand’ drug delivery. J. Am. Chem. Soc. 134 , 4355–4362 (2012).
Zhang, C. Y., Gao, J. & Wang, Z. Bioresponsive nanoparticles targeted to infectious microenvironments for sepsis management. Adv. Mater. 30 , 1803618 (2018).
Carryn, S. et al. Intracellular pharmacodynamics of antibiotics. Infect. Dis. Clin. North Am. 17 , 615–634 (2003).
Tulkens, P. & Trouet, A. The uptake and intracellular accumulation of aminoglycoside antibiotics in lysosomes of cultured rat fibroblasts. Biochem. Pharmacol. 27 , 415–424 (1978).
Lutwyche, P. et al. Intracellular delivery and antibacterial activity of gentamicin encapsulated in pH-sensitive liposomes. Antimicrob. Agents Chemother. 42 , 2511–2520 (1998).
Clemens, D. L. et al. Targeted Intracellular delivery of antituberculosis drugs to Mycobacterium tuberculosis -infected macrophages via functionalized mesoporous silica nanoparticles. Antimicrob. Agents Chemother. 56 , 2535–2545 (2012).
Sémiramoth, N. et al. Self-assembled squalenoylated penicillin bioconjugates: an original approach for the treatment of intracellular infections. ACS Nano 6 , 3820–3831 (2012).
Labouta, H. I. et al. Bacteriomimetic invasin-functionalized nanocarriers for intracellular delivery. J. Control. Release 220 , 414–424 (2015).
Castoldi, A. et al. Aspherical and spherical InvA497-functionalized nanocarriers for intracellular delivery of anti-infective agents. Pharm. Res. 36 , 22 (2019).
Langer, R. & Folkman, J. Polymers for the sustained release of proteins and other macromolecules. Nature 263 , 797–800 (1976).
Langer, R. Controlling the movement of molecules. Q. Rev. Biophys. 52 , e5 (2019).
Ostro, M. J., Giacomoni, D. & Dray, S. Incorporation of high molecular weight RNA into large artificial lipid vesicles. Biochem. Biophys. Res. Commun. 76 , 836–842 (1977).
Martinon, F. et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 23 , 1719–1722 (1993).
Kowalski, P. S., Rudra, A., Miao, L. & Anderson, D. G. Delivering the messenger: advances in technologies for therapeutic mRNA Delivery. Mol. Ther. 27 , 710–728 (2019).
Chahal, J. S. et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl Acad. Sci. USA 113 , E4133–E4142 (2016).
Jackson, L. A. et al. An mRNA vccine against SARS-CoV-2—preliminary report. N. Engl. J. Med . https://doi.org/10.1056/NEJMoa2022483 (2020).
Feldman, R. A. et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 37 , 3326–3334 (2019).
Tian, J.-H. et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 12 , 372 (2021).
Pinto-Alphandary, H. et al. Intracellular visualization of ampicillin-loaded nanoparticles in peritoneal macrophages infected in vitro with Salmonella typhimurium . Pharm. Res. 11 , 38–46 (1994).
Toti, U. S. et al. Targeted delivery of antibiotics to intracellular chlamydial infections using PLGA nanoparticles. Biomaterials 32 , 6606–6613 (2011).
Teirlinck, E. et al. Laser-induced vapour nanobubbles improve drug diffusion and efficiency in bacterial biofilms. Nat. Commun. 9 , 4518 (2018).
Shaaban, M. I., Shaker, M. A. & Mady, F. M. Imipenem/cilastatin encapsulated polymeric nanoparticles for destroying carbapenem-resistant bacterial isolates. J. Nanobiotechnol. 15 , 29 (2017).
Nacucchio, M. C., Bellora, M. J., Sordelli, D. O. & D’Aquino, M. Enhanced liposome-mediated activity of piperacillin against staphylococci. Antimicrob. Agents Chemother. 27 , 137–139 (1985).
Schiffelers, R. M. et al. In vivo synergistic interaction of liposome-coencapsulated gentamicin and ceftazidime. J. Pharmacol. Exp. Ther. 298 , 369–375 (2001).
Miles to Go: Closing Gaps, Breaking Barriers, Righting Injustices (United Nations AIDS, 2018); http://www.unaids.org/en/resources/documents/2018/global-aids-update
Beyrer, C., Wirtz, A. L., O’Hara, G., Léon, N. & Kazatchkine, M. The expanding epidemic of HIV-1 in the Russian Federation. PLoS Med. 14 , e1002462 (2017).
HIV in the United States and Dependent Areas (Centers for Disease Control and Prevention, 2019); https://www.cdc.gov/hiv/statistics/overview/ataglance.html
Swindells, S., Flexner, C., Fletcher, C. V. & Jacobson, J. M. The critical need for alternative antiretroviral formulations, and obstacles to their development. J. Infect. Dis. 204 , 669–674 (2011).
Olsthoorn, A. V. et al. Barriers to the uptake of postexposure prophylaxis among Nairobi-based female sex workers. AIDS 1 , 99–103 (2015).
Wheelock, A. et al. Are Thai MSM willing to take PrEP for HIV prevention? An analysis of attitudes, preferences and acceptance. PLoS ONE 8 , e54288 (2013).
Verloes, R. et al. Safety, tolerability and pharmacokinetics of rilpivirine following administration of a long-acting formulation in healthy volunteers. HIV Med. 16 , 477–484 (2015).
Ford, S. L. et al. Lack of pharmacokinetic interaction between rilpivirine and integrase inhibitors dolutegravir and GSK1265744. Antimicrob. Agents Chemother. 57 , 5472–5477 (2013).
Margolis, D. A. et al. Cabotegravir plus rilpivirine, once a day, after induction with cabotegravir plus nucleoside reverse transcriptase inhibitors in antiretroviral-naive adults with HIV-1 infection (LATTE): a randomised, phase 2b, dose-ranging trial. Lancet Infect. Dis. 15 , 1145–1155 (2015).
Sillman, B. et al. Creation of a long-acting nanoformulated dolutegravir. Nat. Commun. 9 , 443 (2018).
Freeling, J. P., Koehn, J., Shu, C., Sun, J. & Ho, R. J. Y. Anti-HIV drug-combination nanoparticles enhance plasma drug exposure duration as well as triple-drug combination levels in cells within lymph nodes and blood in primates. AIDS Res. Hum. Retrov. 31 , 107–114 (2015).
Freeling, J. P., Koehn, J., Shu, C., Sun, J. & Ho, R. J. Y. Long-acting three-drug combination anti-HIV nanoparticles enhance drug exposure in primate plasma and cells within lymph nodes and blood. AIDS 28 , 2625–2627 (2014).
Kovarova, M. et al. Nanoformulations of rilpivirine for topical pericoital and systemic coitus-independent administration efficiently prevent HIV transmission. PLoS Pathog. 11 , e1005075 (2015).
Kirtane, A. R. et al. Development of an oral once-weekly drug delivery system for HIV antiretroviral therapy. Nat. Commun. 9 , 2 (2018).
Giardiello, M. et al. Accelerated oral nanomedicine discovery from miniaturized screening to clinical production exemplified by paediatric HIV nanotherapies. Nat. Commun. 7 , 13184 (2016).
World Malaria Report 2018 (WHO, 2018).
Financing Malaria Strategic Plans in Africa in 2018-2020 (RBM Parternship to End Malaria, 2018).
World Malaria Report 2017 (WHO, 2017); http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/
Fidock, D. A. Priming the antimalarial pipeline. Nature 465 , 297–298 (2010).
Baird, J. K. Effectiveness of antimalarial drugs. N. Engl. J. Med. 352 , 1565–1577 (2005).
Marques, J. et al. Adaptation of targeted nanocarriers to changing requirements in antimalarial drug delivery. Nanomedicine 13 , 515–525 (2017).
Moles, E. et al. ImmunoPEGliposomes for the targeted delivery of novel lipophilic drugs to red blood cells in a falciparum malaria murine model. Biomaterials 145 , 178–191 (2017).
Urban, P. & Fernandez-Busquets, X. Nanomedicine against malaria. Curr. Med. Chem. 21 , 605–629 (2014).
Urbán, P., Estelrich, J., Cortés, A. & Fernàndez-Busquets, X. A nanovector with complete discrimination for targeted delivery to Plasmodium falciparum -infected versus non-infected red blood cells in vitro. J. Control. Release 151 , 202–211 (2011).
Allen, T. M. & Cullis, P. R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 65 , 36–48 (2013).
Joshi, M., Pathak, S., Sharma, S. & Patravale, V. Solid microemulsion preconcentrate (NanOsorb) of artemether for effective treatment of malaria. Int. J. Pharm. 362 , 172–178 (2008).
Mandawgade, S. D., Sharma, S., Pathak, S. & Patravale, V. B. Development of SMEDDS using natural lipophile: application to β-artemether delivery. Int. J. Pharm. 362 , 179–183 (2008).
Melariri, P. et al. Oral lipid-based nanoformulation of tafenoquine enhanced bioavailability and blood stage antimalarial efficacy and led to a reduction in human red blood cell loss in mice. Int. J. Nanomed. 10 , 1493–1503 (2015).
Kumar, S., Singh, R. K., Sharma, R., Murthy, R. S. R. & Bhardwaj, T. R. Design, synthesis and evaluation of antimalarial potential of polyphosphazene linked combination therapy of primaquine and dihydroartemisinin. Eur. J. Pharm. Sci. 66 , 123–137 (2015).
Parashar, D., Aditya, N. P. & Murthy, R. S. R. Development of artemether and lumefantrine co-loaded nanostructured lipid carriers: physicochemical characterization and in vivo antimalarial activity. Drug Deliv. 23 , 123–129 (2016).
RTS,S Clinical Trials Partnership. A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N. Engl. J. Med. 367 , 2284–2295 (2012).
Huang, W.-C. et al. A malaria vaccine adjuvant based on recombinant antigen binding to liposomes. Nat. Nanotechnol . https://doi.org/10.1038/s41565-018-0271-3 (2018).
Global Tuberculosis Report 2018 (WHO, 2018).
Kim, J. Y., Shakow, A., Castro, A. Vande, C. & Farmer, P. Tuberculosis Control: The burden of tuberculosis: Economic burden (2) (World Health Organization, 2003).
O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations (2016).
Jain, R. et al. in Handbook of Nanomaterials for Industrial Applications Ch. 33 (Elsevier, 2018).
Cohen, J. Approval of novel TB drug celebrated—with restraint. Science 339 , 130 (2013).
Singh, H., Jindal, S., Singh, M., Sharma, G. & Kaur, I. P. Nano-formulation of rifampicin with enhanced bioavailability: development, characterization and in-vivo safety. Int. J. Pharm. 485 , 138–151 (2015).
Pandey, R., Sharma, S. & Khuller, G. K. Oral solid lipid nanoparticle-based antitubercular chemotherapy. Tuberculosis 85 , 415–420 (2005).
Ahmad, Z. & Khuller, G. Alginate-based sustained release drug delivery systems for tuberculosis. Expert Opin. Drug Deliv. 5 , 1323–1334 (2008).
Pandey, R. & Khuller, G. K. Nanoparticle-based oral drug delivery system for an injectable antibiotic—streptomycin. Evaluation in a murine tuberculosis model. Chemotherapy 53 , 437–441 (2007).
Pandey, R., Zahoor, A., Sharma, S. & Khuller, G. K. Nanoparticle encapsulated antitubercular drugs as a potential oral drug delivery system against murine tuberculosis. Tuberculosis 83 , 373–378 (2003).
Sharma, A., Pandey, R., Sharma, S. & Khuller, G. K. Chemotherapeutic efficacy of poly (DL-lactide-co-glycolide) nanoparticle encapsulated antitubercular drugs at sub-therapeutic dose against experimental tuberculosis. Int. J. Antimicrob. Agents 24 , 599–604 (2004).
Costa, A. et al. The formulation of nanomedicines for treating tuberculosis. Adv. Drug Deliv. Rev. 102 , 102–115 (2016).
Pandey, R. & Khuller, G. K. Solid lipid particle-based inhalable sustained drug delivery system against experimental tuberculosis. Tuberculosis 85 , 227–234 (2005).
Zahoor, A., Sharma, S. & Khuller, G. K. Inhalable alginate nanoparticles as antitubercular drug carriers against experimental tuberculosis. Int. J. Antimicrob. Agents 26 , 298–303 (2005).
Chuan, J. et al. Enhanced rifampicin delivery to alveolar macrophages by solid lipid nanoparticles. J. Nanopart. Res. 15 , 1634 (2013).
Hawn, T. R. et al. Tuberculosis vaccines and prevention of infection. Microbiol. Mol. Biol. Rev. 78 , 650–671 (2014).
Kaufmann, S. H. E., Weiner, J. & von Reyn, C. F. Novel approaches to tuberculosis vaccine development. Int. J. Infect. Dis. 56 , 263–267 (2017).
Garcia-Contreras, L. et al. Immunization by a bacterial aerosol. Proc. Natl Acad. Sci. USA 105 , 4656–4660 (2008).
Ballester, M. et al. Nanoparticle conjugation and pulmonary delivery enhance the protective efficacy of Ag85B and CpG against tuberculosis. Vaccine 29 , 6959–6966 (2011).
Feng, G. et al. Enhanced immune response and protective effects of nano-chitosan-based DNA vaccine encoding T cell epitopes of Esat-6 and FL against Mycobacterium tuberculosis infection. PLoS ONE 8 , e61135 (2013).
Liu, Q. et al. Preparation and evaluation of antigen/N-trimethylaminoethylmethacrylate chitosan conjugates for nasal immunization. Vaccine 32 , 2582–2590 (2014).
Latent Tuberculosis Infection: Updated and Consolidated Guidelines for Programmtic Management (WHO, 2018).
Van Ginderdeuren, E., Bassett, J., Hanrahan, C., Mutunga, L. & Van Rie, A. Health system barriers to implementation of TB preventive strategies in South African primary care facilities. PLoS ONE 14 , e0212035 (2019).
Sundar, S. & Jaya, J. Liposomal amphotericin B and leishmaniasis: dose and response. J. Glob. Infect. Dis. 2 , 159–166 (2010).
Mitchell, S. L. & Carlson, E. E. Tiny things with enormous impact: nanotechnology in the fight against infectious disease. ACS Infect. Dis . https://doi.org/10.1021/acsinfecdis.8b00138 (2018).
Desai, N. Challenges in development of nanoparticle-based therapeutics. AAPS J. 14 , 282–295 (2012).
Ioannidis, J. P. A., Kim, B. Y. S. & Trounson, A. How to design preclinical studies in nanomedicine and cell therapy to maximize the prospects of clinical translation. Nat. Biomed. Eng. 2 , 797–809 (2018).
Satalkar, P., Elger, B. S., Hunziker, P. & Shaw, D. Challenges of clinical translation in nanomedicine: a qualitative study. Nanomedicine 12 , 893–900 (2016).
Nanotechnology: A Report of the US Food and Drug Administration Nanotechnology Task Force (USFDA, 2007).
Bhatia, P., Vasaikar, S. & Wali, A. A landscape of nanomedicine innovations in India. Nanotechnol. Rev. 7 , 131–148 (2018).
Armstead, A. L. & Li, B. Nanomedicine as an emerging approach against intracellular pathogens. Int. J. Nanomed. 6 , 3281–3293 (2011).
Aly, A. S. I., Vaughan, A. M. & Kappe, S. H. I. Malaria parasite development in the mosquito and infection of the mammalian host. Annu. Rev. Microbiol. 63 , 195–221 (2009).
Pai, M. et al. Tuberculosis. Nat. Rev. Dis. Prim. 2 , 16076 (2016).
Download references
Acknowledgements
A.R.K. received financial support from a PhRMA Foundation postdoctoral fellowship. M.V. was supported in part by an MIT Tata Centre Grant and a National Science Foundation Graduate Research Fellowship. R.L. and G.T. were supported in part by Bill and Melinda Gates Foundation grant number OPP1179091, NIH grant number EB000244 and the MIT Tata Centre. G.T. was supported in part by the Karl van Tassel (1925) Career Development Professorship at MIT, the Department of Mechanical Engineering, MIT and the Division of Gastroenterology, Brigham and Women’s Hospital.
Author information
These authors contributed equally: Ameya R. Kirtane, Malvika Verma.
Authors and Affiliations
David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA, USA
Ameya R. Kirtane, Malvika Verma, Paramesh Karandikar & Robert Langer
Department of Chemical Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA
Ameya R. Kirtane, Paramesh Karandikar & Robert Langer
Division of Gastroenterology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA
Ameya R. Kirtane & Giovanni Traverso
Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA
Malvika Verma
Tata Center for Technology and Design, Massachusetts Institute of Technology, Cambridge, MA, USA
Malvika Verma, Robert Langer & Giovanni Traverso
Department of Global Health and Social Medicine, Harvard Medical School, Boston, MA, USA
Jennifer Furin
Media Lab, Massachusetts Institute of Technology, Cambridge, MA, USA
Robert Langer
Institute of Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA, USA
Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA
Giovanni Traverso
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Giovanni Traverso .
Ethics declarations
Competing interests.
A.R.K., M.V., R.L. and G.T. are co-inventors on multiple patent applications describing gastric resident systems for extended drug release with a focus on global health applications. R.L. has a financial interest in Moderna, Inc, and Alnylam, Inc., which are biotechnology companies focused on RNA therapeutics including vaccines against SARS-CoV-2. R.L. and G.T. have a financial interest in Lyndra Therapeutics, Inc., a biotechnology company focused on the development of encapsulated gastric resident systems for extended drug delivery.
Additional information
Peer review information Nature Nanotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Cite this article.
Kirtane, A.R., Verma, M., Karandikar, P. et al. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 16 , 369–384 (2021). https://doi.org/10.1038/s41565-021-00866-8
Download citation
Received : 14 June 2019
Accepted : 26 January 2021
Published : 22 March 2021
Issue Date : April 2021
DOI : https://doi.org/10.1038/s41565-021-00866-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Nanotechnology’s frontier in combatting infectious and inflammatory diseases: prevention and treatment.
- Yujing Huang
- Xiaohan Guo
Signal Transduction and Targeted Therapy (2024)
Inhalable spray-dried porous microparticles containing dehydroandrographolide succinate phospholipid complex capable of improving and prolonging pulmonary anti-inflammatory efficacy in mice
- Wei-Ya Chen
- Jia-Xing Wei
- Yong-Hong Liao
Drug Delivery and Translational Research (2024)
Efficacy of co-loading Ag nanoparticles and metronidazole in PEG–gelatin-based sponges for the treatment of chronic wounds
- Sibusiso Alven
- S. A. Adeyemi
- B. A. Aderibigbe
Polymer Bulletin (2024)
Bioinspired nanomaterials for the treatment of bacterial infections
- Xiaojing Ma
- Wenjing Tang
Nano Research (2024)
Revisiting the smart metallic nanomaterials: advances in nanotechnology-based antimicrobials
- Ngozi J. Anyaegbunam
- Ifeanyi Elibe Mba
- David B. Olawade
World Journal of Microbiology and Biotechnology (2024)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Elsevier - PMC COVID-19 Collection

Public Health, Communicable Diseases and Global Health
Public health professionals are considering how molecular medicine applications can impact on preventive medicine and health provision enabling interventions to start earlier. There is the potential for molecular medicine to improve the workplace although inappropriate use will lead to discrimination. The public health issue of communicable diseases is being addressed through molecular medicine in more effective and early detection and a better understanding of pathogenesis. A threat from emerging and re-emerging infections requires multiple approaches but early and definitive detection will need nucleic acid based approaches. The non-communicable diseases are now a global world problem, particularly obesity. While molecular medicine is still to make a significant impact on this disorder some interesting findings are emerging about the role of genes in obesity. Ultimately, global health issues must include more efficient and environmentally sustainable strategies. Molecular technologies provide a promising approach for the way forward.
Epidemiology, workplace, diagnostics, resistance, virulence, emerging and re-emerging infections, obesity, bioeconomy
Public Health
Introduction.
Public health is a community-based strategy to improve health and well being, and to prevent disease through research, policy, education and appropriate practice. It is very different to personalized medicine which focuses on the individual. Common to both is the potential for DNA-based information to enhance clinical care. Fundamental to the practice of public health is epidemiology – the study of the causes, distribution, control and prevention of diseases within a population. Until fairly recently epidemiologists relied on traditional approaches and measures of population health. Now an additional dimension is available through molecular (DNA) testing.
The potential to study the interactions between genes and environments is a powerful instrument for those whose research and clinical focus is a population. Tools used by geneticists, such as genome wide association studies (GWAS) and DNA banks are now accepted as legitimate methodologies for research undertaken by public health professionals and epidemiologists. A flavor of what is possible in public health genomics can be found in the Centers for Disease Control and Prevention website [1] .
Preventive Medicine
The concept of prevention is a gold standard in public health, moving the focus from treating an established disease to maintaining well being, and avoiding disease or delaying its onset. As well as research and education, prevention requires appropriate interventions . In the prevention of a disease such as cervical cancer, screening is a core activity, but there is another preventive focus which is directed at finding risk factors , for example elevated blood cholesterol. The social determinants of disease, particularly in developing countries, and the underprivileged, under-resourced or minority groups are also being emphasized and factored into prevention strategies.
There are multiple preventive approaches ( Table 6.1 ), and prevention is not all-or-nothing as interventions are possible during different phases of pre-disease and disease development ( Figure 6.1 ). This expanding view of prevention is considered by some to be a weakening of the concept, and even the terminology is confusing with public health and population health used by some to mean the same thing (the pragmatic approach adopted in Molecular Medicine ) while others distinguish the two [2] . In this complex mix, the application of molecular (DNA) knowledge provides additional options for prevention strategies; from the earliest possible detection of disease development to novel therapies.
Preventive measures can be implemented at different stages of disease [2] .

Progression of disease with burden (Y axis) plotted against time (X axis).
Three different disorders (green, red and yellow) develop over four time periods with gradual increase in burden of disease over this time. Ideally, detection of disease onset should occur at the earliest phase (1) rather than waiting until the burden is such that treatment becomes very difficult (phase 4). Some disorders lead to significant burden of disease (green) while others move along at a relatively lower level of activity (yellow). Preventive steps should be available for all four phases with the goal being to push back from 4 to 3 to 2 and ideally 1 to optimize treatment outcomes. DNA testing (screening) plays an important part in this push-back as it has the potential to detect genetic mutations that predispose to disease (germline DNA) or early signs of disease perhaps in somatic cell DNA in tumors (phases 2 or 3). While the focus is on DNA one should not forget that developments in other omics (particularly proteomics, metabolomics or epigenomics) can identify changes that assist earlier diagnosis.
Population Screening
There are different ways DNA genetic testing can be incorporated into population screening programs once established criteria balancing risk versus benefit are adequately addressed ( Table 6.2 ). Screening programs developed through a public health perspective have as the primary focus the health of the community , while screening programs coming from a genetics viewpoint consider the individual’s rights to be paramount. Hence, the philosophy behind the consent process can be different. This is illustrated by the newborn screening program.
Some criteria that a disease should meet prior to it being considered suitable for population screening [3] .
From a public health perspective, screening newborns to prevent a serious disorder such as congenital hypothyroidism, with its associated severe intellectual impairment, can lead to important clinical outcomes. The screening test itself poses no risk to the newborn (compared to, for example, some vaccinations) and the benefits are significant. Therefore, what type of consent process is needed? The options vary from no consent (if newborn screening is mandated by law) to an opt-out consent process to a fully informed written consent (more on consent in Chapter 10). Some argue that informed consent is essential because the parent(s) must be engaged to ensure they will provide regular supplementation for congenital hypothyroidism in an affected child. Others would say that the health implications of an affected child for parents and society make screening the highest priority. Less seems to be written about the child and who protects his or her right to a healthy life. Different arguments based on a public health versus a genetics approach are possible [4] .
The types of screening programs available through DNA genetic testing are listed in Table 6.3 . The utility of DNA-based strategies, particularly the potential for PCR to test many samples quickly and cheaply, has meant that widespread screening of a population becomes a practical consideration. As more genes are sequenced, the number of mutations identifiable by PCR will increase. Omics-based technologies will continue to expand the options for DNA testing.
Some examples of DNA-based screening strategies.
Two early examples of selective screening programs targeted to at-risk populations illustrate some advantages and disadvantages of this type of testing. Tay Sachs disease is a fatal neurodegenerative disorder of childhood. It is inherited as an autosomal recessive trait. Since the early 1970s, individuals at risk of having children with Tay Sachs have had the opportunity for genetic screening and counseling. As a result, the incidence of Tay Sachs disease has been reduced without the societal problems that developed following the implementation of population screening for sickle cell disease.
Sickle cell disease is an autosomal recessive disorder with around 100 million carriers worldwide, and 2 million in the USA, most of whom are African Americans. There can be considerable morbidity and mortality associated with those who are homozygous affected, although the ultimate outcome is not entirely genetic in origin, as environmental factors are important (more on this in Chapter 2). The US-based sickle cell screening program, which was also started in the early 1970s, targeted the at-risk African American population. The initial version of this program produced more harm than good. Results led to a lowering of self-esteem, overprotection by parents and discrimination. The discrimination came from employers, insurance companies, health insurers and potential spouses. Why did the two screening programs produce different outcomes?
One reason for successful Tay Sachs screening was the nature of the target group, which comprised individuals of Jewish origin who had better educational opportunities and social infrastructure. Another contrast between the two programs was the close community consultation undertaken prior to testing for Tay Sachs. Because of the problems associated with sickle cell screening, changes were made, including the removal of legal compulsion to be screened and improved counseling and education facilities. These enabled more successful testing to be pursued. Experiences with these programs illustrate the necessity for counseling and public education to explain the significance of mass screening results as key ethical considerations in design. Today, there are other population DNA screening dilemmas including: (1) Cystic fibrosis population screening, and (2) Sickle cell trait screening in sport.
Cystic Fibrosis Population Screening
Over 1 500 mutations in the CFTR gene produce cystic fibrosis, although p.Phe508del is the most common found in northern Europeans. Others are much less frequent. So how useful is a test that will not detect all those who are affected? For example, if only the p.PheF508del mutation is sought, false negative results in couples from a population with a frequency for this mutation of 70% will be 0.51 (1−(0.7×0.7)) – i.e. approximately half the couples will not be identified by this approach. Detection of the less common mutations (some of which are only present at a 1–2% frequency in the population) would add to the workload, but would not substantially increase the information gained by the screening program. Additional problems that would need to be resolved before embarking on widespread cystic fibrosis screening include:
- 1. Uncertainty about disease severity for some mutations. Thus, counseling in a number of instances will be difficult and incomplete, and
- 2. Potential for racial profiling as cystic fibrosis is rarely found in some populations, for example Asian, and so detection rates in these will be minimal. Some would argue that one should not exclude or include particular ethnic groups in screening programs because this places undue emphasis on ethnicity and predisposition to genetic diseases. Others would say that disease and ethnic predisposition is a reality and, in the context of personalized healthcare, needs to be considered.
Debate continues about the value of cystic fibrosis mass population screening in contrast to testing individuals or at-risk families (selective screening). Even if laboratory facilities were available, major genetic counseling and public education efforts would be required to ensure that those tested fully understood the implications of the results. The financial resources needed to carry out a mass screening program would be enormous. In view of this, and the inability to detect all mutations with present technology, recommendations vary. In the USA the recommendation is for limited screening – perhaps of pregnant women, or selective screening of groups or families who are at higher risk than the general population [5] . Other countries do not recommend screening of pregnant women. A 2010 European consensus statement on carrier screening provides an in depth overview as well as a framework for what might be possible in member states [6] .
Sickle Cell Screening in Sport
Population screening for sickle cell disease is in place in some newborn screening programs, particularly those involving at-risk populations. Sickle cell disease has potentially fatal consequences, but its effects can be ameliorated or avoided by early medical intervention including the use of antibiotics. DNA testing for the sickle cell trait is used in at-risk couples or populations, since offspring of an at-risk couple have a 1 in 4 chance of inheriting sickle cell disease.
As will be discussed below under Workplace, DNA testing can be used to screen selected populations to detect individuals who are at risk of a work-related illness. In this context, work can include sport. Since hypoxia is one precipitant for an acute attack in sickle cell disease, one might see justification in screening players involved in a sport likely to lead to hypoxia. What to do with this information could be problematic, but the issue is already facing some sporting bodies, as exemplified by the case of a 19 year old university student who died as a result of a rare complication of sickle cell trait , and the subsequent court action. In this case, the organization responsible for student sports at this level determined that sickle cell trait screening would become mandatory despite the trait, in contrast to the disease, rarely leading to serious medical complications [7] .
Screening for a trait is another example of the public health versus the genetic approach, with the latter considering sickle cell trait to be a good trait since it has evolved with time to protect against malaria. Therefore, care is taken to avoid discrimination against or stigmatization of carriers. In contrast, the public health (or more likely in this case the medico-legal) perspective views the trait as a risk factor that needs to be screened for, to identify those who might need appropriate interventions or, more problematic, exclusion from a sport. It will be interesting to see how this controversial screening program for an autosomal recessive trait unfolds.
Newborn Screening
Taking blood from the newborn’s heel to test for treatable and/or preventable medical disorders has been in place since the early 1960s. Initially this was undertaken with biochemical testing and then DNA analysis was added. Next, tandem mass spectrometry (Chapter 4) became possible, allowing metabolomic-type approaches to screening for amino acids, organic acids and fatty acid metabolism to be included [8] .
Today, there is little dispute that screening newborns for treatable disorders such as phenylketonuria and congenital hypothyroidism are important public health initiatives. Less clear is the value of newborn screening for a variety of other conditions, including the hemoglobinopathies, galactosemia, maple syrup urine disease, homocystinuria, biotinidase deficiency, congenital adrenal hyperplasia and cystic fibrosis [5] . The options for screening have been further expanded by tandem mass spectrometry, with its potential to detect many metabolites both normal and abnormal [8] . The former is an important consideration, since false positive results from screening will place additional pressure on the health system as well as increasing the worried well ( Table 6.1 ). The debate about informed consent, presumed consent or even legal compulsion in public health measures such as newborn screening will continue for some time.
Changing Behavior
The applications of molecular medicine in public health practice have introduced new options for preventive programs and interventions. However, changes will only occur if health professionals (starting with medical students) understand the implications and basis for molecular medicine and incorporate this knowledge into their work.
Will DNA based knowledge lead to better health choices by members of the community? Data on this are only now starting to be gathered. One review found evidence that DNA genetic testing for rare genetic variants such as the BRCA1 and BRCA2 genes in breast cancer does lead to changes, such as follow-up mammograms [9] . Less clear was whether this knowledge influenced the behavior of other at-risk family members. The health literacy of the population remains a critical factor in whether behaviors change. If so, statistics emerging from the same review are worrying; more than a third of US adults have limited health literacy and only about 12% have sufficient health literacy skills to understand this type of information [9] (see Chapter 10 for more discussion of education).
Familial Hypercholesterolemia
It is worthwhile concluding this section with a scenario discussing from the laboratory to the bedside , although in today’s philosophy of avoiding hospitalization and expensive medical interventions we should be saying from the laboratory to the community . The example is familial hypercholesterolemia (FH), an autosomal dominant Mendelian disorder which is reasonably common in many populations, affecting about 1 in 500 people in a country like the UK. Familial hypercholesterolemia is clinically important, as >50% of affected men will develop coronary artery disease by the age of 50, and >30% of women will do so by the age of 60 [10] . Heart UK also estimates that of the 120 000 predicted to be affected in the UK, only 15 000 have been identified [11] . Can public health measures utilizing DNA testing help to bridge this gap? Presently the standard criteria of family history, clinical examination and serum cholesterol measurement are insufficient, particularly if familial hypercholesterolemia needs to be detected earlier to optimize the effect of anti-cholesterol drug therapy.
Our molecular understanding of familial hypercholesterolemia started in 1972, when M. Brown and J. Goldstein used biochemical and cell culture approaches to study this disorder. Subsequently they showed that cholesterol metabolism was controlled by a receptor called LDL (low density lipoprotein) and abnormalities in it would lead to familial hypercholesterolemia. They were awarded the Nobel Prize in Physiology or Medicine in 1985 for their work. Once the LDLR gene for this disorder was isolated, DNA tests for a variety of purposes (diagnosis, prediction and screening) could be developed.
The addition of DNA testing in the management of familial hypercholesterolemia now improves the diagnostic accuracy, and the same test can be used to identify at-risk family members. However, this comes at a cost. DNA testing is not simple, as the LDLR gene is large and mutations are often family-specific. Therefore, DNA sequencing is needed and any changes found are not necessarily pathogenic in nature, but can be variants of unknown significance (Chapter 3). Mutations in other genes can also produce a similar clinical picture (phenotype). These include APOB , ARH and PCSK9 which interfere with the cholesterol pathway. Finally, environmental factors such as diet, smoking and hormones also impact on the cholesterol level. Thus, the costs and considerable work involved would need to be balanced against the clinical benefits of earlier diagnosis for individuals, families and the broader community.
Failure to make a diagnosis of familial hypercholesterolemia might have been less of an issue before cholesterol-lowering drugs such as the statins became available. Today, treating an individual with elevated cholesterol is very effective, and it is generally believed that intervening early avoids cardiovascular and related complications of familial hypercholesterolemia. In 2008 NICE (the UK’s N ational I nstitute for Health and C linical E xcellence) published guidelines for a new approach to the treatment and diagnosis of familial hypercholesterolemia, which included personalized medicine through DNA testing of individuals and at-risk family members detected by cascade testing. In the Netherlands there is an ongoing, community-based, familial hypercholesterolemia screening service run by specialized nurses. It has produced some impressive detection rates which are expected to reduce morbidity and mortality in the longer term. The NICE guidelines allow a similar approach in other countries. It will be important to evaluate the clinical effectiveness of this preventive measure utilizing DNA testing.
DNA testing in the workplace could be undertaken for:
- 1. Detecting predisposition to disease or injury because of genetic susceptibility;
- 2. Detecting exposure to toxins;
- 3. Litigation, and
- 4. Identity checks [12] .
Detecting Predisposition to Disease or Injury
This is the most contentious of the four applications, since it implies that DNA genetic testing can predict who will develop an illness or an injury in a particular work environment. One example of the approach is beryllium exposure, which occurs in industries such as defense, aerospace, nuclear power, electronics and dental prostheses. Even if a worker is not directly dealing with beryllium, secondary exposure can occur via airborne particles. Family members exposed to dust carried on clothing or footwear may also be at risk. Individuals sensitized to beryllium are at risk of developing acute or chronic disorders of the skin and lung, with the most serious consequences being carcinoma of the lung or chronic granulomatous lung disease (chronic beryllium disease).
Research has shown that genetic variants of the HLA-DPB1 gene, particularly HLA-DPB1 E69 are found more often in exposed workers who go on to develop a cell-mediated, type IV, delayed hypersensitivity reaction, leading to chronic beryllium lung disease. Mortality associated with this complication is around 36–62% [13] . However, it is important to note that the HLA genotype per se is insufficient to lead to disease and within the environment there are modifying factors such as the type of job; e.g. machining is more risky.
Will testing for HLA-DPB1 variants predict which workers are likely to develop beryllium related disease? Despite the odds of lung disease associated with the glutamic acid 69 variant being high (84% of workers with chronic beryllium disease versus 36% in exposed workers without this disorder), the DNA test would not be particularly helpful, because the prevalence of HLA-DPB1 Glu69 in the normal population is high (40%) while the prevalence of disease among beryllium workers is relatively low (5%) so the positive predictive value of 11.7% is not high enough to make DNA testing a worthwhile screen [13] .
Other examples highlighting ethical and legal dilemmas include the APOE4 DNA marker and predisposition to dementia following head injury in boxing ( Box 6.1 ). Another genetic link between sport and illness is autosomal dominant familial hypertrophic cardiomyopathy, which is caused by mutations in muscle sarcomere genes. This disorder may initially present as sudden cardiac death following strenuous physical activity. Although the molecular DNA defects underlying this disorder are known, their number and complexity make it impractical to screen professional sportsmen and women, unless there are reasons such as a family history, unexplained syncopal attacks, or cardiac findings during clinical examination. Generally, an individual with this type of inherited cardiomyopathy is warned against playing competitive sports as strenuous activity is associated with sudden cardiac death. Those with the disorder can have their heart rates monitored electronically, or have defibrillators implanted to instantly revert ventricular arrhythmias that arise.
Genes and Sport.
The APOE4 gene variant described earlier (Chapter 2) is associated with a greater risk of developing Alzheimer disease, and the risk appears to be further increased in boxers – presumably as a consequence of chronic brain trauma. In a recently reported study, 50% of individuals with chronic traumatic encephalopathy were shown to carry at least one APOE4 allele (one was homozygous for this marker) compared to the general population carrier rate of 15% [14] . Although considering only a small sample size, a 2006 report suggested that the APOE4 variant was also associated with poorer cognitive and behavioral outcomes following moderate and severe traumatic brain injury [14] . Should an individual who has the APOE4 marker (particularly someone who is homozygous for this marker) avoid boxing? Would an employer or trainer be at risk of litigation for not advising a boxer to have their APOE4 status determined? Should someone with this genetic marker be excluded from boxing? Hypothetical questions such as these continue to be asked, but there are no clear answers. If genetic testing is used for screening for susceptibility to work related conditions it should show:
- 1. Strong evidence for linking the working environment and the disorder;
- 2. The disorder has serious implications for the health or safety of employees;
- 3. The test has the appropriate sensitivity, specificity and other parameters, and
- 4. Privacy and the potential for inappropriate discrimination are addressed.
Detecting Exposure to Toxins
There are many potential toxins in the workplace. Genetic monitoring has been used in circumstances involving radiation and genotoxic chemical exposures. Detecting damage to DNA is important but difficult, especially at low exposures where health effects may not become apparent until well into the future. As was shown after the Chernobyl nuclear power reactor accident in 1986, chromosomal damage in workers exposed to significant γ radiation in the clean-up operation was an important indicator of damage. However, age and smoking habits were confounding factors for genetic damage, and the costs of FISH assays for detecting chromosomal abnormalities were too high for large scale population studies [12] .
A new approach to detecting DNA damage might be possible with Next Generation (NG) DNA sequencing, which is interesting since detecting radiation-induced DNA damage was one of the early reasons for initiating the Human Genome Project (Box 1.2). The potential for quantitating cellular and tissue damage is illustrated by the use of this to study genomes of patients with lung cancer caused by cigarette smoking. Tobacco smoke contains more than 60 carcinogens, and damage results from chemical modification of purines by mutagens, inability of the DNA repair mechanisms to correct this damage and incorrect nucleotide incorporation opposite the distorted base during DNA replication [15] . NG DNA sequencing allows the DNA signatures of tissue damage and DNA repair to be cataloged. It may show sufficient specificity to permit monitoring of the environment (by screening workers) or detect when damage has been caused and by what particular toxin (screening workers with illness).
Quantifying the evidence of exposure is a significant hurdle in a tort action (called toxic tort if the wrongful act involves exposure to a toxic substance). It is not easy for a plaintiff to prove that exposure to a toxic substance has occurred and that the toxic substance was the cause of illness or injury. Conversely, a defendant in a toxic tort may have difficulty disproving a claim because of doubtful or minimal evidence. However, exposure to xenobiotics (compounds that are foreign to the body) will provoke changes in gene expression in any biological system. This is the rationale behind the use of transcriptomics to identify or characterize changes that result from exposure to toxins. There is potential for toxicogenomics to provide a new and more definitive evidence of exposure to a toxic substance by looking for particular cellular responses before and after exposure to it.
Workplace DNA testing to establish identity is used in the military and the police. The purpose is to have on record a reference DNA profile for identifying, if necessary, body parts (war, fighting or terrorism) or to assess crime scene contamination. These aims are not controversial but concerns include:
- 1. Security of the DNA sample, and who has access to it;
- 2. Will the DNA sample, or more likely the DNA profile, be included in the databases which are used to search for criminal activities?
- 3. How long are the DNA samples/profiles kept – i.e. are they destroyed once the individual is no longer in the military/police, and
- 4. Is this a voluntary or compulsory part of the employment agreement?
Communicable Diseases
There are many applications of molecular medicine in the communicable diseases caused by bacteria, viruses, fungi, parasites and in a rare example by an abnormal protein. As well as the known infectious agents, there are the newly emerging (or re-emerging) infections and an increasing number of immunocompromized patients exist. To this mix the development of therapy-resistant organisms and bioterror can be added. In such a changing environment, no single therapeutic or preventive approach will be sufficient. What is certain is the ongoing requirement for rapid and accurate detection of infectious agents, which is best undertaken by molecular-based diagnostics. In infectious diseases, these are usually known as NAT (nucleic acid testing) because they involve both DNA and RNA.
Previous editions of Molecular Medicine gave an in depth overview of how knowledge of DNA could be used to improve the detection of infectious agents for patient care. This detail is no longer necessary because DNA testing is now used routinely in clinical management and public health strategies. The various diagnostic tests derived from the traditional phenotypic tests to DNA-based genotypic tests are summarized in Table 6.4 .
Two approaches to laboratory testing in microbiology are the traditional phenotypic tests or the new genotypic DNA or RNA tests (NATs).
As already noted, the utility of DNA sequencing, particularly for viral infections (because their genomes are relatively small), has expanded rapidly and now contributes key data for investigating new outbreaks. Just as occurred in genetics, an omics approach will become increasingly preferred – already the concept of infectomics is being touted. More sophisticated bioinformatics is being developed to deal with metagenomics (Chapter 4) and this will ensure that new software will allow the sequence information from complex mixes of organisms (even those in clinical specimens) to be analyzed and separated into distinct organisms.
Evaluating a NAT is based on traditional measures:
- 1. Sensitivity;
- 2. Specificity;
- 3. Positive predictive value (PPV), and
- 4. Negative predictive value (NPV) (Table 3.6).
Tests with high PPVs are needed for infections where a false negative will have significant clinical or psychological consequences, for example, tests for sexually transmitted infections. Tests with high NPVs are required when it is essential that positives are not missed, for example blood screening.
Taxonomy and Comparative Genomics
The first microorganism to be sequenced was H. influenzae in 1995. Since, there have been large numbers of microbial and viral sequences deposited in databases, including both pathogens and non-pathogens. Completed, whole-genome sequences exist for around 3 000 bacteria, 41 eukaryotes (19 of these being fungi) and 2 675 viruses. In addition, 40 000 and 300 000 partial sequences for influenza and HIV-1, respectively, have been completed [16] . The numbers of sequenced microorganisms will continue to grow exponentially and metagenomic approaches will allow the detection of many novel organisms (Chapter 4). The larger databases available for study will ensure sophisticated comparative genomics can be undertaken for research and clinical applications. DNA-based information is adding a new dimension to taxonomic classification, as described below for viruses.
As multiple de novo sequences of the same organism are obtained, it has become apparent that there is a pangenome. This means that different strains of an organism have:
- 1. The same core genes;
- 2. A number of genes that are variable and used for adaptation to particular environments, and

The pangenome comprises all genomes in a group of organisms [16] , [17] .
The pangenome is divided into: (1) Core genes – essential for basic function; (2) Variable genes – these reflect the environment that the organism needs to deal with, and (3) Unknown genes – found on DNA sequencing but function is unknown. The relative sizes are not drawn to scale but are meant to show a smaller core, with large numbers of genes with unknown function.
The pangenome varies between organisms, for example, all genes for B. anthracis appear to be present in only four species. In contrast, for E. coli , it is likely that the pangenome will require hundreds of these bacteria to be sequenced. Apart from providing further insights into the structure and function of organisms, knowledge of the pangenome is likely to be more informative than any individual genome when considering new virulent forms or the development of drug resistance. To study and understand the pangenome requires an omics approach. It is also apparent that while microbial genomes are small compared to eukaryotes (Table 1.7) they are relatively rich in protein-coding genes (humans 1–2%, microbes 90%) [16] , [17] .
Unlike all other cells that have DNA as their genetic material, viruses are considerably more diverse in what they use. This is reflected in a molecular classification that defines seven different viral classes on the basis of their genetic material and replication strategies. ds – double stranded; ss – single stranded; (+) – positive-sense or plus strand; (−) – negative-sense or complementary strand:
- • dsDNA – example is adenovirus
- • ssDNA – adeno-associated virus
- • dsRNA – rotavirus
- • ss(+) RNA – poliovirus
- • ss(−) RNA – rabies virus
- • ss(+) RNA plus reverse transcriptase – retrovirus
- • DNA plus reverse transcriptase – HBV.
Viruses are the smallest organisms, and have genome sizes measured in kilobases. The International Committee on Taxonomy of Viruses (ICTV) develops an agreed taxonomy and nomenclature. It maintains an official index and publishes this information. In its 2009 release, the ICTV recognized six orders of viruses with another group yet to be placed into an order. There were 87 families, 19 subfamilies, 348 genera and 2 285 species confirming further the heterogeneity found in viruses. The building of an accurate taxonomic classification has many advantages, including new insights into the biology of the viruses and their evolutionary relatedness which provide important clues when dealing with new infections [18] ( Box 6.2 ).
Integrated DNA and RNA in the Genome.
The genomes of vertebrates contain many copies of retroviral sequences acquired during evolution. These could function to protect the host from viral infection, and possibly as a source or natural reservoir for the virus to persist and transmit. However, it is now apparent that it is not only retroviruses that can integrate a copy of their RNA into the host’s somatic and germline genome, which is the necessary first step before replication can occur. The genome of some bees has been shown to contain sequences from a positive (+) strand RNA Dicistroviridae that infects insects. These bees are resistant to infection by the virus. Following this observation, a comparative bioinformatics study of genomic sequences from 48 vertebrate species using sequence data from non-retroviruses containing single-stranded (ss) RNA genomes was undertaken. Surprisingly, it was shown that about half the vertebrates had integrated non-retrovirus sequences into their genomes. The next unexpected finding was that these integrations came mostly from two groups of RNA viruses from the negative (−) strand RNA Mononegavirales order. These were either Ebola and Marburg viruses – Filoviridae family associated with lethal hemorrhagic fevers – or Bornavirus – Bornaviridae family are associated with neurological and psychiatric disorders which can be fatal. The vertebrates that had the integrations suggested these events had occurred over 40 million years ago. Therefore, the conservation of sequences coding for virus-like proteins is thought to have some selective advantage, possibly increasing the host’s resistance to infection. Conversely, continued integration and persistence might provide viruses with a natural reservoir for future infections. An example would be bats, which are now thought to be natural reservoirs for the Ebola and Marburg viruses. Sequences from these viruses are detectable in some bats with some having open reading frames [19] .
Applications of DNA sequencing in virology include:
- 1. Identifying the function of viral proteins to allow a better understanding of how viruses evade host immune responses or promote their own migration and spread;
- 2. Defining regulatory controls or proteins that might become targets for new anti-virals;
- 3. Developing rapid diagnostics and detecting the identity of new viral outbreaks, and
- 4. Understanding evolution and hence relatedness for molecular epidemiologic strategies investigating outbreaks of old and new viruses, and monitoring drug resistance [18] .
Nosocomial Infections and Drug Resistance
Nosocomial, or hospital acquired, infections are usually associated with medical devices such as catheters, or surgical procedures. Apart from wound and urinary tract infections they lead to life-threatening pneumonia and septicemia. Some statistics on these types of infections include:
- 1. They were the sixth leading cause of death in the USA in 2002 with approximately 99 000 deaths;
- 2. Estimated cost to the US Healthcare budget is over $5 billion annually;
- 3. Approximately one third are preventable, and
- 4. Gram negative bacteria are involved in more than 30% of infections [20] .
The convergence of gram negative bacteria that are increasingly antibiotic resistant and a reduction in drug development programs has produced a gloomy scenario for hospital acquired infections.
Causes for antibiotic resistance are many including:
- 1. Unnecessary or inappropriate use of antibiotics in humans;
- 2. Availability of antibiotics over the counter;
- 3. Use in the food industry including meat, agriculture, aquaculture;
- 4. Poor patient compliance in taking prescribed drugs;
- 5. Transmission by farm or pet animals treated with antibiotics, and
Some therapy resistant multidrug resistant organisms a [21] , [22] , [23] , [24] , [25] .
New drugs are not being developed as quickly as they are needed because of high production costs, the time required for clinical trials, regulatory demands and a concern that products will become obsolete once resistance develops. Apart from rapid diagnosis of the causative microorganism, improved detection of antibiotic resistance strains is also needed. These requirements can be met by a NAT approach, although this is only the first step of a more comprehensive internationally coordinated plan to address the issues of antibiotic resistance.
The urgency of this matter is well illustrated by tuberculosis (TB), where global control of this increasingly problematic public health challenge requires better and faster diagnosis of the primary infection as well as early detection of drug resistance. The traditional phenotypic culture methods to diagnose TB are slow. Similarly, the first generation of molecular DNA diagnostic tests is complex, requiring sophisticated laboratory expertise and resources [26] . Multidrug resistant TB (defined as infections that are resistant to at least isoniazid and rifampicin) is emerging globally, particularly in India and China. Cases of extensively drug-resistant TB now exist, meaning that TB is also resistant to a number of the second line anti-TB drugs. Failure to detect resistant cases of TB is the rule rather than the exception, particularly where laboratory resources are limited. This means that new, DNA-based, detection kits, especially those that can be multiplexed and automated are eagerly awaited.
A fully automated NAT method to detect both TB and rifampicin resistance was reported in 2010. This uses uncultured sputum and can be completed in less than 2 hours with impressive sensitivities and specificities even in patients with TB and HIV, where smear-negative disease is more common. Since it is fully automated, it does not require sophisticated hands-on expertise. Although the NAT only detects resistance to rifampicin it shows the way ahead, particularly if omics-based diagnostics including microarrays are developed [26] .
Public Health Testing – Blood Transfusion Services
Viruses such as HIV, HBV and HCV assume added notoriety when they are implicated in transfusion-derived infections involving blood and plasma-derived products. Previously, blood transfusion services based their donor and blood screening programs on detecting antibodies or antigens in the donor or blood supply. However, this has proved to be inadequate, and an important addition to the screening protocols is the use of PCR to identify viral DNA or RNA. The advantages of a NAT include higher sensitivity and greater reliability during the window period – which is the time between a blood donor becoming infectious and donor screening tests becoming positive, i.e. seroconversion has occurred. The use of NAT, better serologic-based assays and more effective regulatory controls have made contemporary blood products considerably safer. Ultimately, transfusion services must balance safety against access to blood and its products. What is screened for will depend on the types of infections found within a geographic region as well as affordability of the screening tests.
NAT-based assays for screening blood donations can be used to screen pools of donations, for example, 16–24 donations simultaneously or individual ones. The former is more rapid and cheaper, but rare instances of HIV, HBV or HCV can be missed. The testing of individual donations is the method of choice but until recently was too expensive. Today, as new analytic platforms allow rapid and automated multiplexing NATs to be used, the screening of individual donations becomes more cost effective.
Blood transfusion services test blood and donors for a range of infectious agents depending on national requirements. The WHO recommends mandatory screening for HIV-1, HIV-2, HBV, HCV and syphilis, while the requirement for HTLV-I, HTLV-II (HTLV – human lymphotropic virus) and malaria are decided on a regional basis [27] . Other infectious agents that can be screened for include West Nile virus, dengue and emerging infections. Screening can also be undertaken in selected cases, for example, CMV free-blood for immunosuppressed patients, the fetus or the neonate. The risk of prion diseases is considered below.
Ease of international travel means a potential donor could become infected elsewhere. This contingency is covered by donor questionnaires that allow self-exclusion (particularly for infections that are not routinely sought). For example, to prevent transmission of prion diseases through blood, some transfusion services have excluded donors who have lived in the UK over certain time periods (see below). Other reasons for deferral include fever with headaches the week before donation (a risk of West Nile and other viruses) or travel to certain regions (a risk of malaria).
Awaiting Better Diagnostics – Prion Diseases
A rare but important form of communicable dementia is found in the prion diseases (also called transmissible spongiform encephalopathy, or TSE). These diseases affect humans and a number of animals used for meat including cattle, deer, sheep and goats. The term prion comes from protein and infectious and was coined by S. Prusiner who was awarded the Nobel Prize in Physiology or Medicine in 1997 for his work on prions. The important components of prion disease include the PRNP gene and its cellular product PrP c ( pr ion p rotein c ellular) which can become the infectious protein product PrP Sc ( pr ion p rotein sc rapie). The normal PrP c is a cell surface glycoprotein found in a wide range of animals, having a function that as yet remains unknown. PrP c needs to change its conformation to its isoform PrP Sc to be infectious. No nucleic acid is involved in this process, highlighting the novel way in which prion disease arises and is propagated [28] . The disease leads to widespread neurodegeneration with cognitive and motor impairment. It is fatal and there is no treatment ( Box 6.3 ). Work continues to develop an early diagnostic marker for this disease. This is a priority for screening blood and its products.
Prion Disease.
Prion disease may be sporadic, inherited, iatrogenic or transmissible from animal to human via infected meat and now human to human via blood products. The dementia that results includes sporadic, iatrogenic, inherited and variant Creutzfeldt-Jakob disease (CJD) in humans, b ovine s pongiform e ncephalopathy or BSE in cattle (related to the 1986 epidemic of mad cow disease in the United Kingdom), and scrapie in sheep and goats. In 1996, the emergence of variant CJD (vCJD) in humans is thought to have arisen from transmission across the species of the BSE agent. vCJD is characterized by an early age of onset ( Figure 6.3 ). Mutations in the PRNP gene account for the inherited forms of CJD. However, in the vast majority of sporadic cases, there are no detectable DNA mutations, and the change from PrP c to the abnormal PrP Sc is thought to occur because of somatic mutations or other, as yet unknown genetic or environmental factors. Risk factors for developing vCJD include young age, residence in the United Kingdom especially between 1985 and 1990, and intriguingly, homozygosity for a codon 129 polymorphism in the PRNP gene. At this position there is either a methionine or a valine. In normal individuals, the combinations of methionine/methionine, methionine/valine and valine/valine are present. However, in patients with vCJD, the homozygous methionine is always found, suggesting that this may lead to genetic predisposition. Most patients developing iatrogenic CJD after receiving pituitary extracts for growth hormone are also homozygous for methionine. If this is correct, some have hypothesized that a second wave of vCJD will occur in the future involving those who are methionine/valine heterozygotes or homozygotes for the valine allele because a longer incubation period is needed to develop prion disease without the additional genetic risk factor. Other less well characterized polymorphisms in this gene have been detected and may represent additional genetic modifiers [28] . Prion disease remains a challenge for the future, particularly to explain how the infectious forms occur without any apparent conventional infectious agents being involved. Better diagnostics and some form of therapy are needed for this rare but fatal infection.

Human prion diseases [28] .
There are different types of Creutzfeldt-Jakob diseases (CJD), and two other prion-related diseases. The most recently described is vCJD which is thought to have occurred as a result of direct animal to human spread through contaminated beef products. Now there is evidence for human to human spread via transplants or blood products.
Pathogenesis
The pathogenesis of many infections has been determined from studies utilizing light/electron microscopy, cell culture or immunoassays. To these can now be added nucleic acid (DNA, RNA) based methodologies. Advantages provided by nucleic acid techniques include the ability to detect latent (non-replicating) viruses, and to localize their genomes to nuclear or cytoplasmic regions within cells. Tissue integrity remains preserved during in situ nucleic acid hybridization and so histological evaluation can also be undertaken. NAT can be manipulated to enable a broad spectrum of serotypes to be detectable. This is particularly valuable in emerging infections where the underlying serotypes are unknown. Today, a very powerful application of NAT is the ability to sequence whole genomes, and so identify a pathogen or what it is likely to be. From its genomic sequence it becomes possible to:
- 1. Predict its role in disease pathogenesis;
- 2. Find regions in the genome suitable for rapid diagnostics via NAT, and
- 3. Consider how new treatment options including vaccines can be developed.
Virulence Factors
Microorganisms have developed a range of virulence factors to allow them to invade a host ( Figure 6.4 ). The best known are toxins, which are broadly divided into:
- 1. Exotoxins – usually proteins secreted by both gram positive and gram negative bacteria. They can be deadly, for example, tetanus exotoxin and diphtheria exotoxin, and
- 2. Endotoxins – usually heat stable lipopolysaccharides found in the gram negative bacterial cell wall.

Virulence mechanisms used by bacteria.
Four mechanisms can be used by bacteria to invade a host. Which predominates will vary for each microorganism. (1) Adhesins allow bacteria to attach to host cells. This is the first step in the infective process. Some bacteria have appendages such as pili and flagella to facilitate attachment; (2) Many toxins are produced and have been well characterized both biochemically and molecularly; (3) Bacteria ultimately need to secrete their products into the host cell through specific secretory systems. A number have been described and are needle-like to allow the passage of toxins from the bacteria into the host, and (4) Implied in the concept of a pangenome is a complex bacterial genome to orchestrate the various changes needed to infect a host and produce the appropriate effects. The regulatory environment for this will need some common pathways and specific ones when comparing different bacterial species.
Nevertheless, killing the host is not beneficial to the invading organism and in some circumstances it is essential that the host does not die. This is exemplified by H. pylori , which has sophisticated virulence factors including VacA and CagA allowing it to invade and cause damage to the host. However, the same organism has also evolved to ensure its continued survival by modulating its cell killing capacity because the CagA protein while cytotoxic per se counters some of the effects of the VacA toxin [29] .
Toxins have many different actions, and using broad spectrum antimicrobials to inactivate them might not always suffice ( Table 6.6 ). Nevertheless, the potential for this approach to treating or preventing infection is illustrated by B. anthracis – a bacteria causing anthrax. It achieved added notoriety because of an attempt at bioterror using postal letters in 2001 (Box 9.5). The attenuated anthrax bacteria (Pasteur strain) used for immunization lacks its toxin confirming the latter’s importance in disease causation. Animal studies also suggest that antibodies that inhibit the anthrax toxin from binding to host receptors might provide protection, at least in emergencies [30] . A better understanding of how toxins work and function as targets for new drugs is coming from molecular studies.
Some bacterial toxins in the gastrointestinal tract [30] , [31] , [32] .
The traditional targets for conventional antimicrobials (usually antibiotics) include components of the bacteria that are essential for survival, such as the cell wall, the cell cycle, DNA replication and protein synthesis. This approach kills (bacteriocidal) or inhibits growth (bacteriostatic) of most bacteria, but invariably allows some residual subpopulations with natural immunity to be positively selected for, and hence the development of antimicrobial resistance will follow. Therefore, focus has now shifted to developing the next generation of antimicrobials, which target virulence factors. This would overcome the pathogenicity of the organisms without necessarily killing them and so avoids setting up an environment for resistance strains to emerge [30] .
Host Resistance
Microorganisms have developed sophisticated ways in which to invade a host, but hosts have evolved many protective mechanisms ( Figure 6.5 ). The host’s response in terms of genetic modifications is particularly relevant to molecular medicine. In humans, evidence for a genetic component influencing the outcome of an infectious disease comes from the following observations: (1) Not all exposed to HIV-1 get infected, and those who do progress to AIDS show different responses, and (2) Some ethnic groups are more resistant or susceptible to infections, e.g. resistance to malaria in some Black Africans.

Host mechanisms to protect against invasion by microorganisms.
Various protective mechanisms allow the host to escape or modulate invasion by a microorganism. (1) Microbiota in the host ( microbiota – normal microbial flora; metagenome (Chapter 4) – the genetic (DNA/RNA) material isolated from an uncultured microbial environment); (2) Physical barriers such as skin or mucosa, pH, temperature and secretions; (3) Chemical barriers particularly the immune response, and (4) Genetic adaptations which evolve over a long period of time but provide an effective mechanism to protect against certain pathogens.
HIV-AIDS: The main HIV co-receptor involved in the infection process is CCR5. Naturally occurring mutations in this receptor – such as a 32 base deletion present in up to 20% of European populations (about 1–2% are homozygous) – allow these individuals to be highly resistant (homozygotes) or partially resistant (heterozygotes) to HIV-1 infection and disease progression [33] . Studies are now underway with anti-HIV drugs that target the CCR5 receptor and a bone marrow transplant approach is described in Chapter 8.
Malaria: The two most common forms of malaria ( P. falciparum and P. vivax ) produce severe anemia. P. falciparum is also associated with cerebral malaria, respiratory and metabolic complications. This spectrum is partly explained by P. falciparum being able to invade a large proportion of red blood cells, whereas P. vivax can only invade the reticulocytes. Another explanation is the mode of entry of these parasites into red blood cells; P. falciparum has a number of routes of invasion, whereas P. vivax can only enter red blood cells that carry the Duffy blood group. This parasite is not seen in West Africa because the populations there are Duffy negative.
Host genetic factors that provide some protection from malaria have been identified. These include single gene effects seen in the hemoglobinopathies such as sickle mutation (HbS), HbE, α thalassemias and β thalassemias. The hemoglobinopathy protective effect results from abnormal red blood cells that quickly lyse when invaded by parasites and so the parasites die. In the case of the sickle mutation this occurs because of the sickling effect while with HbE and thalassemias it reflects the small and poorly hemoglobininized red blood cells.
There are many different hemoglobinopathies, but usually one type predominates in a given population; for example, black Africans will have HbS, South East Asians HbE and Mediterranean populations will have different thalassemias. Each protects against malaria but co-inheritance can cancel out this effect. Thus, HbS co-inherited with α thalassemia removes the malaria protection because it makes the red blood cell abnormality less severe [34] .
Genetic factors may also enhance the risk of infection. These are more subtle as they are thought to involve multiple genetic effects; i.e. QTLs (quantitative trait loci) that are difficult to detect. They have been sought by association (case control) studies and now by GWAS (genome wide association studies) (Chapters 2, 3Chapter 2Chapter 3). These studies have identified predisposition genetic loci to N. meningitidis meningitis, tuberculosis, HCV, leprosy and HBV. In the case of HBV it is the HLA locus that seems to be the key factor in predisposition and it is perhaps not coincidental that non-response following vaccination with HBV vaccine is more likely to occur in those with certain HLA types such as DRB1*03 and DRB1*07 HLA types [33] .
The three RNA influenza viruses (A, B, C) are distinguished by their internal group-specific ribonucleoprotein. Only influenza A and B are medically significant, since epidemics or pandemics have not occurred with influenza C. Influenza A has the potential to produce pandemics because it infects other species apart from humans, including birds, pigs and horses. Influenza B only infects humans and so its antigenic structure does not become sufficiently different to cause pandemics. In contrast, viruses such as measles undergo minimal antigenic variation with one infection giving life-long immunity.
The subtyping of the influenza A virus is based on its outer viral proteins, which include two important and distinct antigenic glycoproteins: Hemagglutinin (H – composed of 16 different types) and neuraminidase (N – nine different types) ( Figure 6.6 ). Although the envelope antigens are capable of producing many different combinations (as seen in water birds), a smaller number are found in humans. To date only a few have been implicated in human to human spread (H1N1, H2N2, H3N2, H1N2, H5N1, H9N2 and H7N7) with highly pathogenic avian influenza subtypes found only in H5 and H7 subtypes ( Figure 6.7 ) [35] .

Structure of the Influenza virus.
This RNA virus has two key surface glycoproteins: (1) Hemagglutinin (HA or H) – facilitates the entry of virus into host cells through attachment to sialic acid receptors, and (2) Neuraminidase (NA or N) – involved in the release of progeny virions from infected cells. The HA is the major determinant against which are directed neutralizing antibodies, and so also the target for influenza vaccines. In contrast, the NA is an important target for antiviral agents.

Major animal–human and human–human influenza outbreaks.
Since the 1918 pandemic, a number of important outbreaks have been recorded (subtypes and dates are given as well as hosts involved). A worrying trend is the increasing numbers of new subtypes in humans, as well as an expanding animal involvement since 1997, in particular the domestic chicken.
As the influenza A virus passes through its hosts, the most important of which in terms of global spread are the water birds, it undergoes genetic changes. In the past 100 years there have been four influenza pandemics:
- 1. 1918 H1N1;
- 2. 1957 H2N2;
- 3. 1968 H3N2, and
- 4. 2009 H1N1.
A fifth outbreak (H5N1) has not been declared a pandemic but remains a concern.
Avian influenza (avian flu, bird flu, H5N1, 1997 and re-emergence in 2003): This remains a worldwide threat to health, with some regarding a H5N1 pandemic as being potentially more devastating than the 1918 Spanish flu outbreak. In 1997, the first cases of human infection from exposure to sick birds or their droppings were reported in Hong Kong, indicating that this virus subtype had jumped the species barrier. Eighteen patients were admitted to hospital and six died. Fortunately, the timely culling of over a million chickens controlled this particular outbreak. Today, H5N1 still causes outbreaks in chickens, and sporadic human infections continue to be reported, with a mortality of over 50%. In contrast to H1N1 swine flu and SARS (Severe Acute Respiratory Syndrome) that have been spread from human to human and through travel, the H5N1 bird flu remains relatively contained because spread is predominantly through chickens or other birds.
The common human influenza virus (H3N2) is highly contagious but rarely lethal. Avian flu in chickens (H5N1) is a particularly virulent type that can kill rapidly and causes widespread organ damage. Fortunately, it is not easily transmitted from birds to humans, and more importantly, human to human spread is poor. However, swapping genetic material, should an individual be co-infected with both, might produce a hybrid H5 (avian flu) N2 (human flu) virus with devastating effects. DNA sequencing of the viral genome from various outbreaks has shown that the virus continues to mutate. This has implications for pathogenicity, as well as antiviral drug resistance, and having the right vaccine ready if needed. In this unpredictable environment, the value of rapid NAT diagnostics is crucial to detect early cases and for public health surveillance. The genes of the virus that caused the 1918 pandemic have been studied to better understand what makes an influenza virus virulent and capable of producing a pandemic [35] .
Spanish influenza (H1N1, 1918): The virus from this pandemic, which killed about 40 million people, had not been isolated. Without a virus little research was possible, then the viral RNA sequence was determined using material from archival tissue, including formalin-fixed autopsy material. The sequence itself did not provide clues for why the Spanish influenza virus was so virulent, and so the next step was to reconstruct the viral coding segments and clone them into plasmids. Individual genes from the H1N1 1918 virus were then introduced into a common laboratory viral strain and pathogenicity sought. Although the H and N glycoproteins were factors in the virulence of this virus, it was also shown that one of the RNA polymerase subunits known as PB1 was involved. Another subunit (PB2) was then found to be important for viral transmissibility [35] .
Swine influenza (H1N1, later called H1N1(09), 2009): After the appearance and then rapid disappearance of SARS ( Box 6.4 ), followed by the concerns regarding the possibility of a H5N1 pandemic that did not occur (so far), the world in 2009 was faced with another possible serious influenza outbreak. This outbreak was described as swine flu, because it was a well-recognized cause of influenza in pigs. The virus is related to the H1N1 virus that caused the Spanish flu, and can spread from person to person. The WHO declared a swine flu pandemic in June 2009. Vaccines were rapidly developed and stockpiles of antiviral drugs, particularly the two mentioned in Table 6.5 , were released to the public. Rapid NATs requiring RT-PCR because it is an RNA virus were developed (see Table 3.3). This flu was a little unusual because it tended to be more severe in younger people, including children and pregnant women whereas deaths from seasonal flu involve mostly the elderly. Despite early concerns expressed by public health officials and considerable media hype, the WHO declared the H1N1 pandemic over in August 2010.
SARS (Severe Acute Respiratory Syndrome).
This infection attracted a lot of publicity and provoked considerable fear when it emerged in China and then Hong Kong in 2003. SARS subsequently spread to many countries, producing around 700 deaths in the first half of 2003. This was at one time described as the first pandemic of the 21st century, but it never progressed beyond an epidemic because of effective public health measures effected by mid 2003 [36] . The social and economic impacts of this infection were considerable, including major disruptions to international travel. SARS was shown to be caused by a novel coronavirus (CoV) which was thought to have crossed the species barrier, although the animal reservoir for SARS took a while to find. It is now thought to be:
- 1. Masked palm civets – used for exotic food dishes in China, and
- 2. Horseshoe bats [37] .
Traditional approaches such as viral culture, electron microscopy and serology helped to characterize the SARS virus. Nevertheless, SARS illustrated the value of NAT approaches in dealing with an emerging virus. Molecular testing enabled the following to be possible in a very short time frame:
- 1. Typing of the virus from two different countries (Taiwan and Hong Kong) showed that human to human spread had occurred;
- 2. Rapid whole genome sequencing of viral RNA enabled the development of PCR based diagnostic assays, and
- 3. In searching for animal reservoirs, RT-PCR based techniques were used. These allowed SARS-CoV to be detected, as well as identifying genetic differences between the human and animal virus.
The outbreak ended just as quickly as it started. Only occasional cases were reported in early 2004, and none after the end of April that year. However, there remain many unanswered questions including the inconsistent human to human transmission which might have been due to super-spreaders.
Another observation was the relatively large numbers of health workers who became infected. This became an issue when two of the nine persons infected in China in 2004 worked in a reference laboratory conducting research into the virus. A similar scenario was reported earlier in Singapore. The latter case was documented on RNA sequencing of the virus to be due to a contaminated laboratory culture that the scientist had been working with three days before showing signs of the infection. The WHO subsequently flagged the importance of laboratory containment when dealing with the SARS virus.
Emerging and Re-Emerging Infections
Emerging (newly discovered, for example SARS – Box 6.4 ) and re-emerging (previously known, for example dengue virus) infections have increased significantly in the past 20 years. Many factors contribute including:
- • Globalization, particularly increased travel and trade;
- • Changes in human behavior, poverty and social inequality;
- • Economic development, changes in the environment, weather and land use;
- • Lapses in public health measures including those due to poverty or war;
- • Complacency by communities or government;
- • Mutations, selection and genetic reassortment in organisms;
- • Bioterror.
Very few emerging infections represent novel pathogens. Most are re-emerging infections resulting from a change in the epidemiology or virulence of a pathogen, or secondary to microbial adaptation. A review of the major infections in history provides some background to the emerging ones. They are:
- 1. Plague of Athens 430 BC;
- 2. Black death ( Y. pestis ) in 1340s;
- 3. French pox (syphilis) 1494;
- 4. Small pox 1520;
- 5. European cattle epidemics including anthrax, foot and mouth disease 1700s;
- 6. American plague (yellow fever) 1793;
- 7. Cholera pandemic in Paris 1832;
- 8. Measles outbreak in Fiji 1875;
- 9. Spanish influenza 1918, and
- 10. HIV-AIDS from 1981 [38] .
Most emergent viruses are zoonotic – i.e. they are acquired from animals that are reservoirs of infection. This is particularly relevant in the modern world, where the consequences of easy migration, deforestation, agricultural practices, dam building and urbanization are making, and will continue to make, a major impact on the ecology of animals. For example, yellow fever is thought to have emerged in the New World as a result of the African slave trade which brought the mosquito Aedes aegypti in ships’ water containers. More recently, Aedes albopictus , a potential vector for dengue virus, has become established in the USA following its conveyance from South East Asia in old car tires. With this, the threat of dengue in the North American continent has become real. Humans have populated rural areas to an increasing extent, as well as pursuing more outdoor recreational activities. There is also a growing trend for exotic animals to be kept as household pets. Changes in global climate may also contribute directly, through their effects on vegetation, insect and rodent populations.
Table 6.7 lists a number of zoonoses that have become established as new infectious diseases, or are emerging as problems for the future. Some of these are newly acquired in the west, while others remain endemic to specific countries. However, any disease may be spread through international travel, or the mass dislocation of large populations through civil unrest. There is also an increasing possibility that a number of pathogens could be used for bioterrorism. Some of the zoonoses associated with a viral hemorrhagic clinical picture can be confused with other clinical infections including malaria, leptospirosis, and N. meningitidis and in these potentially fatal conditions, a rapid screening test is essential. In terms of bioterrorism and the differential diagnosis of hemorrhagic fevers, NAT assays are presently the only option to allow rapid and sensitive diagnostic tests to be developed. If new therapeutics are required, the first step will be nucleic acid sequence analysis of the microorganisms’ genomes so that it can be classified and identified. Next, potential targets for vaccines or drug therapies can be established.
Some examples of zoonoses resulting in new human infections a .
Global Health
In an era of personalized medicine, one should not lose sight of how molecular medicine can be used to improve global health. Cheaper drugs and vaccines for all communities is an important benefit that should come from molecular-based technologies. Another would be better NATs. In this respect it is intriguing to recall how direct-to-consumer DNA testing (Chapter 5) makes effective use of the Internet. Could the Internet be one way to improve accessibility for disadvantaged communities or those in rural and remote regions? Consideration of how genomics can play a part in the bioeconomy, with its potential to generate income, improve food production and sustain a better environment, are some of the challenges now being taken up by bodies such as the OECD.
Non-Communicable Diseases
A large part of this chapter has dealt with infectious diseases and how these impact on individuals, communities and ultimately global health. To complete the story, it is necessary to consider non-communicable diseases since, apart from their primary effect on health and well being, they can also contribute to a communities’ vulnerability to infectious diseases ( Table 6.8 ).
Some global health challenges [21] , [40] , [41] .
A Perspective on global non-communicable diseases makes some sobering observations including:
- 1. 60% of all deaths are due to chronic diseases, with most occurring in low to middle income countries with a disproportionate number of young people dying during their productive years;
- 2. Non-communicable diseases are likely to have a more detrimental effect on global economic development than fiscal crises, natural disasters or pandemic influenza;
- 3. In the next 10 years, it is projected that China (as one example) will lose $558 billion in national income because of preventable heart disease, stroke and diabetes, and
- 4. To address these problems it is essential to have better evidence-based decision making, more effective regulation and behavioral interventions that are known to work.
The need to shift focus more to community-based prevention and concentrate less on attempting to cure a problem once it is established has already been highlighted [40] .
A number of the non-communicable health problems listed in Table 6.8 have obesity as a contributing factor. In the USA obesity continues to be a major health challenge; 2003–2004 estimates indicated that 66% of the US population was overweight, and 32% obese, as defined by a BMI≥30 kg/m 2 . Another estimate is that 50% of the adults in the USA will be clinically obese by 2030 [42] , [43] .
Current understanding is that most cases of obesity are caused by a mix of genetic and environmental factors, although their relative contributions remain to be determined. The rapid development of obesity worldwide can only be an environmental effect. Nevertheless, many people in the same environment have not developed obesity and so genes must play a role. Comparisons between monozygotic and dizygotic twins, as well as other studies, show greater concordance for the BMI (a surrogate measure for obesity), i.e. there is an important genetic component to obesity, with estimates indicating that this is a strong effect (around 80%) [43] .
One hypothesis, which has been around for 50 years, captures both genes and environment. It suggests that genes important for metabolism in humans evolved over time to respond to periods of famine. These so called thrifty genes allowed hunter-gatherer populations to process food into fat deposits during times of plenty, so that they could survive when food was not available. Today, these same genes respond inappropriately when food is readily available all year round, and so obesity results. Evidence for this genetic evolutionary effect is still awaited. Other hypotheses include:
- 1. Fetal programming (perhaps via epigenetic changes) with maternal nutrition a key factor in how the child will grow postnatally;
- 2. Sedentary lifestyle, i.e. diet and lifestyle are the main contributors and from the genetics perspective this would put the focus onto metabolic enzymes;
- 3. Increased reproductive fitness, since the number of offspring is positively correlated with the BMI of women – i.e. adiposity increases fertility, and
- 4. Many others [43] .
The public health response to the obesity epidemic is focused on eating less, avoiding fast foods and exercising more. However, this approach is not working. Can a more personalized genomics strategy help? Will a scientifically plausible understanding of how diet, the environment and obesity interact allow governments and individuals to take a more effective approach? One way to pursue this would be to know more about the genes involved in obesity.
The Genetics of Obesity
Our current understanding of genes and obesity is still rudimentary, so medical or motivational interventions cannot be tested. At the genetic level, obesity can be considered in three groups:
Genes and Obesity.
Apart from the MC4R example given, other genes associated with obesity have a recessive mode of inheritance. They include mutations causing deficiency in leptin and its receptor ( LEP , LEPR ) which act via the hypothalamus to control appetite and energy expenditure. One report, concerning a child with congenital leptin deficiency, described how a sustained reduction in weight occurred following treatment with recombinant human leptin. Other genes in the leptin-melanocortin pathway are also implicated including POMC and PCSK1 . A human gene FTO was shown to be implicated strongly with the BMI (body mass index) in a genome wide association study involving subjects with type II diabetes. This has been replicated in other studies and appears to be reflecting common SNP polymorphisms in intron 1, with the risk allele highly prevalent in the general population. European carriers who are homozygous for the risk allele weigh on average 3 kg more. Some clues to FTO gene function include:
- 1. Fto null mice are protected from obesity by increased energy expenditure;
- 2. FTO expression in humans is highest in the brain, particularly the cerebral cortex, and
- 3. Duplication of a chromosomal region containing FTO (and other genes) was associated with mild obesity and mental retardation in a case study.
It was reported recently that a reduction in brain volume in healthy elderly individuals was also associated with the same FTO allele for obesity. Perhaps this is not surprising since obesity is also a risk factor in cognitive decline and dementia. Very rare monogenic causes of obesity include mutations in genes associated with hypothalamic function such as SIM1 , BDNF and NTRK2. These may lead to abnormalities in energy balance resulting in hyperphagia and a net positive energy intake [43] , [44] .
- 2. Syndromal disorders such as Prader-Willi syndrome, Bardet-Biedl syndrome and Pseudohypoparathyroidism type 1A, and
- 3. Complex but common forms of obesity for which the traditional association or GWAS have been used to identify risk alleles [44] .
Genes or gene loci implicated in obesity have been listed in a Human Obesity Gene Map last updated in 2005 [45] . This map provides a summary of published data that are not necessarily confirmed or authenticated but gives a flavor of the rich genetic heterogeneity expected with a complex phenotype such as obesity. Observations made about the 2005 human obesity map include:
- 1. 176 cases involving obesity in humans are due to single gene mutations in 11 genes;
- 2. 253 genetic loci have been reported for obesity from genome wide scans;
- 3. There are 426 findings of positive associations with 127 candidate genes;
- 4. Association studies in 22 genes have been replicated at least five times, and
- 5. There are putative obesity loci on all chromosomes except Y.
Microbiome and Obesity
It is intriguing to recall the observation in Chapter 4 that the gut metagenome shows a characteristic alteration in obese subjects, and so the microbial flora may play a role in obesity that is independent of net calorie intake. In obese humans and animals (mouse, rat and pig) the ratio of the two major bacterial divisions in the gut shows a predominance of Firmicutes over the Bacteroides . This is likely to be a primary rather than secondary effect, because when germ-free mice were fed the microbioata derived from lean or obese mice, the phenotype of the recipient mice moved towards that of the donor mouse – i.e. the obese or lean phenotype was transmissible via the microbiome. One mechanism for this observation may be that the obese microbiome can extract more energy from food [46] . New targets for interventions may be found as the metagenomics story unfolds and more is found about the gut flora and its effects on a range of issues including obesity and inflammation.
Nutrigenetics and Nutrigenomics
Nutrition is a key environmental variable and so any starting point in understanding obesity must encompass nutrition, including its various genetic components. There is a parallel here with pharmacogenetics. Conventional dietary guidelines take consideration of age, sex, height, weight and level of physical activity but not genetic variability. Many of these parameters are used to determine drug dosage, although it is now clear that genetic variability also plays an important role (Chapter 3). A more personalized approach becomes possible through nutrigenetics – how individuals respond differently (because of genetic variation) to the same diet, for example, through changes in blood pressure or serum cholesterol, and nutrigenomics – the role of nutrients and bioactive food compounds in gene expression. The ultimate goal is the development of personalized nutrition options to ensure health and prevent disease [47] . Overarching these goals is the incredible diversity of genetic, cultural and environmental considerations in diet. Nutrigenomics can be approached through many of the omics including genomics, epigenomics, transcriptomics, proteomics, metabolomics and so on.
Diet and Cancer
One can be sure of controversy and robust debate when the influence of diet, nutriceuticals (nutrition + pharmaceutical), complementary medicines or food additives are discussed in relation to cancer development. Knowledge of the link between cancer and diet is not new and numerous research studies provide conflicting data. This is not surprising since individual genetic variability will make the small, multiple but cumulative effects of diet on DNA damage difficult to measure or even replicate, just as association-based studies looking for genetic factors in complex diseases produce conflicting results.
One example is vitamin D deficiency, which is said to cause cancer, although this is very controversial. The US National Cancer Institute confirms a knowledge gap here, stating it does not recommend for or against the use of vitamin D supplements in reducing the risk of cancer. The D2 and D3 forms of vitamin D need to be metabolized to the active 1,25-dihydroxyvitamin D and this involves a number of enzymes (including cytochrome P450 discussed earlier in relation to drug metabolism in Chapter 3). The role of vitamin D in cancer may be better understood through a molecular approach. This is important in view of the successful public health campaigns in reducing the risk of sun-related skin cancers. Interventions recommended include the generous application of sunscreens, avoidance of sun and the wearing of wide brimmed hats, particularly in children. While successful in preventing skin cancers, there is concern (although this is controversial) that vitamin D deficiency may result. If so, there are risks to consider in terms of rickets and related bone problems, and potentially cancer.
The nutrition of cancer cells is also an area of interest. A relevant observation is known as the Warburg effect . O. Warburg was awarded the 1931 Nobel Prize in Physiology or Medicine for discovery of cytochrome C oxidase. He also showed that cancer cells produce lactic acid from glucose even under non-hypoxic conditions; an observation that now bears his name. This is considered to reflect abnormal regulation of glycolysis, since this pathway is very active compared to normal cells, even in the presence of sufficient oxygen [48] . This finding might have implications for new cancer therapy targets and help us to understand better how genes are involved in cancer causation.
The OECD broadly defines bioeconomy as “ the set of economic activities relating to the invention, development, production and use of biological products and processes” . It makes the prediction that biotechnology (in primary production, health and industry) can offer solutions that will lead to the emergence of a bioeconomy. The OECD as an economy-based organization considers greater social benefits globally will come from improving sustainable growth without depleting resources, and labor productivity. The latter can be enhanced through innovation, which is particularly suited to genomics as many of the future developments will be delivered in silico (Chapter 4) and so expensive infrastructure is not necessary. Some examples of how the bioeconomy will benefit from genomics and other omics can be found in Table 6.9 .
Delivering growth and labor productivity through genomics [49] .
The expectation is that the bioeconomy can be used to make substantial socioeconomic contributions to OECD and non-OECD countries, and from this will come better health outcomes, improved productivity of agriculture and industrial processes and enhanced environmental sustainability. In an attempt to optimize the potential of the bioeconomy, the OECD has published a long term (2030) policy agenda [50] .
Note: All web-based references accessed on 21 Feb 2012.
Phone Numbers
Routine and emergency care.
Companion Animal Hospital in Ithaca, NY for cats, dogs, exotics, and wildlife
Equine and Nemo Farm Animal Hospitals in Ithaca, NY for horses and farm animals
Cornell Ruffian Equine Specialists, on Long Island for every horse
Ambulatory and Production Medicine for service on farms within 30 miles of Ithaca, NY
Animal Health Diagnostic Center New York State Veterinary Diagnostic Laboratory
General Information
Cornell University College of Veterinary Medicine Ithaca, New York 14853-6401

Baker Institute for Animal Health
Dedicated to the study of veterinary infectious diseases, immunology, cancer, reproduction, genomics and epigenomics, canine parvovirus.
Canine parvovirus (CPV) is a highly contagious viral disease of dogs that commonly causes acute gastrointestinal illness in puppies. The disease most often strikes in pups between six and 20 weeks old, but older animals are sometimes also affected. A rare variant of the disease may be seen in very young (neonatal) puppies is myocarditis (an inflammation of the heart muscle).
Cause Symptoms and complications Tests and diagnosis Treatment Prevention Additional resources Baker Institute and canine parvovirus
What causes parvovirus infection?
The virus that causes the disease known as “parvo”, canine parvovirus type 2 (CPV), first emerged among dogs in Europe around 1976. By 1978 the virus had spread unchecked, causing a worldwide epidemic of myocarditis and inflammation in the intestines (gastroenteritis). We now know the virus is not limited to dogs, but is capable of causing infections in wild canines such as coyotes and wolves, and other wild animals, including foxes, raccoons and skunks. CPV is closely related to feline panleukopenia virus (FPV), a virus that has been know since the 1920s to infect cats and mink and other animals. CPV probably arose as the result of 2 or 3 genetic mutations in FPV that allowed it to expand its host range to infect dogs.
Three decades after its first appearance, CPV strikes puppies with deadly disease much less frequently due to the development of effective vaccines in the late 1970s, but outbreaks still occur frequently, and vaccinating your dog is of the utmost importance. Puppies and adolescent dogs are especially susceptible to parvovirus, and you should avoid bringing your puppy to public places where there is likely to be lots of virus (animal shelters and kennels) until after their vaccinations are complete.
Why and how might my dog become infected?
Canine parvovirus can be found in almost any environment, but not every dog who comes into contact with the virus becomes infected. Several factors come into play in infection, including the immune status of the dog and the number of viruses the dog is exposed to. If the combination of factors is just right and a dog does become infected, a specific sequence of events is begun as the virus attacks the body.
What happens during infection?
Once a dog or puppy is infected, there is an incubation period of three to seven days before the onset of first symptoms. Inside the dog, CPV needs the help of rapidly dividing cells in order to successfully cause disease, and the virus usually begins by attacking the tonsils or lymph nodes of the throat. Once inside the lymph nodes, the virus typically invades lymphocytes (a type of white blood cell) for one or two days, creating many copies of itself. These viruses hitch a ride inside the lymphocytes, where they are sheltered from the host defenses, and enter the bloodstream. Many of these CPV-infected lymphocytes are ultimately killed, causing a reduction in the number of circulating lymphocytes, a condition called lymphopenia.
Once in the bloodstream, the virus again targets rapidly dividing cells, hitting hardest in the bone marrow and in the cells that line the walls of the small intestine. In very young dogs, CPV can also infect the heart, leading to inflammation of heart muscle, poor function, and arrhythmias.
In the bone marrow, the virus weakens the body’s ability to protect itself by destroying young immune cells and causing a drop in the protective white blood cell count. This probably makes it significantly easier for the virus to invade the gastrointestinal tract, where the virus does its worst damage.
The virus causes this destruction by targeting the epithelium of the small intestine, the lining that helps to absorb nutrients and provides a crucial barrier against fluid loss and bacterial invasion from the gut into the body. The cells that make up the epithelial surface are short-lived and are replaced continually by new cells born in the rapidly-dividing areas known as the crypts of Lieberkühn. The virus invades these crypts where new epithelial cells are born and disables the body’s ability to replenish the intestinal surface.
By preventing the replacement of old and dying cells with fresh new cells, the virus leaves the intestinal surface unable to adequately absorb nutrients, prevent fluid loss into the stool, or prevent bacteria from moving from the gut into the body. Severe diarrhea and nausea are the initial result, but eventually the intestinal surface can become so damaged that it begins to break down, and the bacteria that are normally confined to the gut penetrate the intestine walls and enter the bloodstream. This causes both significant fluid loss from diarrhea and widespread infection inside the body. To make matters worse, the body’s immune system is already weakened, as its ability to produce new white blood cells to combat infection has been hampered by the invasion of CPV into the bone marrow. CPV is not always fatal, but when it does kill, death is as a result of either dehydration and shock, along with the effects of septic toxins produced by the intestinal bacteria roaming throughout the bloodstream.
Symptoms and complications
Symptoms often associated with CPV include lethargy, depression, and loss or lack of appetite, followed by a sudden onset of high fever, vomiting, and diarrhea. If your dog is experiencing bouts of bloody diarrhea and/or vomiting, CPV is only one of several potential culprits. Your veterinarian can run several tests to help determine whether your dog is infected with CPV.
Tests and diagnosis
How will my vet diagnose cpv.
By far the most common and most convenient method of testing for the presence of CPV is the fecal ELISA test. ELISA is an acronym for e nzyme- l inked i mmunosorbent a ssay, a technology is similar to that used in home pregnancy tests. In an ELISA test, antibodies to parvovirus are immobilized on the surface of a testing chamber. A fecal sample is added to the chamber, and antibodies attach to parvovirus proteins that may be present in the stool. A color-changing chemical is then added to the chamber, and if parvoviruses have attached to the antibodies, the chemical will change color and indicate a “positive” result. CPV fecal ELISA tests can usually be completed by your veterinarian in less than 15 minutes. Though the ELISA test is fairly accurate, it is can occasionally produce false positive or false negative results, so further testing may be necessary to confirm a diagnosis.
Veterinarians may also rely on a test that uses a techniques called polymerase chain reaction (PCR) to diagnose CPV from fecal samples. The CPV fecal PCR test detects small pieces of viral DNA that are specific to CPV in the stool of an infected dog. This test is very accurate (more so than CPV fecal ELISA), but requires that a fecal sample be sent to a laboratory that specializes in performing PCR-based testing, so it generally requires more time than a CPV fecal ELISA.
A simple measure of white blood cell count is often the clincher for a CPV diagnosis. Because one of the first things the parvovirus infects is the bone marrow, a low white blood cell count can be suggestive of CPV infection. If a dog has both a positive ELISA reading and a low white blood cell count, a fairly confident diagnosis of CPV may be made.
What are the treatment options for dogs with CPV?
Treatment options for dogs suffering from CPV involve supportive care and management of symptoms. Treatment options will vary, depending on how sick the dog is, but certain aspects are considered vital for all patients.
A hospital stay is often necessary so that the dog can receive intravenous fluids and nutrients to replace the vast quantities lost via vomiting and diarrhea. An intravenous drip is preferred because the digestive tract of stricken dogs is usually in distress and can’t tolerate or absorb what the dog needs. Blood transfusions may also be helpful to boost low blood cell counts that may result from CPV infecting the bone marrow.
Antibiotics may be appropriate therapy for a dog suffering from CPV, administered either intravenously or as injections, to help fight the infection if intestinal bacteria have entered the bloodstream. In addition, medications to control nausea and diarrhea are sometimes useful. Many dogs will respond to medical therapy if it is initiated in a timely fashion, and those dogs that recover from CPV infection retain lifelong protective immunity against the strain that infected them.
How do I vaccinate my pet against CPV?
Since the advent of a number of effective canine vaccinations for CPV, this infectious disease has become much less of a threat to dogs. This does not mean, however, that CPV does not remain a serious problem, and vaccination of your dog should not be considered an option – it is a must.
Veterinarians usually administer the CPV vaccine as part of a combination shot which includes, among others, the distemper, canine adenovirus, and parainfluenza vaccines. These shots are given every 3 to 4 weeks from the time a puppy is 6 weeks old until he is at least 16 weeks of age. A booster vaccination is recommended one year later, and then at one at three year intervals thereafter.
How else can I help prevent the disease?
The tiny parvovirus is extraordinarily hardy. They are capable of surviving for months outside an animal, even through the winter, and are resistant to most household cleaning products. Infected dogs can shed vast numbers of viruses, making it difficult to disinfect an area once it has been exposed to an infected dog. These facts highlight the importance of isolating any dog that is infected with CPV from other dogs. Given the fact that most environments (including dog parks, lawns, and even homes) are not cleaned with disinfecting products regularly, a puppy can be exposed to CPV without any warning, making the vaccine protection all the more important.
If your home and yard have been contaminated by an infected dog, there are steps you can take to disinfect them before introducing a new dog or puppy. Despite its relative resistance to cleaning agents, we do know that CPV can be inactivated by bleach. Cleaning with a solution of one part bleach mixed with approximately 30 parts water is an acceptable method for disinfecting any indoor area (including bedding, food/water bowls, and all surfaces) that once housed an infected dog. There is evidence suggesting that CPV loses some of its ability to infect an animal after one month in an indoor environment. Outside, you cannot (and should not) bleach your lawn, but rain or watering can dilute the concentration of the virus over time. This dilution, combined with the sanitizing effects of sunlight can bring the numbers of viruses down to an acceptable level in a few weeks.
Additional Resources
The AVMA brochure on parvovirus provides a brief overview of what pet owners can expect in canine parvovirus infections.
A more detailed resource for owners can be found at VeterinaryPartner.com .
For veterinarians, the Merck Veterinary Manual provides a comprehensive chapter on parvoviral infection .
Baker Institute and canine parvovirus
The Baker Institute for Animal Health has a long history of working to prevent and treat canine parvovirus infection. The virus first emerged in the United States, Europe, Asia and Australia in 1978, when a virus similar to feline panleukopenia virus crossed over from cats to cause a new type of disease among domestic dogs. Within two years the virus had spread worldwide, killing thousands of dogs and possibly infecting millions more. Baker Institute scientists, including Drs. Leland Carmichael and Max Appel, first isolated the virus later that same year, and by 1979 had developed the first vaccine for parvo. By 1981, Baker Institute scientists had created an improved attenuated vaccine for the disease. Today, Dr. Colin Parrish continues to study the virus and its evolution in order to determine whether existing vaccines provide adequate protection from modern strains of CPV.
For more on Parrish's recent research, such as the following paper recently published, visit our PUBLICATIONS .
Voorhees Ian EH, Lee Hyunwook, Allison Andrew B, Lopez-Astacio Robert, Goodman Laura B, Oyesola Oyebola O, Omobowale Omobowale, Fagbohun Olusegun, Dubovi Edward J, Hafenstein Susan L, Holmes Edward C, Parrish Colin R . Limited Intrahost Diversity and Background Evolution Accompany 40 Years of Canine Parvovirus Host Adaptation and Spread. J Virol. 2019 Dec 12;94(1).
Connect with the Baker Institute on Twitter and Facebook:

IMAGES
VIDEO
COMMENTS
The same can now be said of the field of infectious diseases, particularly with the tools we now have for responding to emerging infectious diseases, such as the rapid and high-throughput ...
This points to a possible new era of infectious disease, ... but this remains conjectural and an important area for research. The changing global context may allow existing human pathogens to both ...
A new partnership between researchers at the University of Kentucky will combat the threat of infectious diseases through research directed at prevention and treatment. The Consortium for Understanding and Reducing Infectious Diseases in Kentucky (CURE-KY) will foster multidisciplinary collaborations to address the burden of infectious diseases ...
NIAID conducts and supports research—at NIH, throughout the United States, and worldwide—to study the causes of infectious and immune-mediated diseases, and to develop better means of preventing, diagnosing and treating these illnesses. News releases, fact sheets and other NIAID-related materials are available on the NIAID website.
The twenty-first century has already recorded more than ten major epidemic or pandemic virus emergence events, including the ongoing and devastating coronavirus disease 2019 (COVID-19) pandemic.
Subjects. Vaccines have revolutionized modern medicine by preventing infectious diseases and safeguarding public health. This Collection showcases cutting-edge research on advancements in vaccine ...
Infectious diseases caused by organisms (bacteria, viruses, fungi, etc.) are among the top leading causes of death. The emergence of new bacteria and new viruses, such as SARS-Cov-2, has been identified as a threat to global public health with high economic and social impacts. Although SARS-CoV-2 primarily targets the respiratory system, other lasting health effects, such as heart ...
Infectious Diseases. Explore the latest in infectious diseases, including community-acquired and nosocomial disease, antibiotic use and stewardship, and more. This cohort study examines whether systemic fluoroquinolone use is associated with increased risk of acute uveitis or retinal detachment. A 43-year-old man presented with a destructive ...
An HIV vaccine candidate elicited trace levels of HIV broadly neutralizing antibodies and high levels of other key immune cells in an early-stage clinical trial. This immune response is an important signal that, if antibody levels can be further amplified, the vaccination strategy might be able to prevent HIV.
The treatment of infectious diseases has a pivotal effect on the life quality of infected individuals, ... new human infectious diseases are usually caused by pathogens that originally circulated only in wild animals and, ... the role of IFN is an important target for basic research and clinical trials ...
In a paper titled "tRNA modification reprogramming contributes to artemisinin resistance in Plasmodium falciparum", published in the journal Nature Microbiology, researchers from SMART's Antimicrobial Resistance (AMR) interdisciplinary research group documented their discovery: A change in a single tRNA, a small RNA molecule that is involved in translating genetic information from RNA to ...
Emerging Infectious Diseases is a peer-reviewed, monthly journal published by the Centers for Disease Control and Prevention (CDC). It offers global health professionals the latest scientific information on emerging infectious diseases and trends. Articles provide the most up-to-date information on infectious diseases and their effects on global health.
This points to a possible new era of infectious disease, defined by outbreaks of emerging, re-emerging and endemic pathogens that spread quickly, aided by global connectivity and shifted ranges owing to climate change (Fig. 1d ). Fig. 1. Human connectivity and infectious disease outbreaks in premodern and modern times.
DOI: 10.1056/NEJMra1108296. VOL. 366 NO. 5. Interactive Graphic. Selected Infectious Diseases of Importance over 200 Years. Among the many challenges to health, infectious diseases stand out for ...
Overview. Universal health coverage (UHC) and the primary health care (PHC) approach are essential to reduce the enormous global burden of communicable diseases. Efforts to reduce the prevalence of these diseases, which include malaria, tuberculosis, HIV/AIDS, hepatitis, sexually transmitted infections and neglected tropical diseases (NTDs ...
The coronavirus (COVID-19) pandemic has highlighted that individuals with behavioural risk factors commonly associated with non-communicable diseases (NCDs), such as smoking, harmful alcohol use, obesity, and physical inactivity, are more likely to experience severe symptoms from COVID-19. These risk factors have been shown to increase the risk of NCDs, but less is known about their broader ...
A 2020 WHO survey found that the ongoing COVID-19 pandemic was disrupting non-communicable disease (NCD) services in 122 (77%) of 159 countries surveyed. COVID-19 is disproportionately affecting people living with NCDs, exacerbating inequalities1 and limiting interventions to control tobacco and alcohol use, create healthy diets, and promote physical activity. NCDs are already responsible for ...
One of the major functions of the immune system is to — through antibodies — fight infections. New findings from The Feinstein Institutes for Medical Research bioelectronic medicine scientists show that neurons that help sense pain and prevent illness, called sensory neurons, play an important role in regulating the production of antibodies.. The study published today in the journal ...
LEXINGTON, Ky. (May 29, 2024) — A new partnership between researchers at the University of Kentucky will combat the threat of infectious diseases through research directed at prevention and treatment. The Consortium for Understanding and Reducing Infectious Diseases in Kentucky (CURE-KY) will foster multidisciplinary collaborations to address ...
The modern-day practice of surveillance has a broader interest of public health. Public health surveillance is important for the prevention and control of both communicable and noncommunicable diseases. It is a vital tool in the immediate detection of epidemics, in the determination of risk factors and population at risk, and in assessing the ...
Emerging infectious diseases can be defined as infectious diseases that have newly appeared in a population or have existed but are rapidly increasing in incidence or geographic range. Research on microbial- or plant-derived toxins or antimicrobial resistance may also be included. Bacteria. Bacillus anthracis (anthrax) Bordetella pertussis (new ...
Antimicrobial resistance is an increasing threat to public health and encouraging the development of new antimicrobials is one of the most important ways to address the problem. This Roadmap ...
At the National Institute of Allergy and Infectious Diseases (NIAID) at NIH, we have increased funding for emerging diseases from $39.3 million in fiscal year 1993 to an estimated (president's budget) $85.0 million in fiscal 1999 ().Approximately 21% of the NIAID non-AIDS infectious diseases budget is devoted to emerging infectious diseases.
Summary. A communicable disease is one that spreads from one person or animal to another or from a surface to a person. They are the result of pathogens, such as viruses and bacteria. Communicable ...
For the eight million people across the world, including the 100,000 in the U.S., who have sickle cell disease, these gene therapies could be life-changers. Yet immediately after the approvals ...
A drug approved to treat certain autoimmune diseases and cancers successfully alleviated symptoms of a rare genetic syndrome called autoimmune polyendocrine syndrome type 1 (APS-1). Researchers identified the treatment based on their discovery that the syndrome is linked to elevated levels of interferon-gamma (IFN-gamma), a protein involved in immune system responses, providing new insights ...
At a celebration event on the sidelines of the 77th World Health Assembly (WHA), partners and donors gathered to commemorate 50 years since the WHA passed a resolution that called for the establishment of a research programme now known as TDR. Distinguished guests included the Minister of Public Health of Cuba, the Director General of Ethiopia's Armauer Hansen Research Institute and the ...
Nanotechnology has the potential to transform both detection and treatment of a wide range of diseases. These technologies, involving systems with a diameter of about one-thousandth of the ...
Introduction. Public health is a community-based strategy to improve health and well being, and to prevent disease through research, policy, education and appropriate practice. It is very different to personalized medicine which focuses on the individual. Common to both is the potential for DNA-based information to enhance clinical care. Fundamental to the practice of public health is ...
Canine parvovirus (CPV) is a highly contagious viral disease of dogs that commonly causes acute gastrointestinal illness in puppies. The disease most often strikes in pups between six and 20 weeks old, but older animals are sometimes also affected. A rare variant of the disease may be seen in very young (neonatal) puppies is myocarditis (an inflammation of the heart muscle).