- Open access
- Published: 29 October 2020

The genome editing revolution: review
- Ahmad M. Khalil ORCID: orcid.org/0000-0002-1081-7300 1
Journal of Genetic Engineering and Biotechnology volume 18 , Article number: 68 ( 2020 ) Cite this article
38k Accesses
121 Citations
Metrics details
Development of efficient strategies has always been one of the great perspectives for biotechnologists. During the last decade, genome editing of different organisms has been a fast advancing field and therefore has received a lot of attention from various researchers comprehensively reviewing latest achievements and offering opinions on future directions. This review presents a brief history, basic principles, advantages and disadvantages, as well as various aspects of each genome editing technology including the modes, applications, and challenges that face delivery of gene editing components.
Genetic modification techniques cover a wide range of studies, including the generation of transgenic animals, functional analysis of genes, model development for diseases, or drug development. The delivery of certain proteins such as monoclonal antibodies, enzymes, and growth hormones has been suffering from several obstacles because of their large size. These difficulties encouraged scientists to explore alternative approaches, leading to the progress in gene editing. The distinguished efforts and enormous experimentation have now been able to introduce methodologies that can change the genetic constitution of the living cell. The genome editing strategies have evolved during the last three decades, and nowadays, four types of “programmable” nucleases are available in this field: meganucleases, zinc finger nucleases, transcription activator-like effector nucleases, and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) (CRISPR/Cas-9) system. Each group has its own characteristics necessary for researchers to select the most suitable method for gene editing tool for a range of applications. Genome engineering/editing technology will revolutionize the creation of precisely manipulated genomes of cells or organisms in order to modify a specific characteristic. Of the potential applications are those in human health and agriculture. Introducing constructs into target cells or organisms is the key step in genome engineering.
Conclusions
Despite the success already achieved, the genome editing techniques are still suffering certain difficulties. Challenges must be overcome before the full potential of genome editing can be realized.
In classical genetics, the gene-modifying activities were carried out selecting genetic sites related to the breeder’s goal. Subsequently, scientists used radiation and chemical mutagens to increase the probability of genetic mutations in experimental organisms. Although these methods were useful, they were time-consuming and expensive. Contrary to this, reverse genetics goes in the opposite direction of the so-called forward genetic screens of classical genetics. Reverse genetics is a method in molecular genetics that is used to help understanding the function of a gene by analyzing the phenotypic effects of specific engineered gene sequences. Robb et al. [ 68 ] defined and compared the three terms: “genome engineering”, “genome editing”, and “gene editing”. Genome engineering is the field in which the sequence of genomic DNA is designed and modified. Genome editing and gene editing are techniques for genome engineering that incorporate site-specific modifications into genomic DNA using DNA repair mechanisms. Gene editing differs from genome editing by dealing with only one gene.
This review briefly presents the evolution of genome editing technology over the past three decades using PubMed searches with each keyword of genome-editing techniques regarding the brief history, basic principles, advantages and disadvantages, as well as various aspects of each genome editing technology including the modes, future perspective, applications, and challenges.
Genome-wide editing is not a new field, and in fact, research in this field has been active since the 1970s. The real history of this technology started with pioneers in genome engineering [ 36 , 59 ]. The first important step in gene editing was achieved when researchers demonstrated that when a segment of DNA including homologous arms at both ends is introduced into the cell, it can be integrated into the host genome through homologous recombination (HR) and can dictate wanted changes in the cell [ 10 ]. Employing HR alone in genetic modification posed many problems and limitations including inefficient integration of external DNA and random incorporation in undesired genomic location. Consequently, the number of cells with modified genome was low and uneasy to locate among millions of cells. Evidently, it was necessary to develop a procedure by which scientists can promote output. Out of these limitations, a breakthrough came when it was figured out that, in eukaryotic cells, more efficient and accurate gene targeting mechanisms could be attained by the induction of a double stranded break (DSB) at a specified genomic target [ 70 ].
Furthermore, scientists found that if an artificial DNA restriction enzyme is inserted into the cell, it cuts the DNA at specific recognition sites of double-stranded DNA (dsDNA) sequences. Thus, both the HR and non-homologous end joining (NHEJ) repair can be enhanced [ 14 ]. Various gene editing techniques have focused on the development and the use of different endonuclease-based mechanisms to create these breaks with high precision procedures [ 53 , 78 ] (Fig. 1 ). The mode of action of what is known as site-directed nucleases is based on the site-specific cleavage of the DNA by means of nuclease and the triggering of the cell’s DNA repair mechanisms: HR and NHEJ.
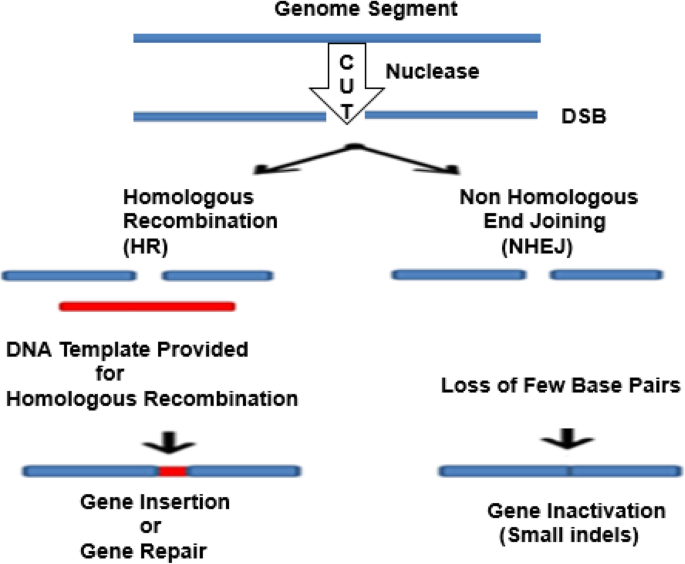
Genome editing outcomes. Genome editing nucleases induce double-strand breaks (DSBs). The breaks are repaired through two ways: by non-homologous end joining (NHEJ) in the absence of a donor template or via homologous recombination (HR) in the presence of a donor template. The NHEJ creates few base insertions or deletion, resulting in an indel, or in frameshift that causes gene disruption. In the HR pathway, a donor DNA (a plasmid or single-stranded oligonucleotide) can be integrated to the target site to modify the gene, introducing the nucleotides and leading to insertion of cDNA or frameshifts induction. (Adapted from [ 78 ])
One of the limitations in this procedure is that it has to be activated only in proliferating cells, adding that the level of activity depends on cell type and target gene locus [ 72 ]. Tailoring of repair templates for correction or insertion steps will be affected by these differences. Several investigations have determined ideal homology-directed repair (HDR) donor configurations for specific applications in specific models systems [ 67 ]. The differences in the activities of the DNA repair mechanisms will also influence the efficiency of causing indel mutations through NHEJ or the classical microhomology-mediated end joining (c-MMEJ) pathway, and even the survival of the targeted cells. The production of such repair in the cell is a sign of a characteristic that errors may occur during splicing the ends and cause the insertion or deletion of a short chain. Simply speaking, gene editing tools involve programmed insertion, deletion, or replacement of a specific segment of in the genome of a living cell. Potential targets of gene editing include repair of mutated gene, replacement of missing gene, interference with gene expression, or overexpression of a normal gene.
The human genome developments paved the way to more extensive use of the reverse genetic analysis technique. Nowadays, two methods of gene editing exist: one is called “targeted gene replacement” to produce a local change in an existing gene sequence, usually without causing mutations. The other one involves more extensive changes in the natural genome of species in a subtler way.
In the field of targeted nucleases and their potential application to model and non-model organisms, there are four major mechanisms of site-specific genome editing that have paved the way for new medical and agricultural breakthroughs. In particular, meganucleases (MegNs), zinc finger nucleases (ZFNs), transcription activator-like effector nuclease (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) (CRISPR/Cas-9) (Fig. 2 ).
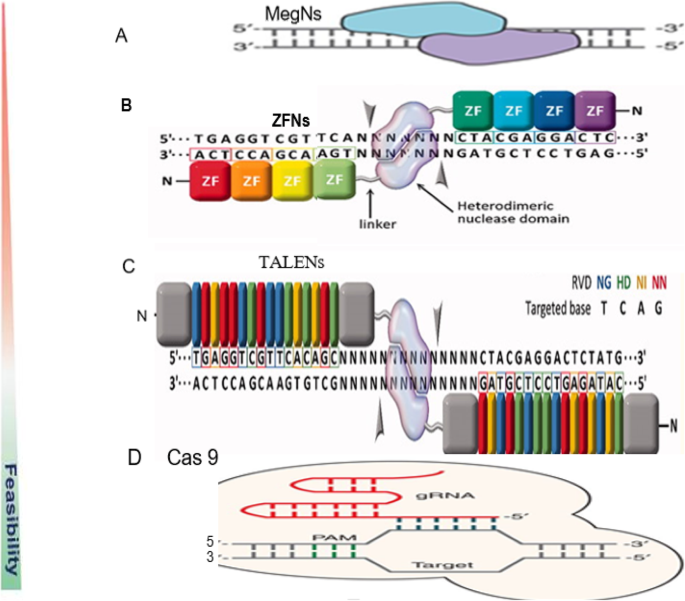
Schematic diagram of the four endonucleases used in gene editing technologies. a Meganuclease (MegN) that generally cleaves its DNA substrate as a homodimer. b Zinc finger nuclease (ZFN) recognizes its target sites which is composed of two zinc finger monomers that flank a short spacer sequence recognized by the FokI cleavage domain. c Transcription activator-like effector nuclease (TALEN) consists of two monomers; TALEN recognizes target sites which flank a fok1 nuclease domain to cut the DNA. d CRISPR/Cas9 system is made of a Cas9 protein with two nuclease domains: human umbilical vein endothelium cells (HuvC) split nuclease and the HNH, an endonuclease domain named for the characteristic histidine and asparagine residue, as well as a single guide RNA (sgRNA). (Adapted from [ 1 , 51 ]; Gaj et al., 2016 [ 53 ];)
Meganucleases (MegNs)
Meganucleases (MegNs) are naturally occurring endodeoxyribonucleases found within all forms of microbial life as well as in eukaryotic mitochondria and chloroplasts. The genes that encode MegNs are often embedded within self-splicing elements. The combination of molecular functions is mutually advantageous: the endonuclease activity allows surrounding introns and inteins to act as invasive DNA elements, while the splicing activity allows the endonuclease gene to invade a coding sequence without disrupting its product. The high specificity of these enzymes is based on their ability to cleave dsDNA at specific recognition sites comprising 14–40 bp (Fig. 2 a). Unlike restriction enzymes, which provide defenses to bacteria against invading DNA, MegNs facilitate lateral mobility of genetic elements within an organism. This process is referred to as “homing” and gives the name homing endonucleases to these enzymes. The high DNA specificity of MegNs makes them a powerful protein scaffold to engineer enzymes for genome manipulation. A deep understanding of their molecular recognition of DNA is an important prerequisite to generate engineered enzymes able to cleave DNA in specific desired genome sites. Crystallographic analyses of representatives from all known MegNs families have illustrated both their mechanisms of action and their evolutionary relationships to a wide range of host proteins. The functional capabilities of these enzymes in DNA recognition vary widely across the families of MegNs. In each case, these capabilities, however, make a balance between what is called orthogonal requirements of (i) recognizing a target of adequate length to avoid overt toxicity in the host, while (ii) accommodating at least a small amount of sequence drift within that target. Indirect readout in protein-DNA recognition is the mechanism by which the protein achieves partial sequence specificity by detecting structural features on the DNA.
Several homing endonucleases have been used as templates to engineer tools that cleave DNA sequences other than their original wild-type targets.
Meganucleases can be divided into five families based on sequence and structure motifs: LAGLIDADG, GIY-YIG, HNH, His-Cys box, and PD-(D/E) XK [ 74 ]. I-CreI is a homodimeric member of MegNs family, which recognizes and cleaves a 22-bp pseudo-palindromic target (5′-CAAAACGTCGTGAGACAGTTTG-3′). The important role of indirect readout in the central region of the target DNA of these enzymes I-CreI suggested that indirect readout may play a key role in the redesign of protein-DNA interactions. The sequences of the I-CreI central substrate region, four bp (± 1 and ± 2) called 2NN, along with the adjacent box called 5NNN, are key for substrate cleavage [ 64 ]. Changes in 2NN significantly affect substrate binding and cleavage because this region affects the active site rearrangement, the proper protein-DNA complex binding, and the catalytic ion positioning to lead the cleavage.
An exhaustive review of each MegN can be found in Stoddard [ 75 ] as well as in Petersen and Niemann [ 63 ]. Several MegNs have been used as templates to engineer tools that cleave DNA sequences other than their original wild-type targets. This technology have advantages of high specificity of MegNs to target DNA because of their very long recognition sites, ease in delivery due to relatively small size, and giving rise to more recombinant DNA (i.e., more recombinogenic for HDR) due to production of a 3′ overhang after DNA cleavage. This lowers the potential cytotoxicity [ 53 , 78 ].
Meganucleases have several promising applications; they are more specific than other genetic editing tools for the development of therapies for a wide range of inherited diseases resulting from nonsense codons or frameshift mutations. However, an obvious drawback to the use of natural MegNs lies in the need to first introduce a known cleavage site into the region of interest. Additionally, it is not easy to separate the two domains of MegNs: the DNA-binding and the DNA-cleavage domains, which present a challenge in its engineering. Another drawback of MegNs is that the design of sequence-specific enzymes for all possible sequences is time-consuming and expensive. Therefore, each new genome engineering target requires an initial protein engineering step to produce a custom MegN. Thus, in spite of the so many available MegNs, the probability of finding an enzyme that targets a desired locus is very small and the production of customized MegNs remains really complex and highly inefficient. Therefore, routine applications of MegNs in genome editing is limited and proved technically challenging to work with [ 24 ].
Zinc finger nucleases (ZFNs)
The origin of genome editing technology began with the introduction of zinc finger nucleases (ZFNs). Zinc finger nucleases are artificially engineered restriction enzymes for custom site-specific genome editing. Zinc fingers themselves are transcription factors, where each finger recognizes 3–4 bases. Zinc finger nucleases are hybrid heterodimeric proteins, where each subunit contains several zinc finger domains and a Fok1 endonuclease domain to induce DSB formation. The first is zinc finger, which is one of the DNA binding motifs found in the DNA binding domain of many eukaryotic transcription factors responsible for DNA identification. The second domain is a nuclease (often from the bacterial restriction enzyme FokI) [ 6 ]. When the DNA-binding and the DNA-cleaving domains are fused together, a highly specific pair of “genomic scissors” is created (Fig. 2b ). In principle, any gene in any organism can be targeted with a properly designed pair of ZFNs. Zinc finger recognition depends only on a match to DNA sequence, and mechanisms of DNA repair, both HR and NHEJ, are shared by essentially all species. Several studies have reported that ZFNs with a higher number of zinc fingers (4, 5, and 6 finger pairs) have increased the specificity and efficiency and improved targeting such as using modular assembly of pre-characterized ZFs utilizing standard recombinant DNA technology.
Since they were first reported [ 41 ], ZFN was appealing and showed considerable promise and they were used in several living organisms or cultured cells [ 11 ]. The discovery of ZFNs overcame some of the problems associated with MegNs applications. They facilitated targeted editing of the gene by inducing DSBs in DNA at specific sites. One major advantage of ZFNs is that they are easy to design, using combinatorial assembly of preexisting zinc fingers with known recognition patterns. This approach, however, suffered from drawbacks for routine applications. One of the major disadvantages of the ZFN is what is called “context-dependent specificity” (how well they cleave target sequence). Therefore, these specificities can depend on the context in the adjacent zinc fingers and DNA. In other terms, their specificity does not only depend on the target sequence itself, but also on adjacent sequences in the genome. This issue may cause genome fragmentation and instability when many non-specific cleavages occur. It only targets a single site at a time and as stated above. Although the low number of loci does not usually make a problem for knocking-out editing, it poses limitation for knocking in manipulation [ 32 ]. In addition, ZFNs cause overt toxicity to cells because of the off-target cleavages. The off-target effect is the probability of inaccurate cut of target DNA due to single nucleotide substitutions or inappropriate interaction between domains.
Transcription activator-like effector nucleases (TALENs)
The limitations mentioned in the previous section paved the way for the development of a new series of nucleases: transcription activator-like effector nucleases (TALENs), which were cheaper, safer, more efficient, and capable of targeting a specified region in the genome [ 13 ].
In principle, the TALENs are similar to ZFNs and MegNs in that the proteins must be re-engineered for each targeted DNA sequence. The ZFNs and TALENs are both modular and have natural DNA-binding specificities. The TALEN is similar to ZFN in that it is an artificial chimeric protein that result from fusing a non-specific FokI restriction endonuclease domain to a DNA-binding domain recognizing an arbitrary base sequence (Fig. 2c ). This DNA-binding domain consists of highly conserved repeats derived from transcription activator-like effectors (TALE). When genome editing is planned, a pair of TALEN is used like ZFNs. The TALE protein made of three domains: an amino-terminal domain having a transport signal, a DNA-binding domain which is made of repeating sequences of 34 amino acids arranged in tandem, and a carboxyl-terminal domain having a nuclear localization signal and a transcription activation domain. Of the 34 amino acids, there is a variable region of two amino acid residues located at positions 12 and 13 called repeat variable di-residues (RVD). This region has the ability to confer specificity to one of the any four nucleotide bps [ 15 ].
Unlike ZFNs, TALENs had advantages in that one module recognizes just one nucleotide in its DNA-binding domain, as compared with 3 bps recognized by the first single zinc finger domains [ 39 ]. So, interference of the recognition sequence does not occur even when several modules are joined. In theory, because cleavage of the target sequence is more specific than ZFN, it became possible to target any DNA sequence of any organism genome. This difference facilitates creation of TALEN systems which recognize more target sequences. Another benefit of the TALEN system over ZFN’s for genome editing is that the system is more efficient in producing DSBs in both somatic cells and pluripotent stem cells [ 35 ]. In addition, TALENs exhibit less toxicity in human cell lines due to off-target breaks that result in unwanted changes and toxicity in the genome. Another advantage of TALENs is a higher percentage of success in genome editing through cytoplasmic injection of TALEN mRNA in livestock embryos than observed with ZFN induction [ 39 ]. In addition, TALENs have been more successfully used in plant genome engineering [ 88 ]. It is hoped that TALENs will be applied in the generation of genetically modified laboratory animals, which may be utilized as a model for human disease research [ 24 , 39 ].
The TALEN-like directed development of DNA binding proteins was employed to improve TALEN specificity by phage-assisted continuous evolution (PACE). The improved version was used to create genetically modified organisms [ 34 ]. Nucleases which contain designable DNA-binding sequences can modify the genomes and have the promise for therapeutic applications. DNA-binding PACE is a general strategy for the laboratory evolution of DNA-binding activity and specificity. This system can be used to generate TALEN with highly improved DNA cutting specificity, establishing DB-PACE as a diverse approach for improving the accuracy of genome editing tools. Thus, similar to ZFN, TALEN is used for DSBs as well as for knocking in/knocking out. In comparison with the ZFN, two important advantages for this editing technique have been reported: first, the simple design, and second, the low number of off-target breaks [ 35 ].
In spite of the improvement and simplification of the TALEN method, it is complicated for whom not familiar with molecular biological experiments. Moreover, it is confronted with some limitations, such as their large size (impeding delivery) in comparison to ZFN [ 24 , 39 ]. The superiority of TALEN relative to ZFN could be attributed to the fact that in the TALEN each domain recognizes only one nucleotide, instead of recognizing DNA triplets in the case of ZEF. The design of TALEN is commonly more obvious than ZNF. This results in less intricate interactions between the TALEN-derived DNA-binding domains and their target nucleotides than those among ZNF and their target trinucleotides [ 35 , 39 ].
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)
The CRISPR/Cas system is the most recent platform in the field of genome editing. The system was developed in 2013 and is known as the third generation genomic editing tools. The clustered regularly interspaced short palindromic repeats, which are sometimes named “short regularly spaced repeats” were discovered in the 1980s. Computational analysis of these elements showed they were found in more than 40% of sequenced bacteria and 90% of archaea [ 37 , 56 ]. The acronym CRISPR was suggested, and a group of genes adjacent to the CRISPR locus, which was termed “CRISPR-associated system”, or Cas was established [ 37 ]. Cas proteins coded by these genes carry functional domains similar to endonucleases, helicases, polymerases, and nucleotide-binding proteins. In addition, the role of CRISPRs as bacterial and archaeal adaptive immunity system against invading bacteriophages and other and in DNA repair was realized [ 17 , 77 ].
Unlike the two previous technologies (ZFN and TALEN), in which the recognition of the DNA site was based on the sequence recognition by artificial proteins requiring interaction between protein and DNA, the DNA recognition of the CRISPR/Cas system is based on RNA-DNA interactions. This offers several advantages over ZFNs and TALENs. These include easy design for any genomic targets, easy prediction regarding off-target sites, and the probability of modifying several genomic sites simultaneously (multiplexing). CRISPR-Cas systems are diverse and have been classified thus far into two classes, six types, and over 20 subtypes based on locus arrangement and signature cas genes [ 33 , 44 , 51 ]. Types I, III, and IV, with multiprotein crRNA-effector complexes, are class 1 systems; types II, V, and VI, with a single protein-crRNA effector complex, are class 2. All CRISPR-Cas systems require Cas proteins and crRNAs for function, and CRISPR- cas expression is a prerequisite to acquire new spacers, process pre-crRNA, and assemble ribonucleoprotein crRNA interference complexes for target degradation. Herein, we will focus on the CRISPR-Cas9 technology, the reader should keep in mind other available variants of the system such as CRISPR-Cas6 [ 5 ], CRISPR-Cas12a, -Cas12b [ 42 ], as well as the most recently discovered c2c2 (Cas13a) and c2c6 (Cas13b [ 19 , 69 ]. The CRISPR/Cas9 system is made of Cas9 nuclease and single-guide RNA (sgRNA). The sgRNA is an engineered single RNA molecule containing crispr RNA and tracr RNA parts. The sgRNA recognizes the target sequence by standard Watson-Crick base pairing. It has to be followed by a DNA motif called a protospacer adjacent motif (PAM). The commonly used wild-type Streptococcus pyogenes Cas (SpCas9) protein has a specific PAM sequence, 5’-NGG-3’, where “N” can be any nucleotide base followed by two guanine (“G”) nucleobases. This sequence is located directly downstream of the target sequence in the genomic DNA, on the non-target strand. Targeting is constrained to every 14 bp (12 bp from the seed sequence and 2 bp from PAM) [ 15 ]. SpCas9 variants may increase the specificity of genome modifications at DNA targets adjacent to NGG PAM sequences when used in place of wild-type SpCas9.
DNA cleavage is performed by Cas9 nuclease and can result in DSB in the the case of a wild-type enzyme, or in a SSB when using mutant Cas9 variants called nickases (Fig. 2d ). It should be emphasized that the utilization of this approach in editing eukaryotes’ genome only needs the manipulation of a short sequence of RNA, and there is no need for complicated manipulations in the protein domain. This enables a faster and more cost-effective design of the DNA recognition moiety compared with ZFN and TALEN technologies. Applications of CRISPR-Cas9 systems are variable like those for ZFNs, TALENs, and MegNs. But, because of the relative simplicity of this system, its great efficiency and high tendency for multiple functions and library construction, it can be applied to different species and cell types [ 35 ].
As shown in Fig. 3 , in all CRISPR/Cas systems, immunity occurs in three distinct stages [ 77 , 81 ]: (1) adaptation or new spacer acquisition, (2) CRISPR transcription and processing (crRNA generation), and (3) interference or silencing. The advantages of the CRISPR/Cas system superseded those of both of the TALEN and ZFN tools, the ZFN in particular. This is due to its target design simplicity since the target specificity depends on ribonucleotide complex formation and non-protein/DNA recognition. In addition, the CRISPR/Cas approach is more efficient because changes can be introduced directly by injecting RNAs that encode the Cas protein and gRNA into developing embryos. Moreover, multigene mutations can be induced simultaneously by injecting them with multiple gRNAs. This is an example that explains the rapid spread of CRISPR/Cas 9 application in various fields. Still, the system has certain drawbacks. Although the CRISPR/Cas9 is much less complicated than TALEN, in terms of execution and construction, the off-target effect in CRISPR/Cas9 is higher than TALEN. Since the DSB results only after accurate binding of a pair of TALEN to the target sequence, the off-target effect problem is considered to be low. These two are different in restriction of target sequence. CRISPR/Cas9 is much more efficient than TALEN in multiple simultaneous modification. Table 1 compares the three main systems of site-directed synthetic nuclease employed in genome editing: ZFN, TALEN, and CRISPR/Cas9.
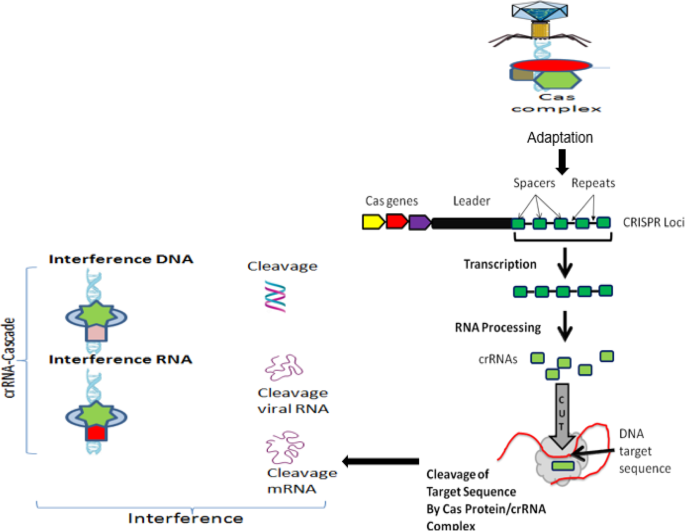
Schematic representation of CRISPR loci and targeting of DNA sequence, which include Cas genes, a leader sequence, and several spacer sequences derived from engineered or foreign DNA that are separated by short direct repeat sequences. The three major steps of CRISPR-Cas immune systems. In the adaptation phase, Cas proteins excise specific fragments from foreign DNA and integrate it into the repeat sequence neighboring the leader at the CRISPR locus. Then, CRISPR arrays are transcribed and processed into multiple crRNAs, each carrying a single spacer sequence and part of the adjoining repeat sequence. Finally, at the interference phase, the crRNAs are assembled into different classes of protein targeting complexes (cascades) that anneal to, and cleave, spacer matching sequences on either invading element or their transcripts and thus destroy them. (Adapted from [ 3 , 53 , 78 ])
The off-target effect is an essential subject for future studies if CRISPR/Cas9 is to achieve its promises as a powerful method for genome editing. Non-specific and unintended genetic modifications (off-target effect) can result from the use of CRISPR/Cas9 system which is one of the drawbacks of this tool. Therefore, this point should be considered for use in researches. One strategy to reduce the off-target activity is to replace the Streptococcus pyogenes Cas9 enzyme (SpyCas9) for a mutant Cas9 nickase (nSpyCas9; ncas9), which cleaves a single strand through the inactivation of a nuclease domain Ruvc or HNH [ 9 ]. Our understanding of off-target effects remains fragmentary. A deeper understanding of this phenomenon is needed. Several approaches that could be followed to characterize the binding domains and consequently Cas9 targeting specificity have been reviewed and summarized [ 83 ].
It has previously been stated that CRISPR/Cas9 system needs both gRNA and PAM to detect its target sequence of interest by integration of a gRNA component that binds to complementary double-stranded DNA sequences. Cell culture studies have shown that off-target effects may be due to the incorrect detection of genomic sequences by sgRNA. This, in turn, affects cleavage when the mismatch is in the vicinity of the PAM (up to 8 bases), but if the PAM is too far apart, these effects will be small [ 4 ], even a slight mismatch between sgRNA and target sequences can lead to a failure. Dependence of this method on specific PAM sequences to act functionally limits the number of target loci, and it can reduce off-target breaks [ 86 ]. For this goal, another type of specific PAM-containing nucleases has been prepared to compensate for this limitation. Genetic engineering and enzyme changing have also been able to overcome the limitation [ 42 ]. For a sgRNA, many similar sequences depending on the genome size of the species may exist [ 86 ]. Interestingly, the initial targeting scrutiny of the CRISPR/Cas9-sgRNA complex showed that not every nucleotide base in the gRNA is necessary to be complementary to the target DNA sequence to effect Cas9 nuclease activity. Regarding that where the similar sequences are found in the genome, their breaks could lead to malignancies or even death [ 86 ]. Various methods have been proposed to prevent off-target breaks, among which the double nicking method, the FokI-dCas9 fusion protein method, and the truncated sgRNA method [ 76 ] (Fig. 4 ).
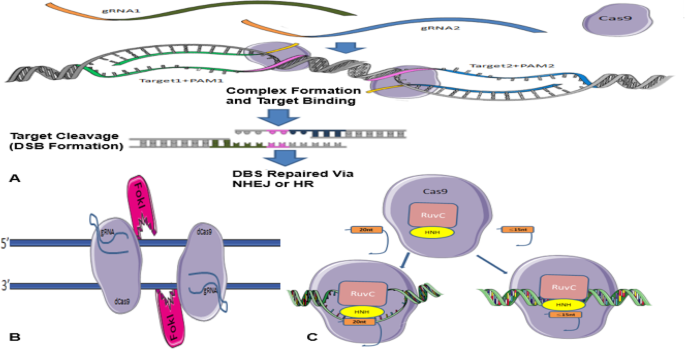
a Summary of the Cas9 nickases methods in efficient genome editing. Two gRNAs target opposite strands of DNA. These double nicks create a DSB that is repaired using non-homologous end joining (NHEJ) or edits via homology-directed repair (HDR) (adapted from www.addgene.org/crispr/nick ). b FokI-dCas 9 fusion protein method. Two FokI-dCas9 fusion proteins are used to adjacent target sites by two different sgRNAs to facilitate FokI dimerization and DNA cleavage. These fusions would have enhanced specificity compared to the standard monomeric Cas9 nucleases and the paired nickase system because they should require two sgRNAs for activity. c Truncated sgRNA method. Cas9 interacting with either a full-length sgRNA (20 nucleotide sequence complementary to target site) or truncated gRNA (less than 15 nucleotide sequence complementary to target site). (Retrieved from blog.addgene.org )
To overcome these problems, researchers explored another generation of base editing technologies, which combine CRISPR and cytidine deaminase (Fig. 5 ). This is a diverse method called CRISPR-SKIP (Fig. 6 ) which uses cytidine deaminase single-base editors to program exon skipping by mutating target DNA bases within splice acceptor sites [ 25 ]. Given its simplicity and precision, CRISPR-SKIP will be widely applicable in gene therapy. Base editing utilizes Cas9 D10A nickases fused to engineered base deaminase enzymes to make single base changes in the DNA sequence without the need of DNA DSB. Also, base editing does not require an external repair template. The Cas9 nickase part of the base editor protein plays a dual function. The first is to target the deaminase activity to the wanted region and the second is to localize the enzyme to certain regions of double-stranded RNA. The deaminase domains in base editors (BEs) occur in two versions: either adenosine deaminase or cytosine deaminase, which catalyze only base transitions (C to T and A to G) and cannot produce base transversions [ 26 , 68 ]. In these base editing tools, the targeted activity of adenosine deaminase can result in an A:T to G:C sequence alteration in a very similar way [ 26 , 68 ].This approach avoided the requirement of breaking DNA to induce an oligonucleotide. In addition, compared to knocking system, it exerted a higher output with lower off-targets [ 40 , 43 ]. Adenosine is deaminated to inosine (I) that is subsequently utilized to repair the nicked strand with a cytosine, and the I:C base pair is resolved to G:C [ 26 ]. More recently, new genome editing technologies have been developed: glycosylase base editors (GBEs), which consist of a Cas9 nickase, a cytidine deaminase, and a uracil-DNA glycosylase (Ung), are capable of transversion mutations by changing C to A in bacterial cells and from C to G in mammalian cells [ 45 , 89 ]. The new BEs can also be designed to minimize unwanted (“off-target”) mutations that could potentially cause undesirable side effects. The novel BE platform may help researchers understand and correct genetic diseases by selective editing of single DNA “alphabets” across nucleobase classes. However, the technique with this new class of transversion BEs is still at an early stage and requires additional optimization, so it would be premature to say this is ready for the clinic applications.
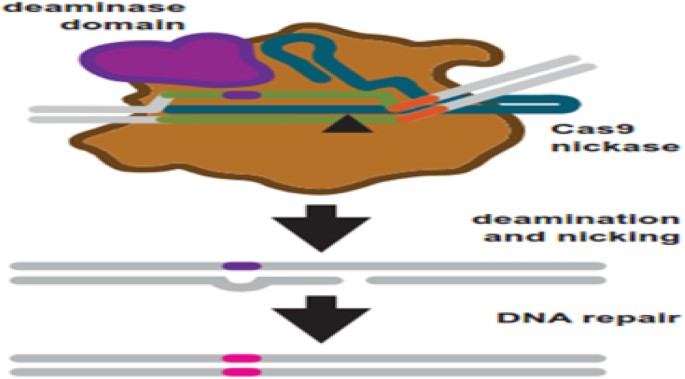
Base editing uses engineered Cas9 variants to induce base changes in a target sequence. Cas9 nickase is fused to a base deaminase domain. The deaminase domain works on a targeted region within the R-loop after target binding and R-loop formation. Simultaneously, the target strand is nicked. DNA repair is started in response to the nick using the strand which contains the deaminated base as a repair template. Repair leads to a transition mutations: C:G to T:A and A:T to G:C for cytosine and adenosine base editors, respectively [ 68 ]
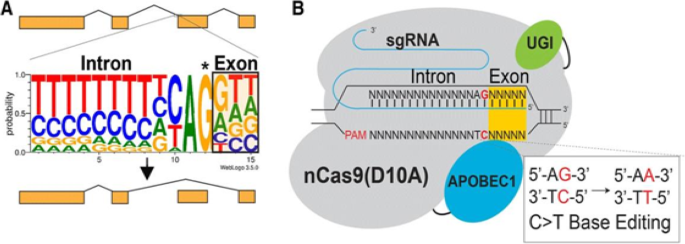
Essential steps in CRISPR-SKIP targeting approach: a Nearly every intron ends with a guanosine (asterisked G). It is hypothesized that mutations that disrupt this highly conserved G within the splice acceptor of any given exon in genomic DNA would lead to exon skipping by preventing incorporation of the exon into mature transcripts base. b In the presence of an appropriate PAM sequence, this G can be effectively mutated by converting the complementary cytidine to thymidine using CRISPR-Cas9 C>T single-base editors. (From [ 25 ])
Gene delivery
From biotechnology’s point of view, the main obstacle that is facing molecular technology is to select the right method that is simple but effective to transfer the gene to the host cell. The components of gene editing have to be transferred to the cell/nucleus of interest using in vivo, ex vivo, or in vitro route. In this regard, several concerns must be considered including physical barriers (cell membranes, nuclear membranes) as well as digestion by proteases or nucleases of the host. Another important issue is the possible rejection by the immune system of the host if the components are delivered in vivo. In general, the gene delivery routes can be categorized in three classes of physical delivery, viral vectors, and non-viral agents. Although the direct delivery of construct plasmids may sound easy and more efficient and specific than the physical and the chemical methods, it proves to be an inappropriate choice because the successful gene delivery system requires the foreign genetic molecule to remain stable within the host cells [ 52 ]. The other possible procedure is to use viruses. However, because plant cells have thick walls, the gene transfer systems for plants involve transient and stable transformation using protoplast-plasmid in vitro [ 54 ]: agrobacterium-mediated transformation, gene gun and viral vectors (transient expression by protoplast transformation), and agro-infiltration [ 1 ]. Viruses may present a suitable vehicle to transfer genome engineering components to all plant parts because they do not require transformation and/or tissue culture for delivering and mutated seeds could easily recovered. For many years, scientists employed different species of Agrobacterium to systematically infect a large number of plant species and generate transgenic plants. These bacterial species have small genome size and this facilitates cloning and agroinfections, and the virus genome does not integrate into plant genomes [ 1 ].
Of the challenges and approaches of delivering CRISPR, it was pointed out [ 18 , 51 ] that although the present genome engineering is in favor of CRISPR tools, TALENs may still be of a primary choice in certain experimental species. For example, TALENs have been utilized in targeted genomic editing in Xenopus tropicalis by knocking-out Klf4 [ 49 , 50 ] or thyroid hormone receptor α [ 23 ]. In addition, TALENs have been utilized to modify genome of human stem cells [ 47 ]. Also TALEN approach has been applied to create amniotic mesenchymal stem cells overexpressing anti-fibrotic interleukin-10 [ 12 ]. Lately, a geminivirus genome has been prepared to deliver various nucleases platforms (including ZFN, TALENs, and the CRISPR/Cas system) and repair template for HR of DSBs [ 62 ].
To deliver the carrying DNA sequence to target cells, non-viral techniques such as electroporation, lipofection, and microinjection can also be used [ 18 ]. In addition, these techniques also reduce off-target cleavages problems. Gene transfer via microinjection is considered the gold standard procedure since its efficiency is approximately 100% [ 85 ]. The advantage of this approach is its high efficacy and less constrains on the size of the delivery. A disadvantage is that it can be employed only in in vitro or ex vivo cargo. Recently, small RNAs, including small interfering RNA (siRNA) and microRNA (miRNA), have been widely adopted in research to replace laboratory animals and cell lines. Development of innovative nanoparticle-based transfer systems that deliver CRISPR/Cas9 constructs and maximize their effectiveness has been tested in the last few years [ 29 , 58 ].
Applications of gene technology
The ability of the abovementioned gene delivery systems to target and manipulate the genome of living organisms has been attractive to many researchers worldwide. Despite all limitations, the interest in this technology has developed its capabilities and enhanced its scope of applications. Genome/gene engineering technology is relatively applicable and has potential to effectively and rapidly revolutionize genome surgery and will soon transform agriculture, nutrition, and medicine. Some of the most important applications are briefly described below.
Plant-based genome editing
The appearance of genome editing has been appealing especially to agricultural experts. One of the major goals for utilizing genome editing tools in plants is to generate improved crop varieties with higher yields and clear-cut addition of valuable traits such as high nutritional value, extended shelf life, stress tolerance, disease and pest resistance, or removal of undesirable traits [ 1 ]. However, several obstacles related to the precision of the genetic manipulations and the incompatibility of the host species have hampered the development of crop improvements [ 2 ]. The use of site-specific nucleases is one of the important promising techniques of gene editing that helped overcome certain limitations by specifically targeting a suitable site in a gene/genome. The employment of the gene editing technologies, including those discussed in this review, seems to be endless ever since their emergence, and several improvements in original tools have further brought accuracy and precision in these methods [ 78 ].
Animal-based genome editing
Recent genome editing techniques has been extensively applied in many organisms, such as bacteria, yeast, and mouse [ 53 , 73 ]. Genetic manipulation tools cover a wide range of fields, including the generation of transgenic animals using embryonic stem cells (ESC), functional analysis of genes, model development for diseases, or drug development. Genome editing techniques have been used in many various organisms. Among the livestock and aquatic species, ZFN is only used for zebrafish, but two other technologies, TALEN and CRISPR, have been used at the cell level in chicken, sheep, pig, and cattle. Engineered endonucleases or RNA-guided endonucleases (RGENs) mediated gene targeting has been applied directly in a great number of animal organisms including nematodes and zebrafish [ 20 , 57 ], as well as pigs [ 71 , 85 ]. Since the first permission to use CRISPR/Cas9 in human embryos and in vivo genome editing via homology-independent targeted integration (HITI), an increasing number of studies have identified striking differences between mouse and human pre-implantation development and pluripotency [ 66 ], highlighting the need for focused studies in human embryos. Therefore, more specific criteria and widely accepted standards for clinical research have to be met before human germline editing would be deemed permissible [ 31 ]. In this regard, results of some research on the human genome editing have been questioned. The “He Jiankui experiments at the beginning of 2019”, which claimed to have created the world’s first genetically edited babies, is simply the most recent example. He Jiankui said he edited the babies’ genes at conception by selecting CRISPR/cas9 to edit the chemokine receptor type 5 (CCR5) gene in cd4+ cells in hopes of making children resistant to the AIDS virus, as their father was HIV-positive. Researchers said He’s actions exposed the twins to unknown health risks, possibly including a higher susceptibility to viral illnesses. For more information on the scientific reactions around the world, the reader may find helpful several excellent sources of information [ 38 , 49 , 79 , 84 ].
- Gene therapy
The original principles of gene therapy arose during the 1960s and early 1970s when restriction enzymes were utilized to manipulate DNA [ 22 ]. Since then, researchers have done great efforts to treat genetic diseases but treatment for multiple mutations is difficult. Different clinical therapy applications have been attempted to overcome these problems. Much of the interest in CRISPR and other gene editing methods revolves around their potential to cure human diseases. It is hoped that eradication of human diseases is not too far to achieve via the CRISPR system because it was employed in other fields of biological sciences such as genetic improvement and gene therapy. It is important to mention that the therapeutic efficiency of gene editing depends on several factors, such as editing efficacy, which varies widely depending on the cell type, senescence status, and cell cycle status of the target [ 69 ]. Other factors that also influence therapeutic effectiveness include cell aptitude, which refers to the feasibility of accomplishing a therapeutic modification threshold, and the efficient transfer of programmable nuclease system to the target tissue, which is only considered to be effective if the engineered nuclease system reaches safely and efficiently to the nucleus of the target cell. Finally, the precision of the editing procedure is another important aspect, which refers to only editing the target DNA without affecting any other genes [ 80 ].
The genome editing tools have enabled scientists to utilize genetically programmed animals to understand the cause of various diseases and to understand molecular mechanisms that can be explored for better therapeutic strategies (Fig. 7 ). Genome editing gives the basis of the treatment of many kinds of diseases. In preliminary experiments, the knocking-in procedure was used to reach this goal. There are examples of gene editing techniques applied in different genetic diseases in cell lines, disease models, and human [ 48 , 53 , 82 ]. These encouraging results suggest the therapeutic capability of these gene editing strategies to treat human genetic diseases including Duchenne muscular dystrophy [ 8 , 28 , 55 ], cystic fibrosis [ 21 ], sickle cell anemia [ 62 ], and Down syndrome [ 7 ]. In addition, this technology has been employed in curing Fanconi anemia by correcting point mutation in patient-derived fibroblasts [ 60 ], as well as in hemophilia for the restoration of factor VIII deficiency in mice [ 61 , 87 ]. The CRISPR tools have also demonstrated promising results in diagnosis and curing fatal diseases such as AIDS and cancer [ 16 , 30 , 84 ].
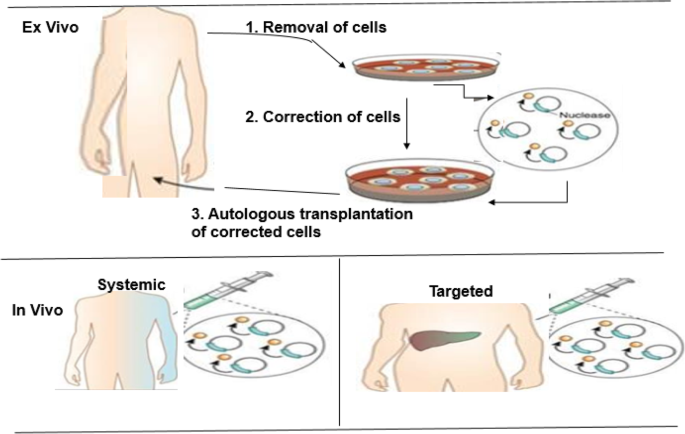
Outline of the ex vivo and in vivo genome editing procedures for clinical therapy. Top: In the ex vivo editing therapy, cells are removed from a patient to be treated, corrected by gene editing and then re-engrafted back to the patient. To achieve therapeutic success, the target cells must be capable of surviving in vitro and autologous transplantation of the corrected cells. Below: In the in vivo editing therapy, designed nucleases are administered using viral or non-viral techniques and directly injected locally to the affected tissue, such as the eye, brain, or muscle. (Adapted from [ 48 ])
Other applications
The applications mentioned above were more about knock out or modification of genes Gapinske et al. [ 25 ]. However due to inactivate nuclease activity nature of the dCas9, CRISPR can be used in other applications as well. By selecting the target sequence, gene expression can be controlled by inhibiting the transcription rate of RNA polymerase II (polII) or inhibiting the transcription factor binding [ 65 ]. Additionally, combining gene expression inhibitors such as Krüppel-associated box with the inactivated Cas9 has led to generate a special kind of gene inhibitors, which are called CRISPR interference (CRISPRi), and downregulate gene expression [ 46 ]. It is also possible to control gene expression by fusing transcription-activating molecule, the transcription-repressing molecule, or the genome-modifying molecule to dCas9 [ 27 ].
Genome editing is a fast-growing field. Editing nucleases have revolutionized genomic engineering, allowing easy editing of the mammalian genome. Much progress has been accomplished in the improvement of gene editing technologies since their discovery. Of the four major nucleases used to cut and edit the genome, each has its own advantages and disadvantages, and the choice of which gene editing method depends on the specific situation. The current genome editing techniques are still buckling up with problems, and it is difficult to perform genome editing in cells with low transfection efficiency or in some cultured cells such as primary cultured cells. Genotoxicity is an inherent problem of enzymes that act on nucleic acids, though one can expect that highly specific endonucleases would reduce or abolish this issue. Exceptional efforts are needed in future to complement and offer something novel approaches in addition to the already existing ones. It is anticipated that research in gene editing is going to continue and tremendously advance. With the development of next-generation sequencing technology, new extremely important clinical applications, such as manufacturing engineered medical products, eradication of human genetic diseases, treatment of AIDS and cancers, as well as improvement of crop and food, will be introduced. Combination of genomic modifications induced by targeted nucleases to their own self-degradation, self-inactivating vectors may help overcoming confronting limitations discussed above to improve the specificity of genome editing, especially because the frequency of off-target modifications. Our understanding of off-target effects remains poor. This is a vital area for continued study if CRISPR/Cas9 is to realize its promise. Regarding gene cargo delivery systems, this remains the greatest obstacle for CRISPR/Cas9 use, and an all-purpose delivery method has yet to emerge. The union between genome engineering and regenerative medicine is still in its infancy; realizing the full potential of these technologies in reprograming the fate of stem/progenitor cells requires that their functional landscape be fully explored in these genetic backgrounds. Humankind can only wait to see what the potential of these technologies will be. One major question is whether or not the body’s immune response will accept or reject the foreign genetic elements within the cells. Another important concern is that along with the revolutionary advances of this biotechnology and related sciences, bioethical concerns and legal problems related to this issue are still increasing in view of the possibility of human genetic manipulation and the unsafety of procedures involved [ 49 , 50 , 66 ]. The enforcement of technical and ethical guidelines, and legislations should be considered and need serious attention as soon as possible.
Availability of data and materials
Not applicable
Abbreviations
CRISPR-associated protein 9
Clustered regularly interspaced short palindromic repeats
Double-stranded break
Embryonic stem cells
Homology-directed repair
Homology-independent targeted integration
Homologous recombination
Human umbilical vein endothelium cells
Intron-encoded endonuclease
- Meganucleases
Microhomology-mediated end joining
Non-homologous end joining
Phage-assisted continuous evolution
Protospacer adjacent motifs
RNA-guided endonucleases
Repeat variable di-residues
Single guide RNA
Streptococcus pyogenes Cas9
Single-strand break
Transcription activator-like effector nuclease
Zinc finger nucleases
Abdallah N, Prakash C, Mchughen A (2015) Genome editing for crop improvement: challenges and opportunities. GM Crops Food 6(4):183–205
Article Google Scholar
Aglawe S, Barbadikar K, Mangrauthia S, Madhav M (2018) New breeding technique “genome editing” for crop improvement: applications, potentials and challenges. 3 Biotech 8:336
Alkhnbashi OS, Fabrizio C, Shah SA, Garrett RA, Saunders SJ, Rolf B (2014) CRISPR strand: predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 30(17):489–496
Anders C, Niewoehner O, Duerst A, Jinek M (2014) Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513:569–573
Bernal-Bernal D, Abellón-Ruiz J, Iniesta AA, Pajares-Martínez E, Bastida-Martínez E, Fontes M et al (2018) Multifactorial control of the expression of a CRISPR-Cas system by an extracytoplasmic function σ/anti-σ pair and a global regulatory complex. Nucleic Acids Res 46(13):6726–6745
Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG et al (2001) Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 21:289–297
Bloh KM, Bialk PA, Gopalakrishnapillai A, Kolb EA, Kmiec EB (2017) CRISPR/Cas9-directed reassignment of the GATA1 initiation codon in K562 cells to recapitulate AML in Down syndrome. Mol Ther Nucleic Acids 7:288–298
Cai A, Kong X (2019) Development of CRISPR-mediated systems in the study of Duchenne muscular dystrophy. Hum Gene Therap Methods https://doi.org/10.1089/hgtb.2018.187
Cao J, Wu L, Zhang SM, Lu M, Cheung WK, Cai W, Gale M et al (2016) An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res 44:e149
Google Scholar
Capecchi MR (1989) Altering the genome by homologous recombination. Science 244:1288–1292
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188:773–782
Choi J, Jeong I, Han J, Cheon S, Kim S (2019) IL-10-secreting human MSCs generated by TALEN gene editing ameliorate liver fibrosis through enhanced anti-fibrotic activity. Biomater Sci 7(3):1078–1087
Christian ML, Demorest ZL, Starker CG, Osborn MJ, Nyquist MD, Zhang Y et al (2012) Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues. PLoS One 7:e45383
Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C et al (1998) I-sceI-induced gene replacement at a natural locus in embryonic stem cells. Mol Cell Biol 18:1444–1448
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Cyranoski D (2016) CRISPR gene-editing tested in a person for the first time. Nature 539:479
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096
Dumeau C-E, Monfort A, Kissling L, Swarts DC, Jinek M, Wutz A (2019) Introducing gene deletions by mouse zygote electroporation of Cas12a/Cpf1. Transgenic Res 28(5–6):525–535
East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, Doudna JA (2017) RNA targeting by functionally orthogonal Type VI-A CRISPR-Cas enzymes. Mol Cell 66(3):373–383
Fernandez JP, Vejnar CE, Giraldez AJ, Rouet R, Moreno-Mateos MA (2018) Optimized CRISPR-Cpf1 system for genome editing in zebrafish. Methods 150:11–18
Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E et al (2015) Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep 12:1385–1390
Friedmann T, Roblin R (1972) Gene therapy for human genetic disease? Science 175(4025):949–955
Fu L, Wen L, Shi Y (2018) Role of thyroid hormone receptor in amphibian development. Methods Mol Biol 1801:247–263
Gaj T, Sirk SK, S-l S, Liu J (2016) Genome-editing technologies: principles and applications. Cold Spring Harb Perspect Biol 8:a023754
Gapinske M, Luu A, Winter J, Woods ES, Kostan KA, Shiva N et al (2018) CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol 19:107 https://doi.org/10.1186/s13059-018-1482-5
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI et al (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551:464–471
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154(2):442–451
Ginn SL, Amaya AK, Alexander IE, Edelstein M, Abedi MR (2018) Gene therapy clinical trials worldwide to 2017: an update. J Gene Med 20:e3015
Givens BE, Naguib YW, Geary SM, Devor EJ, Salem AK (2018) Nanoparticle-based delivery of CRISPR/Cas9 genome-editing therapeutics. AAPS J 20(6):108
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360:439–444
Greely HT (2019) CRISPR’d babies: human germline genome editing in the ‘He Jiankui affair’. J Law Biosci 6(1):111–183
Gupta A, Hall VL, Kok FO, Shin M, McNulty JC, Lawson ND, Wolfe SA (2013) Targeted chromosomal deletions and inversions in zebrafish. Genome Res 23(6):1008–1017
Hille F, Richter H, Wong SP, Bratovic M, Ressel S, Charpentier E (2018) The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259
Hubbard BP, Badran AH, Zuris JA, Guilinger JP, Davis KM, Chen L et al (2015) Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat Methods 12:939
Huo Z, Tu J, Xu A, Li Y, Wang D, Liu M et al (2019) Generation of a heterozygous p53 R249S mutant human embryonic stem cell line by TALEN-mediated genome editing. Stem Cell Res 34:101360
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433
Jansen R, Embden JD, Gaastra W, Schouls LM (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43:1565–1575
Jonlin EC (2020) Informed consent for human embryo genome editing. Stem Cell Rep 14(4):530–537
Khan SH (2019) Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol Ther Nucleic Acids 16:326–334
Kim K, Bang SY, Lee HS, Bae SC (2017) Update on the genetic architecture of rheumatoid arthritis. Nat Rev Rheumatol 13:13–24
Kim Y-G, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93:1156–1160
Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY et al (2020) Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene epigenetic and base editing. Nat Biotechnol 38(7):901
Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW et al (2017) Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3:eaao4774
Koonin EV, Makarova KS, Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78
Kurt IC, Zhou R, Iyer S, Garcia SP, Miller BR, Langner LM, Grünewald J, Joung JK (2020) CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol https://doi.org/10.1038/s41587-020-0609-x
Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS (2013) CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196
Lee J, Termglinchan V, Diecke S, Itzhaki I, Lam C, Garg P et al (2019) Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature 572:335–340
Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X (2020) Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Sig Transduct Target Ther 5(1) https://doi.org/10.1038/s41392-019-0089-y
Li J-R, Walker S, Nie J-B, Xin-qing Zhang X-Q (2019a) Experiments that led to the first gene-edited babies: the ethical failings and the urgent need for better governance. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 20(1):32–38
Li L, Rispoli R, Patient R, Ciau Uitz A, Porcher C (2019b) Etv6 activates vegfa expression through positive and negative transcriptional regulatory networks in Xenopus embryos. Nat Commun 10:1083
Lino CA, Harper JC, Carney JP, Timlin JA (2018) Delivering CRISPR: a review of the challenges and approaches. Drug Deliv 25:1234–1257
Mali S (2013) Delivery systems for gene therapy. Indian J Human Gene 19:3–8
Mandip KC, Steer CJ (2019) A new era of gene editing for the treatment of human diseases. Swiss Med Wkly 149:w20021
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Min Y-L, Bassel-Duby R, Olson EN (2019) CRISPR correction of Duchenne muscular dystrophy. Annu Rev Med 70:239–255
Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60:174–182
Mooney MR, Davis EE, Nicholas Katsanis N (2019) Analysis of single nucleotide variants in CRISPR-Cas9 edited zebrafish exomes shows no evidence of off-target inflation. Front Genet 11. https://doi.org/10.3389/fgene.2019.00949
Mout R, Ray M, Tonga GY, Lee Y-W, Tay T, Sasaki K et al (2017) Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 3:2452–2458
Nakata A, Amemura M, Makino K (1989) Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol 171:3553–3556
Osborn MJ, Gabriel R, Webber BR, deFeo AP, McElroy AN, Jarjour J et al (2015) Fanconi anemia gene editing by the cRISPR/cas9 system. Hum Gene Ther 26:114–126
Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S et al (2015) Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPScs using cRISPR-cas9. Cell Stem Cell 17:213–220
Park JY, Moon BY, Park JW, Thornton JA, Park YH, Seo KS (2017) Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus . Sci Rep 7:44929
Petersen B, Niemann H (2015) Molecular scissors and their application in genetically modified farm animals. Transgenic Res 24:381–391
Prieto J, Redondo P, López-Méndez B et al (2018) Understanding the indirect DNA read-out specificity of I-CreI Meganuclease. Sci Rep 8:10286
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP et al (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Reyes AP, Lanner F (2017) Towards a CRISPR view of early human development: applications, limitations and ethical concerns of genome editing in human embryos. Development 144:3–7
Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34:339–344
Robb GB (2019) Genome editing with CRISPR-Cas: an overview. Curr Protoc Essent Lab Tech 19:e36 https://doi.org/10.1002/cpet.36
Rodrigeuz-Rodrigeuz DR, Ramirez-Solis R, Garza-Elizondo MA, Garza-Rodrigeuz MDL, Barrera-Saldana HA (2019) Genome editing: a perspective on the application of CRISPR/Cas9 to study human diseases (Review). Int J Mol Med 43:1559–1574
Rouet P, Smih F, Jasin M (1994) Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14:8096–8106
Ryu J, Prather RS, Lee K (2018) Use of gene-editing technology to introduce targeted modifications in pigs. J Anim Sci Biotechnol 9:5
Saleh-Gohari N, Helleday T (2004) Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res 32:3683–2688
Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z et al (2013) Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23(5):720–723
Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P et al (2011) Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther 11(1):11–27
Stoddard BL (2014) Homing endonucleases from mobile group I introns: discovery to genome engineering. MobDNA 5:7
Tu Z, Yang W, Yan S, Yin A, Gao J, Liu X et al (2017) Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci Rep 7:42081
Vasebi Y, Khakvar R (2014) CRISPR-Cas: the effective immune systems in the prokaryotes. Int J Mol Clin Microbiol 1:334–344
Walker-Daniels J (2013) CRISPR and genomic engineering. Mater Methods 3:164
Wang H, Yang H (2019) Gene-edited babies: what went wrong and what could go wrong. PLoS Biol 17(4):e3000224
Wang M, Glass ZA, Xu Q (2017) Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther 24:144–150
Wright AV, Nuñez JK, Doudna JA (2016) Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164(1-2):29–44
Wu WH, Tsai YT, Justus S, Cho GY, Sengillo JD, Xu Y et al (2018) CRISPR repair reveals causative mutation in a preclinical model of retinitis pigmentosa: a brief methodology. Retinal Gene Ther 1715:191–205
Wu X, Kriz AJ, Sharp PA (2014) Target specificity of the CRISPR-Cas9 system. Quant Biol 2(2):59–70
Xiao Q, Guo D, Chen S (2019) Application of CRISPR/Cas9-based gene editing in HIV-1/AIDS therapy. Front Cell Infect Microbiol 9:69
Yang H, Wu Z (2018) Genome editing of pigs for agriculture and biomedicine. Front Genet 9:360
Zhang F, Wen Y, Guo X (2014) CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 23:R40–R46
Zhang H, Mccarty N (2016) cRISPR-cas9 technology and its application in haematological disorders. Br J Haematol 175:208–225
Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF (2013) Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161:20–27
Zhao D, Li J, Li S, Xin X, Hu M, Price MA, Rosser SJ, Bi C, Zhang X (2020) Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol 2020:1–6
Download references
Acknowledgements
Not availed any grant either from funding agencies or from any institutions.
Author information
Authors and affiliations.
Department of Biological Sciences, Yarmouk University, Irbid, Jordan
Ahmad M. Khalil
You can also search for this author in PubMed Google Scholar
Contributions
The author read and approved the final manuscript.
Corresponding author
Correspondence to Ahmad M. Khalil .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The author declares that this review was conducted in the absence of any commercial or financial relationships that could be construed as a potential competing interest.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Khalil, A.M. The genome editing revolution: review. J Genet Eng Biotechnol 18 , 68 (2020). https://doi.org/10.1186/s43141-020-00078-y
Download citation
Received : 12 June 2020
Accepted : 22 September 2020
Published : 29 October 2020
DOI : https://doi.org/10.1186/s43141-020-00078-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- CRISPR-Cas system
- Gene editing
- Genome editing
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Review Article
- Open access
- Published: 20 April 2022
Beyond safety: mapping the ethical debate on heritable genome editing interventions
- Mara Almeida ORCID: orcid.org/0000-0002-0435-6296 1 &
- Robert Ranisch ORCID: orcid.org/0000-0002-1676-1694 2 , 3
Humanities and Social Sciences Communications volume 9 , Article number: 139 ( 2022 ) Cite this article
20k Accesses
11 Citations
8 Altmetric
Metrics details
- Medical humanities
- Science, technology and society
Genetic engineering has provided humans the ability to transform organisms by direct manipulation of genomes within a broad range of applications including agriculture (e.g., GM crops), and the pharmaceutical industry (e.g., insulin production). Developments within the last 10 years have produced new tools for genome editing (e.g., CRISPR/Cas9) that can achieve much greater precision than previous forms of genetic engineering. Moreover, these tools could offer the potential for interventions on humans and for both clinical and non-clinical purposes, resulting in a broad scope of applicability. However, their promising abilities and potential uses (including their applicability in humans for either somatic or heritable genome editing interventions) greatly increase their potential societal impacts and, as such, have brought an urgency to ethical and regulatory discussions about the application of such technology in our society. In this article, we explore different arguments (pragmatic, sociopolitical and categorical) that have been made in support of or in opposition to the new technologies of genome editing and their impact on the debate of the permissibility or otherwise of human heritable genome editing interventions in the future. For this purpose, reference is made to discussions on genetic engineering that have taken place in the field of bioethics since the 1980s. Our analysis shows that the dominance of categorical arguments has been reversed in favour of pragmatic arguments such as safety concerns. However, when it comes to involving the public in ethical discourse, we consider it crucial widening the debate beyond such pragmatic considerations. In this article, we explore some of the key categorical as well sociopolitical considerations raised by the potential uses of heritable genome editing interventions, as these considerations underline many of the societal concerns and values crucial for public engagement. We also highlight how pragmatic considerations, despite their increasing importance in the work of recent authoritative sources, are unlikely to be the result of progress on outstanding categorical issues, but rather reflect the limited progress on these aspects and/or pressures in regulating the use of the technology.
Similar content being viewed by others
Genetics experience impacts attitudes towards germline gene editing: a survey of over 1500 members of the public

Between desire and fear: a qualitative interview study exploring the perspectives of carriers of a genetic condition on human genome editing
The interplay of ethics and genetic technologies in balancing the social valuation of the human genome in unesco declarations, introduction.
The ability to alter a sequence of genetic material was initially developed in microorganisms during the 1970s and 1980s (for an overview: Walters et al., 2021 ). Since then, technological advances have allowed researchers to alter DNA in different organisms by introducing a new gene or by modifying the sequence of bases in the genome. The manipulation of the genome of living organisms (typically plants) continues a course that science embraced more than 40 years ago, and may ultimately allow, if not deliberately curtailed by societal decisions, the possibility of manipulating and controlling genetic material of other living species, including humans.
Genetic engineering can be used in a diverse range of contexts, including research (e.g., to build model organisms), pharmacology (e.g., for insulin production) and agriculture (e.g., to improve crop resistance to environmental pressures such as diseases, or to increase yield). Beyond these applications, modern genetic engineering techniques such as genome editing technologies have the potential to be an innovative tool in clinical interventions but also outside the clinical realm. In the clinical context, genome editing techniques are expected to help in both disease prevention and in treatment (Porteus, 2019 ; Zhang, 2019 ). Nevertheless, genome editing technology raises several questions, including the implications of its use for human germline cells or embryos, since the technology’s use could facilitate heritable genome editing interventions (Lea and Niakan, 2019 ). This possible use has fuelled a heated debate and fierce opposition, as illustrated by the moratoriums proposed by researchers and international institutions on the use of the technology (Lander et al., 2019 ; Baltimore et al., 2015 ; Lanphier et al., 2015 ). Heritable human germline modifications are currently prohibited under various legislations (Baylis et al., 2020 ; Ledford, 2015 ; Isasi et al., 2016 ; König, 2017 ) and surveys show public concerns about such applications, especially without clear medical justification (e.g., Gaskell et al., 2017 ; Jedwab et al., 2020 ; Scheufele et al., 2017 ; Blendon et al., 2016 ).
To analyse some implications of allowing heritable genome editing interventions in humans, it is relevant to explore underlying values and associated ethical considerations. Building on previous work by other authors (e.g., Coller, 2019 ; de Wert et al., 2018 ; van Dijke et al., 2018 ; Mulvihill et al., 2017 ; Ishii, 2015 ), this article aims to provide context to the debates taking place and critically analyse some of the major pragmatic, categorical and sociopolitical considerations raised to date in relation to human heritable genome editing. Specifically, we explore some key categorical and sociopolitical considerations to underline some of the possible barriers to societal acceptance, key outstanding questions requiring consideration, and possible implications at the individual and collective level. In doing so, we hope to highlight the predominance of pragmatic arguments in the scientific debate regarding the permissible use of heritable genome editing interventions compared to categorical arguments relevant to broader societal debate.
Human genome editing: a brief history of CRISPR/Cas9
Human genome editing is an all-encompassing term for technologies that are aimed at making specific changes to the human genome. In humans, these technologies can be used in embryos or germline cells as well as somatic cells (Box 1 ). Concerning human embryos or germline cells, the intervention could introduce heritable changes to the human genome (Lea and Niakan, 2019 ; Vassena et al., 2016 ; Wolf et al., 2019 ). In contrast, an intervention in somatic cells is not intended to result in changes to the genome of subsequent generations. It is worth noting that intergenerational effects occur only when the modified cells are used to establish a pregnancy which is carried to term. Thus, a distinction has been made between germline genome editing (GGE), which may only affect in vitro embryos in research activity, and heritable genome editing (HGE), which is used in reproductive medicine (e.g., Baylis et al., 2020 ). HGE could be used to prevent the transmission of serious genetic disease; however, other applications could be imagined, e.g., creating genetic resistance or even augmenting human functions.
In the last decade, prominent technical advances in genome engineering methods have taken place, including the zinc-finger nucleases (ZFNs) and TAL effector nucleases (TALENs), making human genome modification a tangible possibility (Gaj et al., 2013 ; Li et al., 2020 ; Gupta and Musunuru, 2014 ). In 2012, a study showed that the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), combined with an enzyme called Cas9, could be used as a genome‐editing tool in human cell culture (Jinek et al., 2012 ). In 2013, the use of CRISPR/Cas9 in mammalian cells was described, demonstrating the application of this tool in the genome of living human cells (Cong et al., 2013 ). In 2014, CRISPR/Cas9 germline modifications were first used in non-human primates, resulting in the birth of gene-edited cynomolgus monkeys (Niu et al., 2014 ). This was followed in 2015 by the first-ever public reported case of genome modification in non-viable human embryos (tripronuclear zygotes) (Liang et al., 2015 ). This study has caused broad concerns in the scientific community (Bosley et al., 2015 ) with leading journals rejecting publication for ethical reasons. Five years after these initial experiments were conducted, more than 10 papers have been published reporting the use of genome editing tools on human preimplantation embryos (for an overview: Niemiec and Howard, 2020 ).
Compared to counterpart genome technologies (e.g., ZFNs and TALENs), CRISPR/Cas9 is considered by many a revolutionary tool due to its efficiency and reduced cost. More specifically, CRISPR/Cas9 seems to provide the possibility of a more targeted and effective intervention in the genome involving the insertion, deletion, or replacement of genetic material (Dance, 2015 ). The potential applicability of CRISPR/Cas9 technique is considered immense, since it can be used on all type of organisms, from bacteria to plants, non-human cells, and human cells (Barrangou and Horvath, 2017 ; Hsu et al., 2014 ; Doudna and Charpentier, 2014 ; Zhang, 2019 ).
Box 1 Difference associated with germline cells and somatic cells.
For the purposes of the analysis presented in this article, one of the main differences is the heritability of genes associated with either type of cell. Germline cells include spermatozoa, oocytes, and their progenitors (e.g., embryonic cells in early development), which can give rise to a new baby carrying a genetic heritage coming from the parents. Thus, germline are those cells in an organism which are involved in the transfer of genetic information from one generation to the next. Somatic cells, conversely, constitute many of the tissues that form the body of living organisms, and do not pass on genetic traits to their progeny.
Germline interventions: the international debate
As a reaction to the 2015 study with CRISPR/Cas9, several commentaries by scientists were published regarding the future use of the technology (e.g., Bosley et al., 2015 ; Lanphier et al., 2015 ; Baltimore et al., 2015 ). Many of them focused on germline applications, due to the possibility of permanent, heritable changes to the human genome and its implications for both individuals and future generations. These commentaries included position statements calling for great caution in the use of genome editing techniques for heritable interventions in humans and suggested a voluntary moratorium on clinical germline applications of CRISPR/Cas9, at least until a broad societal understanding and consensus on their use could be reached (Brokowski, 2018 ; Baltimore et al., 2015 ; Lander, 2015 ). Such calls for a temporary ban were often seen as reminiscent of the “Asilomar ban” on recombinant DNA technology in the mid-1970s (Guttinger, 2017 ). Other commentaries asked for research to be discouraged or halted all together (Lanphier et al., 2015 ). More firmly, the United States (US) National Institutes of Health (NIH) released a statement indicating that the NIH would not fund research using genome editing technologies on human embryos (Collins, 2015 ).
In December 2015, the first International Summit on Human Gene Editing took place, hosted by the US National Academy of Sciences, the US National Academy of Medicine, the UK Royal Society, and the Chinese Academy of Sciences (NASEM). The organizing committee issued a statement about appropriate uses of the technology that included the following: “It would be irresponsible to proceed with any clinical use of germline editing unless and until (i) the relevant safety and efficacy issues have been resolved, based on appropriate understanding and balancing of risks, potential benefits, and alternatives, and (ii) there is broad societal consensus about the appropriateness of the proposed application” (NASEM, 2015 ).
Following this meeting, initiatives from different national bodies were organized to promote debate on the ethical issues raised by the new genome editing technologies and to work towards a common framework governing the development and permissibility of their use in humans. This included an ethical review published in 2016 by the Nuffield Council on Bioethics, addressing conceptual and descriptive questions concerning genome editing, and considering key ethical questions arising from the use of the technology in both human health and other contexts (Nuffield Council on Bioethics, 2016 ). In 2017, a committee on human genome editing set up by the US National Academy of Sciences (NAS) and the National Academy of Medicine (NAM) carried out a so-called consensus study “Human Genome Editing: Science, Ethics, and Governance” (NASEM, 2017 ). This study put forward a series of recommendations on policies and procedures to govern human applications of genome editing. Specifically, the study concluded that HGE could be justified under specific conditions: “In some situations, heritable genome editing would provide the only or the most acceptable option for parents who desire to have genetically related children while minimizing the risk of serious disease or disability in a prospective child” (NASEM, 2017 ). The report stimulated much public debate and was met with support and opposition since it was seen as moving forward on the permissibility of germline editing in the clinical context (Ranisch and Ehni, 2020 ; Hyun and Osborn, 2017 ).
Following the report in 2016, the Nuffield Council on Bioethics published a second report in 2018. Similar to the NASEM 2017 report, this report emphasizes the value of procreative freedom and stresses that in some cases HGE might be the only option for couples to conceive genetically related, healthy offspring. In this document, the Nuffield Council on Bioethics maintains that there are no categorical reasons to prohibit HGE. However, it highlights three kinds of interests that should be recognized when discussing prospective HGE. They are related to individuals directly affected by HGE (parents or children), other parts of society, and future generations of humanity. In this context, two ethical principles are highlighted as important to guide future evaluations of the HGE use in specific interventions: “(...) to influence the characteristics of future generations could be ethically acceptable, provided if, and only if, two principles are satisfied: first, that such interventions are intended to secure, and are consistent with, the welfare of a person who may be born as a consequence, and second, that any such interventions would uphold principles of social justice and solidarity (…)” (Nuffield Council on Bioethics, 2018 ). This report was met with criticism for (implicitly) advocating genetic heritable interventions might be acceptable even beyond the boundaries of therapeutic uses. This is particularly controversial and goes well beyond the position previously reached by the NASEM report (which limited permissible uses of genome editing at preventing the transmission of genetic variants associated to diseases) (Drabiak, 2020 ). On the other hand, others have welcomed the report and, within it, the identification of explicit guiding ethical principles helpful in moving forward the debate on HGE (Gyngell et al., 2019 ).
As a follow-up to the 2015 conference, a second International Summit on Human Gene Editing was scheduled for November 2018 in Hong Kong (National Academies of Sciences, Engineering, and Medicine, 2019 ). The event, convened by the Hong Kong Academy of Sciences, the UK Royal Society, the US National Academy of Sciences and the US National Academy of Medicine, was supposed to focus on the prospects of HGE. Just before the Summit began, news broke that He Jiankui, a Chinese researcher and invited speaker at the Summit, created the world’s first genetically edited babies resulting from the use of CRISPR/Cas9 in embryos (Regalado, 2018 ; Lovell-Badge, 2019 ). Although an independent investigation of the case is still pending, his experiments have now been reviewed in detail by some scholars (e.g., Greely, 2019 , 2021 ; Kirksey, 2020 ; Davies, 2020 ; Musunuru, 2019 ). These experiments were globally criticized, since they did not follow suitable safety procedures or ethical guidelines (Wang and Yang, 2019 ; Lovell-Badge, 2019 ; Krimsky, 2019 ), nor considered the recommendations previously put forward by international reports (NASEM, 2017 ; Nuffield Council on Bioethics, 2018 ) and legal frameworks (Araki and Ishii, 2014 ; Isasi et al., 2016 ). Different reactions were triggered, including another call by scientists for a global moratorium on clinical human genome editing, to allow time for international discussions to take place on its appropriate uses (Lander et al., 2019 ) or an outright ban on the technology (Botkin, 2019 ). There were also calls for a measured analysis of the possible clinical applications of human genome editing, without the imposition of a moratorium (Daley et al., 2019 ; Dzau et al., 2018 ).
Most countries currently have legal frameworks to ban or severely restrict the use of heritable genome editing technologies (Araki and Ishii, 2014 ; Isasi et al., 2016 ; Baylis et al., 2020 ). However, since He’s experiment, the possibility that researchers might still attempt (with some likelihood of success) to use the technology in human embryos, became a growing concern, particularly since some scientists have already announced their interest in further clinical experiments (Cyranoski, 2019 ). For many, He’s experiments highlighted the ongoing risks associated with the use of modern genome editing technology without proper safety protocols and regulatory frameworks at an international level (Ranisch et al., 2020 ). This has triggered the need to develop clear and strict regulations to be implemented if these tools are to be used in the future. This incident also led to the formation of several working groups, including the establishment of an international commission on the Clinical Use of Human Germline Genome Editing set up by the US National Academy of Medicine, the US National Academy of Sciences, and the UK’s Royal Society. In 2020, the commission published a comprehensive report on HGE, proposing a translational pathway from research to clinical use (National Academy of Medicine, National Academy of Sciences, and the Royal Society, 2020 ). Likewise, a global expert Advisory Committee was established by the World Health Organization (WHO) with the goal of developing recommendations on governance mechanisms for human genome editing. Although the committee insisted in an interim recommendation that “it would be irresponsible at this time for anyone to proceed with clinical applications of human germline genome editing” (WHO, 2019 ), it did not express fundamental concerns on the possibility that some forms of HGE will one day become a reality. In 2021, the WHO’s Advisory Committee issued some publications, including a “Framework for governance” report and a “Recommendations” report (WHO, 2021 ). Building on a set of procedural and substantive values and principles, the “Framework for Governance” report discusses a variety of tools and institutions necessary for developing appropriate national, transnational, and international governance and oversight mechanisms for HGE. Specifically, the report considers the full spectrum of possible applications of human genome editing (including epigenetic editing and human enhancement) and addresses specific challenges associated with current, possible and speculative scenarios. These range from somatic gene therapy for the prevention of serious hereditary diseases to potentially more controversial applications reminiscent of the He Jiankui case (e.g., the use of HGE in reproductive medicine outside regulatory controls and oversight mechanisms). Additionally, the “Recommendations” report proposes among other things whistleblowing mechanisms to report illegal or unethical research. It also highlights the need for a global human genome editing registry, that should also cover basic and preclinical research on different applications of genetic manipulation, including HGE. The report also emphasises the need of making possible benefits of human genome editing widely accessible.
The idea of a human genome editing registry has also been supported by the European Group on Ethics in Science and New Technologies (EGE), an advisory board to the President of the European Commission. After an initial statement on genome editing published in 2016, still calling for a moratorium on editing of human embryos (EGE, 2016 ), the EGE published a comprehensive Opinion in 2021 (EGE, 2021 ). Although the focus of this report is on the moral issues surrounding genome editing in animals and plants, HGE is also discussed. Similar to the WHO Advisory Committee, the EGE recommends for HGE not to be introduced prematurely into clinical application and that measures should be taken to prevent HGE’s use for human enhancement.
Overall, when reviewing reports and initiatives produced since 2015, common themes and trajectories can be identified. A key development is the observation that the acceptance of the fundamental permissibility of such interventions appears to be increasing. This constitutes an important change from previous positions, reflecting the fact that human germline interventions have long been considered a ‘red line’ or at least viewed with deep scepticism (Ranisch and Ehni, 2020 ). In particular, while there is agreement that it would be premature to bring HGE into a clinical context, key concerns expressed by authoritative international bodies and committees are now associated with acceptable uses of the technology, rather than its use per se. Consideration is now being given to the conditions and objectives under which germline interventions could be permissible, instead of addressing the fundamental question of whether HGE may be performed at all. The question of permissibility is often linked to the stage of technological development. These developments are remarkable, since the key ethical aspects of genome editing are now frequently confined to questions of safety or cost–benefit ratios, rather than categorical considerations.
Another common issue can also be found in recent reports: the question of involving society in the debate. There is consensus on the fact that the legitimacy and governance of HGE should not be left solely to scientists and other experts but should involve society more broadly. Since germline interventions could profoundly change the human condition, the need for a broad and inclusive public debate is frequently emphasized (Iltis et al., 2021 ; Scheufele et al., 2021 ). The most striking expression of the need for public engagement and a “broad societal consensus” can be found in the final statement by the 2015 International Summit on Human Gene Editing organizing committee, as previously quoted (NASEM, 2015 ). Furthermore, the EGE and others also stresses the need for an inclusive societal debate before HGE can be considered permissible.
The pleas for public engagement are, however, not free of tension. For example, the NASEM’s 2017 report was criticised for supporting HGE bypassing the commitment for the broad societal consensus (Baylis, 2017 ). Regarding HGE, some argue that only a “small but vocal group of scientists and bioethicists now endorse moving forward” (Andorno et al., 2020 ). Serious efforts to engage the public on the permissibility and uses of HGE have yet to be made. This issue not only lacks elaboration on approaches to how successful public participation can occur, but also how stop short of presenting views on how to translate the public’s views into ethical considerations and policy (Baylis, 2019 ).
Potential uses of heritable genome editing technology
HGE is expected to allow a range of critical interventions: (i) preventing the transmission of genetic variants associated with severe genetic conditions (mostly single gene disorders); (ii) reducing the risk of common diseases (mostly polygenic diseases), with the promise of improving human health; and (iii) enhancing human capabilities far beyond what is currently possible for human beings, thereby overcoming human limitations. The identification of different classes of potential interventions has shifted the debate to the applications considered morally permissible beyond the acceptable use of HGE (Dzau et al., 2018 ). Specifically, there are differences in the limits of applicability suggested by some of the key cornerstone publications discussed above. For example, the NASEM ( 2017 ) report suggests limiting the use of HGE to the transmission of genetic variants linked to severe conditions, although in a very regulated context. In a very similar way, the 2020 report from the International Commission on the Clinical Use of Human Germline Genome Editing suggests that the initial clinical use of HGE should be limited to the prevention of serious monogenic diseases. By contrast, the 2018 Nuffield Council on Bioethics Report does not seem to limit the uses of genome editing to specific applications, though suggests that applications should be aligned with fundamental guiding ethical principles and need to have followed public debate (Savulescu et al., 2015 ). The same report also discusses far-reaching and speculative uses of HGE that might achieve “other outcomes of positive value” (Nuffield Council on Bioethics, 2018 ). Some of these more speculative scenarios include “built-in genetic resistance or immunity to endemic disease”; “tolerance for adverse environmental conditions” and “supersenses or superabilities” (Nuffield Council on Bioethics, 2018 , p. 47).
There have been different views on the value of HGE technology. Some consider that HGE should be permissible in the context of therapeutic applications, since it can provide the opportunity to treat and cure diseases (Gyngell et al., 2017 ). For example, intervention in severe genetic disorders is considered as therapeutic and hence morally permissible, or even obligatory. Others consider HGE to be more like a public health measure, which could be used to reduce the prevalence of a disease (Schaefer, 2020 ). However, others maintain that reproductive uses of HGE are not therapeutic because there is no individual in a current state of disease which needs to be treated, rather a prospective individual to be born with a specific set of negative prospective traits (Rulli, 2019 ).
Below, HGE is discussed in the context of reproductive uses and conditions of clinical advantage over existent reproductive technologies. The HGE applications are explored regarding their potential for modifying one or more disease-related genes relevant to the clinical context. Other uses associated with enhancement of physical and mental characteristics, which are considered non-clinical (although the distinction is sometimes blurred), are also discussed.
Single gene disorders
An obvious application of HGE interventions is to prevent the inheritance of genetic variants known to be associated with a serious disease or condition. Its potential use for this purpose could be typically envisaged through assisted reproduction, i.e., as a process to provide reproductive options to couples or individuals at risk of transmitting genetic conditions to their offspring. Critics of this approach often argue that other assisted reproductive technologies (ARTs) and preimplantation screening technologies e.g., preimplantation genetic diagnosis (PGD), not involving the introduction of genetic modifications to germline cells, are already available for preventing the transmission of severe genetic conditions (Lander, 2015 ; Lanphier et al., 2015 ). These existent technologies aim to support prospective parents in conceiving genetically related children without the condition that affect them. In particular, PGD involves the creation of several embryos by in vitro fertilization (IVF) treatment that will be tested for genetic anomalies before being transferred to the uterine cavity (Sermon et al., 2004 ). In Europe, there is a range in the regulation of the PGD technology with most countries having restrictions of some sorts (Soini, 2007 ). The eligibility criteria for the use of PGD also vary across countries, depending on the range of heritable genetic diseases for which it can be used (Bayefsky, 2016 ).
When considering its effectiveness, PGD presents specific limitations, which include the rare cases in which either both prospective parents are homozygous carriers of a recessive genetic disease, or one of the parents is homozygous for a dominant genetic disease (Ranisch, 2020 ). In these cases, all embryos produced by the prospective parents will be affected by the genetic defect, and therefore it will not be possible to select an unaffected embryo after PGD. Currently, beyond adoption of course, the options available for these prospective parents include the use of a third-party egg or sperm donors.
Overall, given the rarity of cases in which it is not applicable, PGD is thought to provide a reliable option to most prospective parents for preventing severe genetic diseases to be transmitted to their offspring, except in very specific cases. HGE interventions have been suggested to be an alternative method to avoid single gene disorders in the rare cases in which selection techniques such as PGD cannot be used (Ranisch, 2020 ). It has also been proposed to use tools such as CRISPR/Cas9 to edit morphologically suitable but genetically affected embryos, and thus increase the number of embryos available for transfer (de Wert et al., 2018 ; Steffann et al., 2018 ). Moreover, HGE interventions are considered by some as a suitable alternative to PGD, even when the use of PGD could be possible. One argument in this respect is that, although not leading to the manifestation of the disease, the selected embryos can still be carriers of it. In this respect, differently from PGD, HGE interventions can be used to eliminate unwanted, potential future consequences of genetic diseases (i.e., by eliminating the critical mutation carried out in the selected embryo), with the advantage of reducing the risks of further propagation of the disease in subsequent future generations (Gyngell et al., 2017 ).
Overall, HGE interventions are thought to offer a benefit over PGD in some situations by providing a broader range of possible interventions, as well as by providing a larger number of suitable embryos. The latter effect is usually important in the cases where unaffected embryos are small in number, making PGD ineffective (Steffann et al., 2018 ). Whether these cases provide a reasonable ground to justify research and development on the clinical use of HGE remain potentially contentious. Some authors have suggested that the number of cases in which PGD cannot be effectively used to prevent transmission of genetic disorders is so marginal that clinical application of HGE could hardly be justified (Mertes and Pennings, 2015 ). Particularly when analyzing economic considerations (i.e., the allocation of already scarce resources towards clinical research involving expensive techniques with limited applicability) and additional risks associated with direct interventions. In either case of HGE being used as an alternative or a complementary tool to PGD, PGD will most likely still be used to identify those embryos that would manifest the disease and would hence require subsequent HGE.
The PGD technique, however, is not itself free of criticism and possible moral advantages of HGE over PGD have also been explored (Hammerstein et al., 2019 ; Ranisch, 2020 ). PGD remains ethically controversial since, identifying an unaffected embryo from the remaining embryos (which will not be used and ultimately discarded) amounts to the selection of ‘healthy’ embryos rather than ‘curing’ embryos affected by the genetic conditions. On the other hand, given a safe and effective application of the technology, the use of HGE is considered by many morally permissible to prevent the transmission of genetic variants known to be associated with serious illness or disability (de Miguel Beriain, 2020 ). One question that remains is whether HGE and PGD have a differing or equal moral permissibility or, at least, comparable. On issues including human dignity and autonomy, it was argued that HGE and PGD interventions can be considered as equally morally acceptable (Hammerstein et al., 2019 ). This equal moral status was, however, only valid if HGE is used under the conditions of existent gene variants in the human gene pool and to promote the child health’s best interest in the context of severe genetic diseases (Hammerstein et al., 2019 ). Because of selection and ‘therapy’, moral assessments resulted in HGE interventions being considered to some extent preferable to PGD, once safety is carefully assessed (Gyngell et al., 2017 ; Cavaliere, 2018 ). Specifically, PGD’s aim is selective and not ‘therapeutic’, which could be said to contradict the aims of traditional medicine (MacKellar and Bechtel, 2014 ). In contrast to PGD’s selectivity, HGE interventions are seen as ‘pre-emptively therapeutic’, and therefore closer to therapy than PGD (Cavaliere, 2018 ). However, it is also argued that HGE does not have curative aims, and thus it is not a therapeutic application, as there is no patient involved in the procedure to be cured (Rulli, 2019 ). On balance, there appears to be no consensus on which of the approaches, HGE and PGD, is morally a better strategy to prevent the transmission of single gene disorders, with a vast amount of literature expressing diverse positions when considering different scenarios (Delaney, 2011 ; Gyngell et al., 2017 ; Cavaliere, 2018 ; Ranisch, 2020 ; Rehmann-Sutter, 2018 ; Sparrow, 2021 ).
Polygenetic conditions
HGE is also argued to have the potential to be used in other disorders which have a polygenic disposition and operate in combination with environmental influences (Gyngell et al., 2017 , 2019 ). Many common diseases, which result from the involvement of several genes and environmental factors, fall into this category. Examples of common diseases of this type includes diabetes, coronary artery disease and different types of cancers, for which many of the genes involved were identified by studies of genome wide association (e.g., Wheeler and Barroso, 2011 ; Peden and Farral, 2011 ). These diseases affect the lives of millions of people globally, severely impacting health and often leading to death. Furthermore, these diseases have a considerable burden on national health systems. Currently, many of these diseases are controlled through pharmaceutical products, although making healthier life choices about diet and exercise can also contribute to preventing and managing some of them. Despite the interest, the use of PGD in polygenic conditions would hardly be feasible, due to the number of embryos needed to select the preferred genotype and available polygenic predictors (Karavani et al., 2019 ; Shulman and Bostrom, 2014 ).
In theory, HGE could be a potentially useful tool to target different genes and decrease the susceptibility to multifactorial conditions in current and future generations. The application of HGE to polygenic conditions is often argued by noting that the range of applicability of the technique (well beyond single gene disorders) would justify and outweigh the cost needed to develop it. However, to do so, a more profound knowledge of genetic interactions, of the role of genes and environmental factors in diverse processes would be needed to be able to modify such interconnected systems with limited risk to the individual (Lander, 2015 ). Besides, it is now understood that, depending on the genetic background, individuals will have different risks of developing polygenetic diseases (risk-associated variants), but hardly any certainty of it. In other words, although at the population level there would most likely be an incidence of the disease, it is not possible to be certain of the manifestation of the disease in any specific individual. As a result, the benefits of targeting a group of genes associated to a disease in a specific individual would have to be assessed in respect to the probability of incidence of the disease. The risk-benefit ratio for HGE is considerably increased for polygenic conditions compared to monogenic disorders. Additionally, the risks of adverse effects, e.g., off-target effects, increases with the number of genes targeted for editing. The latter effects make the potential benefits of HGE in polygenic diseases more uncertain than in single gene disorders.
Genetic enhancement
A widespread concern regarding the use of HGE is that such interventions could be used not only to prevent serious diseases, but also to enhance desirable genetic traits. Currently, our knowledge on how to genetically translate information into specific phenotypes is very limited and some argue that it might never be technically feasible to achieve comprehensive genetic enhancements using current gene editing technologies (Janssens, 2016 ; Ranisch, 2021 ). Similar to many diseases, in which different genetic and other factors are involved, many of the desirable traits to be targeted by any enhancement will most likely be the result of a combination of several different genes influenced by environment and context. Moreover, the implications for future generations of widespread genetic interventions in the human population and its potential impact on our evolutionary path are difficult to assess (Almeida and Diogo, 2019 ). Nevertheless, others argue that genetic enhancement through HGE could be possible in the near future (de Araujo, 2017 ).
There has been much discussion regarding the meaning of the terms and the conceptual or normative difference between ‘therapy’ and ‘enhancement’ (for an early discussion: Juengst, 1997 ; Parens, 1998 ). There are mainly three different meanings of ‘enhancement’ used in the literature. First, ‘enhancement’ is sometimes used to refer to measures that go beyond therapy or prevention of diseases, i.e., that transcend goals of medicine. Second, ‘enhancement’ is used to refer to measures that equip a human with traits or capacities that they typically do not possess. In both cases, the term points to equally controversial and contrasting concepts: on the one hand, those of ‘health’, ‘disease’ or ‘therapy’, and on the other, those of ‘normality’ or ‘naturalness’. Third, ‘enhancement’ is sometimes also used as an umbrella-term describing all measures that have a positive effect on a person’s well-being. According to this definition, the cure, or prevention of a disease is then also not opposed to an enhancement. Here again, this use refers to the controversial concept of ‘well-being’ or a ‘good life’.
It is beyond the scope of this article to provide a detailed review of the complex debate about enhancement (for an overview: Juengst and Moseley, 2019 ). However, three important remarks can be made: first, although drawing a clear line between ‘enhancement’ and ‘therapy’ (or ‘normality’, etc.) will always be controversial, some cases can be clearly seen as human enhancement. This could include modifications to augment human cognition, like having a greater memory, or increasing muscle mass to increase strength, which are not considered essential for human health (de Araujo, 2017 ).
Second, it is far from clear whether a plausible account of human enhancement would, in fact, be an objectivist account. While authors suggest that there is some objectivity regarding the conditions that constitute a serious disease (Habermas, 2003 ), the same might not be true for what constitutes an improvement of human functioning. It may rather turn out that an enhancement for some might be seen as a dis-enhancement for others. Furthermore, the use of the HGE for enhancement purposes can be considered at both an individual and a collective level (Gyngell and Douglas, 2015 ; Almeida and Diogo, 2019 ), with a range of ethical and biological implications. If HGE is to be used for human enhancement, this use will be in constant dependence on what we perceive as ‘normal’ functioning or as ‘health’. Therefore, factors such as cultural and societal norms will have an impact on where such boundaries are drawn (Almeida and Diogo, 2019 ).
Third, it should be noted that from an ethical perspective the conceptual question of what enhancement is, and what distinguishes it from therapy, is less important than whether this distinction is ethically significant in the first place. In this context, it was pointed out that liberal positions in bioethics often doubt that the distinction between therapy and enhancement could play a meaningful role in determining the limits of HGE (Agar, 1998 ). The consideration of genetic intervention for improving or adding traits considered positive by individuals have raised extreme positions. Some welcome the possibility to ameliorate the human condition, whilst others consider it an alarming attempt to erase aspects of our common human ‘nature’. More specifically, some authors consider HGE a positive step towards allowing humans the opportunity to obtain beneficial traits that otherwise would not be achievable through human reproduction, thus providing a more radical interference in human life to overcome human limitations (de Araujo, 2017 ; Sorgner, 2018 ). The advocates of this position are referred to as ‘bioliberals’ or ‘transhumanists’ (Ranisch and Sorgner, 2014 ), and its opponents are referred to as ‘bioconservatives’ (Fukuyama, 2002 ; Leon, 2003 ; Sandel, 2007 ). Transhumanism supports the possibility of humans taking control of their biology and interfering in their evolution with the use of technology. Bioconservatism defends the preservation and protection of ‘human essence’ and expresses strong concerns about the impact of advanced technologies on the human condition (Ranisch and Sorgner, 2014 ).
For the general public, HGE used in a clinical context seems to be less contentious compared when used as a possible human enhancement tool. Specifically, some surveys indicate that the general-public typically exhibits a reduced support for the use of genome editing interventions for enhancement purposes compared to therapeutic purposes (Gaskell et al., 2017 ; Scheufele et al., 2017 ). In contrast, many technologies and pharmaceutical products developed in the medical context to treat patients are already being used by individuals to ‘enhance’ some aspect of their bodies. Some examples include drugs to boost brain power, nutritional supplements, and brain-stimulating technologies to control mood, even though their efficiency and safety is not clear. This could suggest that views on enhancement may vary depending on the context and on what is perceived as an enhancement by individuals. It may be informative to carry out detailed population studies to explore whether real ethical boundaries and concerns exist, or whether these are purely the result of the way information is processed and perceived.
Heritable genome editing: Mapping the ethical debate
Even though genome editing methods have only been developed in the last decade, the normative implication of interventions into the human germline have been discussed since the second half of the 20th century (Walters et al., 2021 ). Some even argue that, virtually, all the ethical issues raised by genetic engineering were already being debated at that time (Paul, 2005 ). This includes questions about the distinction between somatic and germline interventions, as well as between therapy and enhancement (e.g., Anderson, 1985 ). Nevertheless, as it has been widely noted, it is difficult to draw clear lines between these two categories (e.g., McGee, 2020 ; Juengst, 1997 ), and alternative frameworks have been proposed, particularly in the context of HGE (Cwik, 2020 ). Other questions include the normative status of human nature (e.g., Ramsey, 1970 ), the impossibility of consent from future generations (e.g., Lappe, 1991 ), possible slippery slopes towards eugenics (e.g., Howard and Rifkin, 1977 ), or implications for justice and equality (e.g., Resnik, 1994 ).
When discussing the ethics of HGE, roughly three types of considerations can be distinguished: (i) pragmatic, (ii) sociopolitical, and iii) categorical (Richter and Bacchetta, 1998 ; cf. Carter, 2002 ). Pragmatic considerations focus on medical or technological aspects of HGE, such as the safety or efficacy of interventions, risk–benefit ratio, possible alternatives or the feasibility of responsible translational research. Such considerations largely depend on the state of science and are thus always provisional. For example, if high-risk technologies one day evolve into safe and reliable technologies, some former pragmatic considerations may become obsolete. Sociopolitical aspects, on the other hand, are concerned with the possible societal impact of technologies, e.g., how they can promote or reduce inequalities, support or undermine power asymmetries, strengthen, or threaten democracy. Similar to pragmatic considerations, sociopolitical reasons depend on specific contexts and empirical factors. However, these are in a certain sense ‘outside’ the technology—even though technologies and social realities often have a symbiotic relationship. While sociopolitical considerations can generate strong reasons against (or in favour of) implementing certain technologies, most often these concerns could be mitigated by policies or good governance. Categorical considerations are different and more akin to deontic reasons. They emphasise categorical barriers to conduct certain deeds. It could be argued, for instance, that the integrity of the human genome or the impossibility to obtain consent from future generation simply rule out certain options to modify human nature. Such categorical considerations may persist despite technological advances or changing sociopolitical conditions.
Comparing the bioethical literature on genetic engineering from the last century with the ongoing discussions shows a remarkable shift in the ethical deliberation. In the past, scholars from the field of medical ethics, as well as policy reports, used to focus on possible categorical boundaries for germline interventions and on possible sociopolitical consequences of such scenarios. For instance, the influential 1982 report “Splicing Life” from the US President’s Commission for the Study of Ethical Problems in Medicine and Biomedical and Behavioural Research prominently discussed concerns about ‘playing God’ against the prospects of genetically engineering human beings, as well as possible adverse consequences of such interventions. Although this study addresses potential harms, pragmatic arguments played only a minor role, possibly due to the technical limitations at the time.
With the upcoming availability of effective genome editing techniques, the focus on the moral perspective seems to have been reversed. Increasingly, the analysis of the permissibility of germline interventions is confined to questions of safety and efficacy. This is demonstrated by the 2020 consensus study report produced by an international commission convened by the US National Academy of Medicine, the US National Academy of Sciences, and the UK’s Royal Society, which aimed at defining a translational pathway for HGE. Although the report recognizes that HGE interventions does not only raise pragmatic questions, ethical aspects were not explicitly addressed (National Academy of Medicine, National Academy of Sciences, and the Royal Society, 2020 ).
Similarly, in 2019, a report on germline interventions published by the German Ethics Council (an advisory body to the German government and parliament) emphasizes that the “previous categorical rejection of germline interventions” could not be maintained (Deutscher Ethikrat, 2019 , p. 5). The German Ethics Council continues to address ethical values and societal consequences of HGE. However, technical progress and the development of CRISPR/Cas9 tools seem to have changed the moral compass in the discussion about germline interventions.
For a comprehensive analysis of HGE to focus primarily on pragmatic arguments such as safety or efficacy would be inadequate. In recent years, developments in the field of genome editing have occurred at an incredibly fast pace. At the same time, there are still many uncertainties about the efficacy of the various gene editing methods and unexpected effects in embryo editing persist (Ledford, 2015 ). Social and political implication also remain largely unknown. To date, it has been virtually impossible to estimate how deliberate interventions into the human germline could shape future societies and to conduct a complete analysis of the safety aspects of germline interventions.
Moreover, as the EGE notes, we should be cautious not to limit the complex process of ethical decision-making to pragmatic aspects such as safety. The “‘safe enough’ narrative purports that it is enough for a given level of safety to be reached in order for a technology to be rolled out unhindered, and limits reflections on ethics and governance to considerations about safety” (EGE, 2021 , p. 20). Consequently, the EGE has highlighted the need to engage with value-laden concepts such as ‘humanness’, ‘naturalness’ or ‘human diversity’ when determining the conditions under which HGE could be justified. Even if a technology has a high level of safety, its application may still contradict ethical values or lead to undesirable societal consequences. Efficacy does not guarantee compatibility with well-established ethical values or cultural norms.
While concepts such as ‘safety’ or ‘risk’ are often defined in scientific terms, this does not take away the decision of what is ethically desirable given the technical possibilities. As Hurlbut and colleagues put it in the context of genome editing: “Limiting early deliberation to narrowly technical constructions of risk permits science to define the harms and benefits of interest, leaving little opportunity for publics to deliberate on which imaginations need widening, and which patterns of winning and losing must be brought into view” (Hurlbut et al., 2015 ). Therefore, if public engagement is to be taken seriously, cultural norms and values of those affected by technologies must also be considered (Klingler et al., 2022 ). This, however, means broadening the narrow focus on pragmatic reasons and allowing categorical as well as sociopolitical concerns in the discourse. Given the current attention on pragmatic reasons in current debates on HGE, it is therefore beneficial to revisit the categorical and sociopolitical concerns that remain unresolved. The following sections provide an overview of relevant considerations that can arise in the context of HGE and that underline many of the societal concerns and values crucial for public engagement.
Human genome ‘integrity’
Heritability seems to be one of the foremost considerations regarding germline genome editing, as it raises relevant questions on a ‘natural’ human genome and its role in ‘human nature’ (Bayertz, 2003 ). This follows an ongoing philosophical debate on ‘human nature’, at least as defined by the human genome. This has ensued a long debate on the value of the human genome and normative implications associated with its modification (e.g., Habermas, 2003 ). Although a comprehensive discussion of these topics goes beyond the scope of this paper, the human genome is viewed by many as playing an important role in defining ‘human nature’ and providing a basis for the unity of the human species (for discussion: Primc, 2019 ). Considering the implications for the individual and the collective, some affirm the right of all humans to inherit an unmodified human genome. For some authors, germline modification is considered unethical, e.g., a “line that should not be crossed” (Collins, 2015 ) or a “crime against humanity” (Annas et al., 2002 ).
The Universal Declaration on the Human Genome and Human Rights (UDHGHR) states that “the human genome underlies the fundamental unity of all members of the human family, as well as the recognition of their inherent dignity and diversity. In a symbolic sense, it is the heritage of humanity” (Article 1, UNESCO, 1997 ). The human genome is viewed as our uniquely human collective ‘heritage’ that needs to be preserved and protected. Critics of heritable genetic interventions argue that germline manipulation would disrupt this natural heritage and therefore would threaten human rights and human equality (Annas, 2005 ). Heritable human genome editing creates changes that can be heritable to future generations. For many, this can represent a threat to the unity and identity of the human species, as these modifications could have an impact on the human’s gene pool. Any alterations would then affect the evolutionary trajectory of the human species and, thus, its unity and identity.
However, the view of the human genome as a common heritage is confronted with observations of the intrinsic dynamism of the genome (Scally, 2016 ). Preservation of the human genome, at least in its current form, would imply that the genome is static. However, the human genome is dynamic and, at least in specific periods of environmental pressure, must have naturally undergone change, as illustrated by human evolution (Fu and Akey, 2013 ). The genome of any individual includes mutations that have occurred naturally. Most of them seem to be neither beneficial nor detrimental to the ability of an individual to live or to his/her health. Others can be detrimental and limiting to their wellbeing. It has been shown that, on average, each human genome has 60 new mutations compared to their parents (Conrad et al., 2011 ). At the human population level, a human genome can have in average 4.1–5 million variants compared to the ‘reference’ genome (Li and Sadler, 1991 ; Genomes Project C, 2015 ). The reference genome itself is thus a statistical entity, representing the statistic distribution of the probability of different gene variants in the whole genome. Human genomic variation is at the basis of the differences in the various physical traits present in humans (e.g., eye colour, height, etc.), as well as specific genetic diseases. Thus, the human population is comprised of genomes with a pattern of variants and not of ‘one’ human genome that needs to be preserved (Venter et al. 2001 ). The human genome has naturally been undergoing changes throughout human history. An essentialist view of nature seems to be the basis for calling for the preservation of genome integrity. However, in many ways, this view is intrinsically challenged by the interpretation portrayed by evolutionary biology of our genetic history already more than a century ago. Nevertheless, despite the dynamic state of the human genome, this in itself cannot justify the possibility of modifying the human genome. It is also worth considering that the integrity of the human genome could also be perceived in a ‘symbolic’ rather than biological literal meaning. Such an interpretation would not require a literally static genome over time, but instead suggest a boundary between ‘naturally’ occurring variation and ‘artificially’ induced change. This is rather a version of the ‘natural’/unnatural argument, rather than an argument for a literally unchanged genetic sequence.
The modification of the human genome raises complex questions about the characterization of the human species genome and if there should be limits on interfering with it. The options to modify the human genome could range from modifying only the genes that are part of the human gene pool (e.g., those genes involved in severe genetic diseases such as Huntington’s disease) to adding new variants to the human genome. Regarding variants which are part of the common range of variation found in the human population (although it is not possible to know all the existent variations), the question becomes whether HGE could also be used in any of them (e.g., even the ones providing some form of enhancement) or only in disease-associated variants and thus be restricted to the prevention of severe genetic diseases. In both cases, the integrity of the human genome is expected to be maintained with no disruption to human lineage. However, it could be argued that this type of modification is defending a somewhat conservative human nature argument, since it is considering that a particular genetic make-up is ‘safe’ or would not involve any relevant trade-offs. In contrast, a different conclusion could be drawn on the integrity of the human genome when introducing genotypical and phenotypical traits that do not lie within the common range of variation found in the population (Cwik, 2020 ). In all cases, since the implications of the technology are intergenerational and consequently, it will be important to carry out an assessment of the risks that we, as a species, are willing to take when dealing with disease and promoting health. For this, we will need to explore societal views, values and cultural norms associated with the human genome, as well as possibly existing perceptions of technology tampering with ‘nature’. To support such an assessment, it would be useful to draw on a firm concept of human nature and the values it implies, beyond what is implied by genetic aspects.
Human dignity
In several of the legally binding and non-binding documents addressing human rights in the biomedical field, human dignity is one of the key values emphasized. There are concerns that heritable genome interventions might conflict with the value of human dignity (Calo, 2012 ; Melillo, 2017 ). The concerns are considered in the context of preserving the human genome (Nordberg et al. 2020 ). More specifically, the recommendation on Genetic Engineering by the Council of Europe (1982) states that “ the rights to life and to human dignity protected by Articles 2 and 3 of the European Convention on Human Rights imply the right to inherit a genetic pattern which has not been artificially changed” (Assembly, 1982 ). This is supported by the Oviedo Convention on Human Rights and Biomedicine (1997), where Article 13 prohibits any genetic intervention with the aim of introducing a modification in the genome of any descendants. The Convention is the only international legally binding instrument that covers human germline modifications among the countries which have ratified it (Council of Europe, 1997 ). However, there have been some authors disputing the continued ban proposed by the Oviedo Convention (Nordberg et al. 2020 ). Such authors have focused on the improvements of safety and efficacy of the technology in contrast to authors focusing on its value for human dignity (Baylis and Ikemoto, 2017 ; Sykora and Caplan, 2017 ). The latter authors seem to highlight the concept of human dignity to challenge heritable interventions to the human genome.
But a question in debate has been to demonstrate how ‘human dignity’, described in such norms, relates to heritable genome interventions. The concept of the human genome as common genetic heritage, distinguishing humans from other species seems one of the main principles implied by such norms. In this view, the human genome determines who belongs to the human species and who does not, and thus confers an individual the dignity of being a human by association. This creates an inherent and strong link between the concept of human genome and the concept of human dignity and its associated legal rights (Annas, 2005 ). It could be argued that a genetic modification to an individual may make it difficult for him/her to be recognized as a human being and therefore, preservation of the human genome being important for human dignity to be maintained. This simple approach, or at least interpretation, however, ignores the fact that the human genome is not a fixed or immutable entity, as exemplified by human evolution (as discussed in the previous section). As a result, the view that HGE interventions are inherently inadmissible based on the need to preserve human dignity is contested (Beriain, 2018 ; Raposo, 2019 ). More broadly, the idea that biological traits are the basis for equality and dignity, supporting the need for the human genome to be preserved, is often challenged (Fenton, 2008 ).
It is argued that to fully assess the impact of the HGE interventions on human dignity, it will be necessary to have a better understanding of the concept of human dignity in the first place (Häyry, 2003 ; Cutas, 2005 ). For some, however, human dignity is a value that underlies questions of equality and justice. Thus, the dignity-based arguments could uncover relevant questions in the discussion of ethical implications on modifying the human genome (Segers and Mertes, 2020 ). In the Nuffield Council on Bioethics Report (2018) principles of social justice and solidarity, as well as welfare, are used to guide the debate on managing HGE interventions. Similarly, the concept of human dignity could, therefore, provide the platform upon which consideration of specific values could be discussed, broaden the debate on HGE to values shared by society.
Right of the child: informed consent
In many modern societies, every individual, including children, have the rights to autonomy and self-determination. Therefore, each person is entitled to decide for themselves in decisions relating to their body. These rights are important for protecting the physical integrity of a person. When assessing the implication of allowing individuals to take (informed) decisions relative to the use of heritable genetic interventions on someone else’s body, it is useful to reflect on the maturity of existing medical practices and, more broadly, on the additional complexities associated with the heritability of any such intervention.
In modern health-care systems, informed consent provides the opportunity for an individual to exercise autonomy and make an informed decision about a medical procedure, based on their understanding of the benefits and risks of such procedure. Informed consent is thus a fundamental principle in medical (research) ethics when dealing with human subjects (Beauchamp and Childress, 2019 ).
Heritable genome interventions present an ethical constraint on the impossibility of future generations of providing consent to an intervention on their genome (Smolenski, 2015 ). In other words, future generations cannot be involved in a decision which could limit their autonomy, since medical or health-related decisions affecting them are placed on the present generation (and, in the case of a child to be born, more specifically, on his/her parents). However, many other actions taken by parents of young children also intentionally influence the lives of those children and have been doing so for millennia (Ranisch, 2017 ). Although these actions may not involve altering their genes, many of such actions can have a long-lasting impact on a child’s life (e.g., education and diet). However, it could be argued that they do not have the irreversible effect that HGE will have in the child and future generations. In cases where parents act to expand the life choices of their children by eliminating disease (e.g., severe genetic diseases), this would normally be thought to outweigh any possible restriction on autonomy. In these cases, if assuming HGE benefits will outweigh risks regarding safety and efficacy, the use of HGE could be expected to contribute to the autonomy of the child, as him/her would be able in the future to have a better life, not constrained by the limitations of the disease. As a result, even if it is accepted that these technologies may in one way reduce the autonomy of future generations, some believe that this will often be outweighed by other effects increasing autonomy (Gyngell et al., 2017 ). In other words, it is reasonable to suppose that, when taken by parents based on good information and understanding of risks and impacts, the limitation in the autonomy of unborn children associated with heritable genetic interventions would be compensated by the beneficial effects of increasing their autonomy when born (Gyngell et al., 2017 ).
It has often been emphasized that possible genetic interventions must not curtail the future possibilities of offspring to live their lives according to their own idea of a good life. This view originated in the liberal tradition and is associated with the “right to an open future”, defended by Joel Feinberg ( 1992 ). That is an anticipatory autonomy right that parents can violate, even though the offspring could exercise it only in the future. Feinberg has discussed the right to an open future in the context of religious education. However, various authors have applied this argument to the question of permissible and desirable genetic interventions (Buchanan et al., 2000 ; Glover, 2006 ; Agar, 1998 ). Accordingly, germline modifications or selection would have to allow the offspring to have a self-determined choice of life plans. It would therefore be necessary to provide offspring with genetic endowments that represent the so-called all-purpose goods. These goods are “useful and valuable in carrying out nearly any plan of life or set of aims that humans typically have” (Buchanan et al., 2000 , p. 167). While this claim is certainly appealing, in reality it will be difficult to identify phenotypes that will only broaden and do not narrow the spectrum of life plans. Take, for example, body size: a physique favourable for a basketball player would at the same time be less favourable in successfully riding horses as a professional jockey and vice versa. Increasing some opportunities often means reducing other ones.
The arguments of informed consent and open future need to be explored outside the realm of severe genetic diseases by considering other scenarios (including scenarios of genetic enhancement). Hereby, the effects of the interventions on the autonomy of future generations can be assessed more comprehensively. As for enhancement, decisions outside the realm of health can be more controversial, as the traits that parents see fit to generate enhancement may inadvertently condition a child’s choices in the future in an undesirable way.
If HGE is to be used, questions on how the consent and information should be provided to parents to fully equip them to decide in the best interests of the child will need to be assessed (Evitt et al., 2015 ). This is evident if considering the informed consent used in the study conducted by He Jiankui. One of the many criticisms of the study was the inadequacy of the informed consent process provided to the parents, which did not meet regulatory or ethical standards (Krimsky, 2019 ; Kirksey, 2020 ). This raises questions on how best to achieve ethical and regulatory compliance regarding informed consent in applications of HGE (Jonlin, 2020 ).
Discrimination of people with disabilities
For many years, there has been an effort to develop selective reproduction technologies to prevent genetic diseases or conditions leading to severe disabilities. These forms of reproductive genetic disease prevention are based on effectively filtering and eradicating embryos or foetuses affected by genetic diseases. There are divergent views regarding the use of these technologies. For example, the disability rights movement argues that the use of technologies such as prenatal testing (PNT) and PGD discriminates against people living with a disability (Scully, 2008 ; Asch and Barlevy, 2012 ). The key arguments presented supporting this view are: (i) the limited value of a genetic trait in respect to the life of an embryo (Parens and Asch, 2000 ) and (ii) the ‘expressivist’ argument (Buchanan, 1996 ; Shakespeare, 2006 ). The first argument is based on the critique that a disabling trait is viewed as being more significant than the life of an embryo/foetus. This argument was initially used in the context of prenatal testing and selective termination, and has also been applied in the context of new technologies like PGD (Parens and Asch, 2000 ). The second, the ‘expressivist’ argument, argues that the use of these technologies expresses negative or discriminatory views on the disabling conditions they are targeting and subsequently on the people living with these conditions (Asch and Wasserman, 2015 ). The expressivist argument, however, has been challenged by stressing the importance of differentiating between the disability itself and the people living with disability (Savulescu, 2001 ). The technology’s use is aimed at reducing the incidence of disability, and it does not have a position of value on the people that have a specific condition.
When applying the same arguments to the use of HGE in comparison with other forms of preventing heritable genetic diseases, some important considerations can be made. Regarding the first argument, in contrast to selective reproduction technologies, HGE may allow the removal of the disabled trait with the aim of ensuring survival of the affected embryo. However, most likely, PGD would be used before and after the editing of the embryos to help the identification of the ones requiring intervention and verifying the efficiency of the genetic intervention (de Miguel Beriain, 2018 ; Ranisch, 2020 ). Similarly, the expressivist argument continues to be challenged if the application of human HGE is envisaged in the context of severe genetic diseases (e.g., Tay-Sachs and Huntington’s disease). It has been argued that the choice to live without a specific genotype neither implies discriminating people living with a respective condition nor considering the life of people living with the disease not worth living or less valuable (Savulescu, 2001 ). In other words, the expressivist argument is not a valid or a sufficiently strong ethical argument for prospective parents not to have the option to have a future child without a genetic disease.
It is worth noting that the debate on the use of reproduction technologies for the prevention of genetic diseases is not at all new, and that modern HGE techniques only serve to highlight ethical concerns that have been expressed for a long time. In the case of preventing genetic diseases, the application of both arguments to HGE intervention could be considered not to provide sufficiently strong ethical arguments to limit the use of the technology in the future. However, it is worth exploring whether scientific innovations like HGE are either ameliorating or reinvigorating ethical concerns expressed so far, for example in creating a future that respects or devalues disability as a part of the human condition. Perhaps even more importantly, given their potential spectrum of possible intervention and efficacy, it is important to reflect on whether the broad use of HGE could have an impact on concepts of disability and ‘normality’ as a whole distorting an already unclear ethical line between clinical and non-clinical interventions. Moreover, research work exploring the relationship between disability and identity indicated that personhood with disability can be an important component to people’s identity and interaction with the world. In the case of heritable human genome editing, it is not yet known how this technology will impact the notions of identity and personhood in people who had their germline genome modified (Boardman and Hale, 2018 ). For further progress on these issues public engagement might be important to gather different views and perceptions on the issue.
Justice and equality
Beside the limits of applicability, another common ethical concern associated with the use of genome editing technologies, as with many new technologies, is the question of accessibility (Baumann, 2016 ). Due to the large investments that will need to be made for continuing development of the technology, there is a (perceived) risk of it becoming an expensive technology that only a few wealthy individuals in any population (and/or only citizens in comparatively rich countries) can access. In addition, there is concern that patenting of genome editing technologies will delay widespread access or lead to unequal distribution of corresponding benefits (Feeney et al., 2018 ). This may, consequently, contribute to further increases in existing disparities, since individuals or countries with the means of accessing better health treatments may have economic advantages (Bosley et al., 2015 ). This could enhance inequality at different levels, depending on the limits of applicability of the technology. Taken to its extreme, the use of the technology could allow germline editing to create and distinguish classes of individuals that could be defined by the quality of their manipulated genome.
The concern that the possibility of germline interventions in humans could entrench or even increase inequalities has accompanied the discussion about ethics of genetic interventions from the very beginning until today (e.g. Resnik, 1994 ). In ‘Remaking Eden’ Lee Silver envisioned a divided future society, consisting of a genetically enhanced class, the “genRich”, and a genetic underclass, the “naturals” (Silver, 1997 ). Françoise Baylis recently echoed such concerns regarding future HGE interventions, namely that “unequal access to genome-editing technologies will both accentuate the vagaries of the natural lottery and introduce an unjust genetic divide that mirrors the current unjust economic and social divide between rich and poor individuals” (Baylis, 2019 , p. 67). At the same time, the possibility to genetically intervene in the ‘natural lottery’ has also been associated with the hope of countering natural inequalities and increase equality of opportunities. Robert Sinsheimer may be among the first to envision such a ‘new’ individualistic type of ‘eugenics’ that “would permit in principle the conversion of all of the unfit to the highest genetic level” (Sinsheimer, 1969 , p. 13). More recently, in the book ‘From chance to choice: Genetics and justice’ (2000) it is argued that “equality of opportunity will sometimes require genetic interventions and that the required interventions may not always be limited to the cure or prevention of disease” (Buchanan et al., 2000 , p. 102). When discussing issues related to justice and equality, it will be important to involve a broad spectrum of stakeholders to better evaluate the economic effects of the commercialization of the technology.
Conclusions
With ongoing technological developments and progress with guiding and regulating its acceptable use, the possibility of HGE interventions in the human genome is closer than ever to becoming a reality. The range of HGE applicability can go from preventing the transmission of genetic variants associated with severe genetic conditions (mostly single gene disorders but also, to a lesser extent, polygenic diseases) to genetic enhancements. The permissibility of HGE has often been considered on the basis of possible uses, with therapeutic uses generally considered more acceptable than non-therapeutic ones (including human enhancement). When compared with other technologies with similar therapeutic uses (e.g., PGD) already in use, HGE presents similarities and differences. However, from an ethical acceptability perspective, there is currently no consensus on whether HGE is more or less acceptable than PGD.
An important conclusion of this study is that, along with the technological development of genome germline editing techniques, a shift in the focus of analyses on its applicability has been observed. More specifically, the emphasis on pragmatic considerations seems to have increased substantially compared with the previous emphasis on categorical and sociopolitical arguments. Many of the most recent publications from authoritative advisory committees and institutions discuss the permissibility of HGE interventions primarily on the basis of pragmatic arguments, in which safety and efficacy are the main focus. Since germline interventions could profoundly change the human condition, the need for a broad and inclusive public debate on this topic has also been frequently emphasized. However, limited consideration has been given to approaches to carry out such action effectively, and on how to consider their outcomes in relevant policies and regulations.
It is currently not entirely clear whether: (i) the pragmatic position championed by such authoritative sources builds on the premise that the ethical debate has reached sufficient maturity to allow a turning point; (ii) the lack of progress has somewhat hampered further consideration of issues still considered controversial; (iii) regulatory pressure is somewhat de facto pushing forward the introduction of such technologies despite critical, unresolved ethical issues. Based on the analysis presented in this paper, a combination of the latter factors (ii and iii) seems more likely. In engaging the public in societal debates on the acceptability of such technologies, unresolved questions are likely to re-emerge. Specifically, it is possible that categorical and sociopolitical considerations will gain renewed focus during public engagement. In other words, when involving the public in discussions on HGE, it is possible that cultural values and norms, not only questions of safety and efficacy, will re-emerge as crucial to the acceptance of the technology (What is meant by natural? What is understood by humanity? etc.).
HGE interventions put into question specific biological and moral views of individuals, including views on the value of the human genome, on human dignity, on informed consent, on disability and on societal equality and justice. The range of ethical issues affected by the introduction of such technology, often still characterised by non-convergent, and at times conflicting, positions, illustrate the importance of further consideration of these issues in future studies and public engagement activities. As a result, society’s moral uncertainties will need to be assessed further to support the regulation of HGE technologies and form a well-informed and holistic view on how they can serve society’s common goals and values.
Data availability
This statement is not applicable.
Agar N (1998) Liberal eugenics. Public Aff Q 12(2):137–155
PubMed Google Scholar
Almeida M, Diogo R (2019) Human enhancement: genetic engineering and evolution. Evol Med Public Health 1:183–189. https://doi.org/10.1093/emph/eoz026
Article Google Scholar
Anderson WF (1985) Human Gene Therapy: scientific and ethical considerations. J Med Philos 10(3):275–291. https://doi.org/10.1093/jmp/10.3.275
Article CAS PubMed Google Scholar
Andorno R, Baylis F, Darnovsky M, Dickenson D, Haker H, Hasson K et al. (2020) Geneva statement on heritable human genome editing: the need for course correction. Trends Biotechnol 38(4):351–354
Annas GJ (2005) Bioethics: crossing human rights and health law boundaries. Oxford University Press, New York, NY
Google Scholar
Annas GJ, Andrews LB, Isasi RM (2002) Protecting the endangered human: toward an international treaty prohibiting cloning and inheritable alterations. Am J Law Med 28(2–3):151–178
Article PubMed Google Scholar
Araki M, Ishii T (2014) International regulatory landscape and integration of corrective genome editing into in vitro fertilization. Reprod Biol Endocrinol 12(1):108–120
Article CAS PubMed PubMed Central Google Scholar
Asch A, Barlevy D (2012) Disability and genetics: a disability critique of pre-natal testing and pre-implantation genetic diagnosis. eLS. Wiley, Chichester
Asch A, Wasserman D (2015) Reproductive testing for disability. In: Arras JD, Fenton E, Kukla R (eds.) Routledge companion to bioethics. Routledge, London
Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G et al. (2015) A prudent path forward for genomic engineering and germline gene modification. Science 348(6230):36. https://doi.org/10.1126/science.aab1028
Article ADS CAS PubMed PubMed Central Google Scholar
Baltimore D, Baylis F, Berg P et al. (2015) On human gene editing: international summit statement. News release, December 3, International summit on human gene editing. http://www8.nationalacademies.org/onpinews/newsitem.aspx?RecordID=12032015a
Barrangou R, Horvath P (2017) A decade of discovery: CRISPR functions and applications. Nat Microbiol 2(17092):1–9. https://doi.org/10.1038/nmicrobiol.2017.92
Article CAS Google Scholar
Baumann M (2016) CRISPR/Cas9 genome editing: new and old ethical issues arising from a revolutionary technology. Nanoethics 10:139–59
Bayefsky MJ (2016) Comparative preimplantation genetic diagnosis policy in Europe and the USA and its implications for reproductive tourism. Reprod Biomed Soc Online 3:41–47
Bayertz K (2003) Human nature: how normative might it be? J Med Philos 28(2):131–150. https://doi.org/10.1076/jmep.28.2.131.14210
Baylis F (2017) Human germline genome editing and broad societal consensus. Nat Hum Behav 1:0103
Baylis F (2019) Human genome editing: our future belongs to all of us. Issues Sci Technol 35:42–44
Baylis F, Ikemoto L (2017) The Council of Europe and the prohibition on human germline genome editing. EMBO Rep 8(12):2084–2085. https://doi.org/10.15252/embr.201745343
Baylis F, Darnovsky M, Hasson K, Krahn TM (2020) Human Germ Line and Heritable Genome Editing: the global policy landscape. CRISPR J 3(5):365–377. https://doi.org/10.1089/crispr.2020.0082 . PMID: 33095042
Beauchamp TL, Childress JF (2019) Principles of biomedical ethics. Oxford University Press, USA
Blendon RJ, Gorski MT, Benson JM (2016) The public and the gene-editing revolution. New Engl J Med 374(15):1406–1411. https://doi.org/10.1056/NEJMp1602010
Boardman FK, Hale R (2018) How do genetically disabled adults view selective reproduction? Impairment, identity, and genetic screening. Mol Genet Genom Med 6(6):941–956
Bosley KS, Botchan M, Bredenoord AL, Carroll D, Charo RA, Charpentier E et al. (2015) CRISPR germline engineering: the community speaks. Nat Biotechnol 33(5):478–486. https://doi.org/10.1038/nbt.3227
Botkin JR (2019) The case for banning heritable genome editing. Genet Med 22:487–489
Brokowski C (2018) Do CRISPR germline ethics statements cut it? CRISPR J 1(2):115–125. https://doi.org/10.1089/crispr.2017.0024
Article PubMed PubMed Central Google Scholar
Buchanan A (1996) Choosing who will be disabled: genetic intervention and the morality of inclusion. Soc Philos Policy 13:18–46
Buchanan A, Brock DW, Daniels N, Wikler D (2000) From chance to choice: genetics and justice. Cambridge University Press
Calo Z (2012) Human dignity and health law: personhood in recent bioethical debates. Notre Dame J Law Ethics Public Policy 26:473–499
Carter L (2002) The ethics of germ line gene manipulation—a five dimensional debate. Monash Bioeth Rev 21(4):S66–S81. https://doi.org/10.1007/BF03351288
Cavaliere G (2018) Genome editing and assisted reproduction: curing embryos, society or prospective parents? Med Health Care Phil 21(2):215–25
Coller BS (2019) Ethics of human genome editing. Annu Rev Med 27(70):289–305. https://doi.org/10.1146/annurev-med-112717-094629
Council of Europe (1997) Convention for the protection of human rights and dignity of the human being with regard to the application of biology and medicine: convention on human rights and biomedicine. COE, Oviedo
Collins, F. (2015) Director, National Institutes of Health, https://www.nih.gov/about-nih/who-we-are/nih-director/statements/statement-nih-funding-research-using-gene-editing-technologieshuman-embryos
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F et al. (2011) Variation in genome-wide mutation rates within and between human families. Nat Genet 43(7):712–4. https://doi.org/10.1038/ng.862
Cutas DE (2005) Looking for the meaning of dignity in the bioethics convention and the cloning protocol. Health Care Anal 13(4):303–313
Cwik B (2020) Revising, correcting, and transferring genes. Am J Bioeth 20(8):7–18
Cyranoski D (2019) Russian biologist plans more CRISPR-edited babies. Nature 570(7760):145–147
Article ADS CAS PubMed Google Scholar
Daley GQ, Lovell-Badge R, Steffann J (2019) After the storm—a responsible path for genome editing. N Engl J Med 380:897–899
de Araujo M (2017) Editing the genome of human beings: CRISPR-Cas9 and the ethics of genetic enhancement. J Evol Technol 27(1):24–42. http://jetpress.org/v27.1/araujo.pdf
Dance A (2015) Core concept: CRISPR gene editing. Proc Natl Acad Sci USA 112:6245–6246
Davies K (2020) Editing humanity: the CRISPR revolution and the new era of genome editing. Pegasus Books, New York, NY
Delaney JJ (2011) Possible people, complaints, and the distinction between genetic planning and genetic engineering. J Med Ethics 37(7):410–414
Deutscher Ethikrat, (2019) Intervening in the Human Germline. https://www.ethikrat.org/en/publications/publication-details/?tx_wwt3shop_detail%5Bproduct%5D=119&tx_wwt3shop_detail%5Baction%5D=index&tx_wwt3shop_detail%5Bcontroller%5D=Products&cHash=25e88ad52f8b75d311510a9bf7a8dc86
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096. https://doi.org/10.1126/science.1258096
Drabiak K (2020) The Nuffield Council’s green light for genome editing human embryos defies fundamental human rights law. Bioethics 34:223–227
Dzau VJ, McNutt M, Bai C (2018) Wake-up call from Hong Kong. Science 362(6420):1215. https://doi.org/10.1126/science.aaw3127
de Miguel Beriain I (2020) Gene editing and disabled people: a response to Felicity Boardman. J Community Genet 11(3):241–243
de Miguel Beriain I (2018) Human dignity and gene editing. EMBO Rep 19:e46789
de Wert G, Heindryckx B, Pennings G, Clarke A, Eichenlaub-Ritter U, van El CG (2018) Responsible innovation in human germline gene editing: background document to the recommendations of ESHG and ESHRE. Eur J Hum Genet 26(4):450–470. https://doi.org/10.1038/s41431-017-0077-z
EGE (2016) Statement on gene editing. https://ec.europa.eu/info/sites/default/files/research_and_innovation/ege/gene_editing_ege_statement.pdf
EGE (2021) Ethics of genome editing. https://ec.europa.eu/info/sites/default/files/research_and_innovation/ege/ege_ethics_of_genome_editing-opinion_publication.pdf
Evitt NH, Mascharak S, Altman RB (2015) Human germline crispr-cas modification: toward a regulatory framework. Am J Bioeth 15(12):25–29
Feeney O, Cockbain J, Morrison M, Diependaele L, Van Assche K, Sterckx S (2018) Patenting foundational technologies: lessons from CRISPR and other core biotechnologies. Am J Bioeth 18(12):36–48
Fenton E (2008) Genetic enhancement – a threat to human rights? Bioethics 22(1):1–7 https://doi.org/10.1111/j.1467-8519.2007.00564.x
Feinberg J (1992) The child’s right to an open future. In: Feinberg J (ed.) Freedom and fulfillment: philosophical essays. Princeton University Press, Princeton, pp. 6–97
Fu W, Akey JM (2013) Selection and adaptation in the human genome. Annu Rev Genom Hum Genet 14:467–89
Fukuyama F (2002) Our posthuman future: consequences of the biotechnology revolution. Picador, New York, NY
Gaj T, Gersbach CA, Barbas III CF (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31(7):397–405
Gaskell G, Bard I, Allansdottir A, da Cunha RV, Eduard P, Hampel J et al. (2017) Public views on gene editing and its uses. Nat Biotechnol 35(11):1021–1023. https://doi.org/10.1038/nbt.3958
Genomes Project C et al. (2015) A global reference for human genetic variation. Nature 526(7571):68–74
Article ADS CAS Google Scholar
Greely HT (2019) Human germline genome editing: an assessment. CRISPR J 2(5):253–265. https://doi.org/10.1089/crispr.2019.0038
Glover J (2006) Choosing children: genes, disability, and design. Oxford University Press, Oxford
Book Google Scholar
Greely HT (2021) CRISPR people: the science and ethics of editing humans. MIT Press, Cambridge, MA; London, UK
Gupta RM, Musunuru K (2014) Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Investig 124(10):4154–4161. https://doi.org/10.1172/JCI72992
Gyngell C, Douglas T, Savulescu J (2017) The ethics of germline gene editing. J Appl Philosy 34(4):498–513
Gyngell C, Bowman-Smart H, Savulescu J (2019) Moral reasons to edit the human genome: picking up from the Nuffield report. J Med Ethics 0:1–10. https://doi.org/10.1136/medethics-2018-105084
Gyngell C, Douglas T (2015) Stocking the genetic supermarket: reproductive genetic technologies and collective action problems. Bioethics 29(4):241–250
Guttinger S (2017) Trust in science: CRISPR-Cas9 and the ban on human germline editing. Sci Eng Eth 1–20. https://doi.org/10.1007/s11948-017-9931-1
Habermas J (2003) The future of human nature. Polity Press, Cambridge
Hammerstein AL, Eggel M, Biller-Andorno N (2019) Is selecting better than modifying? An investigation of arguments against germline gene editing as compared to preimplantation genetic diagnosis. BMC Med Eth 83:20
Häyry M (2003) Philosophical arguments for and against human reproductive cloning. Bioethics 17(5–6):447–460
Howard T, Rifkin J (1977) Who should play God? The artificial creation of life and what it means for the future of the human race. Dell Publ. Co
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6):1262–1278. https://doi.org/10.1016/j.cell.2014.05.010
Hurlbut JB, Saha K, Jasanoff S (2015) CRISPR democracy: gene editing and the need for inclusive deliberation. Issues Sci Technol 32(1):25–32
Hyun I, Osborn C (2017) Query the merits of embryo editing for reproductive research now. Nat Biotechnol 35(11):1023–1025. https://doi.org/10.1038/nbt.4000 . PMID: 29121025
Iltis AS, Hoover S, Matthews KRW (2021) Public and stakeholder engagement in developing human heritable genome editing policies: what does it mean and what should it mean? Front Political Sci 3. https://www.frontiersin.org/article/10.3389/fpos.2021.730869
Isasi R, Kleiderman E, Knoppers BM (2016) Genetic technology regulation: editing policy to fit the genome? Science 351(6271):337–339. https://doi.org/10.1126/science.aad6778
Ishii T (2015) Germline genome-editing research and its socio-ethical implications. Trends Mol Med 21(8):473–481. https://doi.org/10.1016/j.molmed.2015.05.006
Janssens AC (2016) Designing babies through gene editing: science or science fiction? Genet Med 18(12):1186–1187. https://doi.org/10.1038/gim.2016.28
Jedwab A, Vears DF, Tse C, Gyngell C (2020) Genetics experience impacts attitudes towards germline gene editing: a survey of over 1500 members of the public. J Hum Genetics65(12):1055–1065
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) Programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Jonlin EC (2020) Informed consent for human embryo genome editing. Stem Cell Rep 14(4):530–537. https://doi.org/10.1016/j.stemcr.2020.03.010
Juengst ET (1997) Can enhancement be distinguished from prevention in genetic medicine? J Med Philos 22(2):125–142
Juengst, ET, Moseley D (2019) Human enhancement. In: Zalta EN (ed.) The Stanford encyclopedia of philosophy (Summer 2019 Edition), Metaphysics Research Lab, Philosophy Department, Stanford University, Stanford
Karavani E, Zuk O, Zeevi D, Barzilai N, Stefanis NC, Hatzimanolis A et al. (2019) Screening human embryos for polygenic traits has limited utility. Cell 179(6):1424–1435
Kirksey E (2020) The Mutant Project: inside the global race to genetically modify humans. St. Martin’s Press
Klingler C, Wiese L, Arnason G, Ranisch R (2022) Public engagement with brain organoid research and application: lessons from genome editing. Am J Bioeth Neurosci 13(2):98–100. https://doi.org/10.1080/21507740.2022.2048733
König H (2017) The illusion of control in germline-engineering policy. Nat Biotechnol 35(6):502–506. https://doi.org/10.1038/nbt.3884
Krimsky S (2019) Ten ways in which He Jiankui violated ethics. Nat Biotechnol 37(1):19–20. https://doi.org/10.1038/nbt.4337
Lander ES (2015) Brave new genome. N Engl J Med 373(1):5–8
Lander ES, Baylis F, Zhang F, Charpentier E, Berg P, Bourgain C et al. (2019) Adopt a moratorium on heritable genome editing. Nature 567(7747):165–168. https://doi.org/10.1038/d41586-019-00726-5
Lanphier E, Urnov F, Haecker SE, Werner M, Smolenski J (2015) Don’t edit the human germ line. Nature 519(7544):410–1. https://doi.org/10.1038/519410a
Lappe M (1991) Ethical issues in manipulating the human germ line. J Med Philos 16(6):621–639. https://doi.org/10.1093/jmp/16.6.621
Lea RA, Niakan KK (2019) Human germline genome editing. Nat Cell Biol 21(12):1479–1489. https://doi.org/10.1038/s41556-019-0424-0
Ledford H (2015) The landscape for human genome editing. Nature 526(7573):310–311
Leon K (2003) Ageless bodies, happy souls: biotechnology and the pursuit of perfection. New Atlantis 1:9–28
Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X (2020) Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther 5(1):1–23
Li WH, Sadler LA (1991) Low nucleotide diversity in man. Genetics 129(2):513–23
Liang P, Xu Y, Zhang X et al. (2015) CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 6(5):363–372
Lovell-Badge R (2019) CRISPR babies: a view from the centre of the storm. Development 146(3):dev175778
MacKellar C, Bechtel C eds. (2014) The ethics of the new eugenics. Berghahn Books, New York, Oxford
McGee A (2020) Using the therapy and enhancement distinction in law and policy. Bioethics 34(1):70–80
Melillo TR (2017) Gene editing and the rise of designer babies. Vand J Trans Law 50:757–790
Mertes H, Pennings G (2015) Modification of the embryo’s genome: more useful in research than in the clinic. Am J Bioeth 15(12):52–53. https://doi.org/10.1080/15265161.2015.1103813
Mulvihill JJ, Capps B, Joly Y, Lysaght T, Zwart HAE, Chadwick R, International Human Genome Organisation Committee of Ethics Law and Society (2017) Ethical issues of CRISPR technology and gene editing through the lens of solidarity. Br Med Bull 122(1):17–29. https://doi.org/10.1093/bmb/ldx002
Musunuru K (2019) The CRISPR generation: the story of the world’s first gene-edited babies. BookBaby.
National Academies of Sciences, Engineering, and Medicine (2015) International summit on human gene editing: a global discussion. The National Academies Press, Washington
National Academy of Sciences, National Academy of Medicine (2017) Human genome editing: science, ethics and governance. The National Academies Press, Washington, DC
National Academy of Medicine, National Academy of Sciences, and the Royal Society (2020) Heritable human genome editing. The National Academies Press, Washington, DC
Niemiec E, Howard HC (2020) Ethical issues related to research on genome editing in human embryos. Comput Struct Biotechnol J 18:887–896
National Academies of Sciences, Engineering, and Medicine (2019) Second International Summit on human genome editing: continuing the global discussion: proceedings of a workshop in brief. The National Academies Press, Washington
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L et al. (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156(4):836–843
Nordberg A, Minssen T, Feeney O, de Miguel Beriain I, Galvagni L, Wartiovaara K (2020) Regulating germline editing in assisted reproductive technology: an EU cross‐disciplinary perspective. Bioethics 34(1):16–32
Nuffield Council on Bioethics (2016) Genome Editing: an ethical review. https://www.nuffieldbioethics.org/publications/genome-editing-an-ethical-review
Nuffield Council on Bioethics (2018) Genome editing and human reproduction: social and ethical issues. http://nuffieldbioethics.org/project/genome-editing-human-reproduction
Parens E, Asch A (2000) The disability rights critique of prenatal testing: reflections and recommendations. In: Parens E, Asch A (eds.) Prenatal testing and disability rights. Georgetown University Press, Washington
Parliamentary Assembly (1982) Recommendation on genetic engineering. In Recommendation 934. Council of Europe
Paul D (2005) Genetic engineering and eugenics: the uses of history. In: Baillie HW, Casey TK (eds.) Is human nature obsolete? Genetics, bioengineering, and the future of the human condition. MIT Press, Cambridge Mass
Peden JF, Farrall M (2011) Thirty-five common variants for coronary artery disease: the fruits of much collaborative labour. Hum Mol Genet 20:R198–205
Porteus MH (2019) A new class of medicines through DNA editing. New Engl J Med 380(10):947–959. https://doi.org/10.1056/NEJMra1800729
Parens ET (Ed.) (1998) Enhancing human traits: ethical and social implications. Georgetown University Press, Washington, DC
Primc N (2020) Do we have a right to an unmanipulated genome? The human genome as the common heritage of mankind Bioethics 34(1):41–48
Article MathSciNet PubMed Google Scholar
Ramsey P (1970) Fabricated man: the ethics of genetic control. Yale University Press, New Haven
Ranisch R (2017) Germline genome editing and the functions of consent. Am J Bioeth 17(12):27–29
Ranisch R (2020) Germline genome editing versus preimplantation genetic diagnosis: Is there a case in favour of germline interventions? Bioethics 34:60–69. https://doi.org/10.1111/bioe.12635
Ranisch R, Ehni HJ (2020) Fading red lines? Bioethics of germline genome editing. Bioethics 34(1):3–6. https://doi.org/10.1111/bioe.12709
Ranisch R, Rudolph T, Cremer HJ, Knoepffler N (2020) Ordo-responsibility for germline gene editing. CRISPR J 3(1):37–43
Ranisch R (2021) When CRISPR meets fantasy: transhumanism and the military in the age of gene editing. In: Transhumanism: the proper guide to a posthuman condition or a dangerous idea?. Springer, Cham, pp. 111–120
Ranisch R, Sorgner SL (2014) Introducing post-and transhumanism. In: Ranisch & Sorgner (eds.) Post-and transhumanism: an introduction. pp. 7–27. Peter Lang Group AG, Switzerland
Raposo VL (2019) Gene editing, the mystic threat to human dignity. Bioeth Inq 16:249–257. https://doi.org/10.1007/s11673-019-09906-4
Regalado A (2018) EXCLUSIVE: Chinese scientists are creating CRISPR babies. MIT Technol Rev. https://www.technologyreview.com/s/612458/exclusive-chinese-scientists-are-creating-crispr-babies/ . Accessed 4 Aug 2021
Rehmann-Sutter C (2018) Why human germline editing is more problematic than selecting between embryos: ethically considering intergenerational relationships. New Bioeth 24(1):9–25. https://doi.org/10.1080/20502877.2018.1441669
Resnik D (1994) Debunking the slippery slope argument against human germ-line gene therapy. J Med Philos 19(1):23–40. https://doi.org/10.1093/jmp/19.1.23
Richter G, Bacchetta MD (1998) Interventions in the human genome: some moral and ethical considerations. J Med Philos 23(3):303–317. https://doi.org/10.1076/jmep.23.3.303.2581
Rulli T (2019) Reproductive CRISPR does not cure disease. Bioethics 33:1072–1082
Sandel M (2007) The case against perfection: ethics in the age of genetic engineering. The Belknap Press of Harvard University Press, Cambridge
Savulescu J (2001) Procreative beneficence: why we should select the best children. Bioethics 15(5–6):413–26
Savulescu J, Pugh J, Douglas T, Gyngell C (2015) The moral imperative to continue gene editing research on human embryos. Protein Cell 6:476–479
Scally A (2016) The mutation rate in human evolution and demographic inference. Curr Opin Genet Dev 41:36–43
Schaefer GO (2020) Can reproductive genetic manipulation save lives? Med Health Care Philos 23(3):381–386
Scheufele DA, Xenos MA, Howell EL, Rose KM, Brossard D, Hardy BW (2017) U.S. attitudes on human genome editing. Science 357(6351):553–554. https://doi.org/10.1126/science.aan3708
Scheufele DA, Krause NM, Freiling I, Brossard D (2021) What we know about effective public engagement on CRISPR and beyond. Proc Natl Acad Sci USA 118(22). https://doi.org/10.1073/pnas.2004835117
Scully JL (2008) Disability and genetics in the era of genomic medicine. Nat Rev Genet 9(10):797–802
Segers S, Mertes H (2020) Does human genome editing reinforce or violate human dignity? Bioethics 34(1):33–40
Sermon K, Van Steirteghem A, Liebaers I (2004) Preimplantation genetic diagnosis. Lancet 363(9421):1633–1641. https://doi.org/10.1016/S0140-6736(04)16209-0
Shakespeare T (2006) Disability rights and wrongs. Routledge, London
Shulman C, Bostrom N (2014) Embryo selection for cognitive enhancement: curiosity or game-changer? Glob Policy 5(1):85–92. https://doi.org/10.1111/1758-5899.12123
Silver LM (1997) Remaking Eden: cloning and beyond in a brave new world. William Morrow, New York, NY
Sinsheimer RL (1969). The prospect for designed genetic change. Am Sci, 57(1):134–142. http://www.jstor.org/stable/27828443
Smolenski J (2015) Crispr/cas9 and germline modification: new difficulties in obtaining informed consent. Am J Bioeth 15(12):35–37
Soini S (2007) Preimplantation genetic diagnosis (PGD) in Europe: diversity of legislation a challenge to the community and its citizens. Med Law 26(2):309–323
ADS CAS PubMed Google Scholar
Sorgner SL (2018) Genes, CRISPR/Cas 9, and posthumans. In: Sinaci M and Sorgner SL (eds.) Ethics of emerging biotechnologies, pp. 5–17. Trivent Publishing
Sparrow R (2021) Human germline genome editing: on the nature of our reasons to genome edit Am J Bioeth 19:1–12
Steffann J, Jouannet P, Bonnefont JP, Chneiweiss H, Frydman N (2018) Could failure in preimplantation genetic diagnosis justify editing the human embryo genome? Cell Stem Cell 22(4):481–482. https://doi.org/10.1016/j.stem.2018.01.004
Sykora P, Caplan A (2017) The Council of Europe should not reaffirm the ban on germline genome editing in humans. EMBO Rep 18:1871–1872. https://doi.org/10.15252/embr.201745246
UNESCO (1997) Universal declaration on the human genome and human rights. UNESCO, Paris
van Dijke I, Bosch L, Bredenoord AL, Cornel M, Repping S, Hendriks S (2018) The ethics of clinical applications of germline genome modification: a systematic review of reasons Hum Reprod 33(9):1777–1796
Vassena R, Heindryckx B, Peco R, Pennings G, Raya A, Sermon K, Veiga A (2016) Genome engineering through CRISPR/Cas9 technology in the human germline and pluripotent stem cells. Hum Reprod Update 22(4):411–419. https://doi.org/10.1093/humupd/dmw005
Venter JC et al. (2001) The sequence of the human genome. Science 291(5507):1304–51
Walters L, Cook-Deegan RM, Adashi EY (2021) Governing heritable human genome editing: a textual history and a proposal for the future. CRISPR J 4(4):469–476
Wang H, Yang H (2019) Gene-edited babies: what went wrong and what could go wrong. PLoS Biol 17(4). https://doi.org/10.1371/journal.pbio.3000224
Wheeler E, Barroso I (2011) Genome-wide association studies and type 2 diabetes. Brief Funct Genom 10:52–60
WHO (2019) Statement on governance and oversight of human genome editing. https://www.who.int/news/item/26-07-2019-statement-on-governance-and-oversight-of-human-genome-editing
WHO (2021) Human genome editing: position paper. https://www.who.int/publications/i/item/9789240030404
Wolf DP, Mitalipov PA, Mitalipov SM (2019) Principles of and strategies for germline gene therapy. Nat Med 25(6):890–897
Zhang F (2019) Development of CRISPR-Cas systems for genome editing and beyond. Q Rev Biophys 52. https://doi.org/10.1017/s0033583519000052
Download references
Acknowledgements
This work was supported by Fundação para Ciência e a Tecnologia (FCT) of Portugal [UIDP/00678/2020 to M.A]. We thank Dr. Michael Morrison for his comments and Dr. Gustav Preller for his proofreading of this manuscript.
Author information
Authors and affiliations.
Centro de Filosofia das Ciências da Universidade de Lisboa, Campo Grande, 1749-016, Lisbon, Portugal
Mara Almeida
Faculty of Health Sciences Brandenburg, University of Potsdam, Karl-Liebknecht-Str. 24-25, House 16, 14476, Potsdam, Golm, Germany
- Robert Ranisch
Research Unit “Ethics of Genome Editing”, Institute for Ethics and History of Medicine, University of Tübingen, Gartenstr. 47, 72074, Tübingen, Germany
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Mara Almeida .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Ethical approval
An ethical approval is not applicable.
Informed consent
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Almeida, M., Ranisch, R. Beyond safety: mapping the ethical debate on heritable genome editing interventions. Humanit Soc Sci Commun 9 , 139 (2022). https://doi.org/10.1057/s41599-022-01147-y
Download citation
Received : 06 August 2021
Accepted : 28 March 2022
Published : 20 April 2022
DOI : https://doi.org/10.1057/s41599-022-01147-y
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Clinical translation of wireless soft robotic medical devices.
- Tianlu Wang
- Metin Sitti
Nature Reviews Bioengineering (2024)
An analysis of different concepts of “identity” in the heritable genome editing debate
- Ying-Qi Liaw
Medicine, Health Care and Philosophy (2024)
Initial heritable genome editing: mapping a responsible pathway from basic research to the clinic
- Katharina Trettenbach
- Gardar Arnason
Medicine, Health Care and Philosophy (2023)
“What if” should precede “whether” and “how” in the social conversation around human germline gene editing
- Diewertje Houtman
- Wendy Geuverink
- Sam Riedijk
Journal of Community Genetics (2023)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Search Menu
- Volume 12, Issue 1, 2024 (In Progress)
- Volume 11, Issue 1, 2023
- Advance articles
- Editor's Choice
- Virtual Issues
- Clinical Briefs
- Author Guidelines
- Submission Site
- Open Access
- Calls for Papers
- Why submit?
- About Evolution, Medicine, and Public Health
- About the International Society for Evolution, Medicine and Public Health
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Self-Archiving Policy
- For Reviewers
- Journals on Oxford Academic
- Books on Oxford Academic


Article Contents
Introduction, human enhancement, genetic engineering, conclusions.
- < Previous
Human enhancement: Genetic engineering and evolution
- Article contents
- Figures & tables
- Supplementary Data
Mara Almeida, Rui Diogo, Human enhancement: Genetic engineering and evolution, Evolution, Medicine, and Public Health , Volume 2019, Issue 1, 2019, Pages 183–189, https://doi.org/10.1093/emph/eoz026
- Permissions Icon Permissions
Genetic engineering opens new possibilities for biomedical enhancement requiring ethical, societal and practical considerations to evaluate its implications for human biology, human evolution and our natural environment. In this Commentary, we consider human enhancement, and in particular, we explore genetic enhancement in an evolutionary context. In summarizing key open questions, we highlight the importance of acknowledging multiple effects (pleiotropy) and complex epigenetic interactions among genotype, phenotype and ecology, and the need to consider the unit of impact not only to the human body but also to human populations and their natural environment (systems biology). We also propose that a practicable distinction between ‘therapy’ and ‘enhancement’ may need to be drawn and effectively implemented in future regulations. Overall, we suggest that it is essential for ethical, philosophical and policy discussions on human enhancement to consider the empirical evidence provided by evolutionary biology, developmental biology and other disciplines.
Lay Summary: This Commentary explores genetic enhancement in an evolutionary context. We highlight the multiple effects associated with germline heritable genetic intervention, the need to consider the unit of impact to human populations and their natural environment, and propose that a practicable distinction between ‘therapy’ and ‘enhancement’ is needed.
There are countless examples where technology has contributed to ameliorate the lives of people by improving their inherent or acquired capabilities. For example, over time, there have been biomedical interventions attempting to restore functions that are deficient, such as vision, hearing or mobility. If we consider human vision, substantial advances started from the time spectacles were developed (possibly in the 13th century), continuing in the last few years, with researchers implanting artificial retinas to give blind patients partial sight [ 1–3 ]. Recently, scientists have also successfully linked the brain of a paralysed man to a computer chip, which helped restore partial movement of limbs previously non-responsive [ 4 , 5 ]. In addition, synthetic blood substitutes have been created, which could be used in human patients in the future [ 6–8 ].
The progress being made by technology in a restorative and therapeutic context could in theory be applied in other contexts to treat non-pathological conditions. Many of the technologies and pharmaceutical products developed in a medical context to treat patients are already being used by humans to ‘enhance’ some aspect of their bodies, for example drugs to boost brain power, nutritional supplements, brain stimulating technologies to control mood or growth hormones for children of short stature. Assistive technology for disabled people, reproductive medicine and pharmacology, beside their therapeutic and restorative use, have a greater potential for human ‘enhancement’ than currently thought. There are also dual outcomes as some therapies can have effects that amount to an enhancement as for example, the artificial legs used by the South African sprinter Oscar Pistorius providing him with a competitive advantage.
This commentary will provide general ethical considerations on human enhancement, and within the several forms of so-called human biomedical enhancement, it will focus on genetic engineering, particularly on germline (heritable) genetic interventions and on the insights evolutionary biology can provide in rationalizing its likely impact. These insights are a subject often limited in discussions on genetic engineering and human enhancement in general, and its links to ethical, philosophical and policy discussions, in particular [ 9 ]. The rapid advances in genetic technology make this debate very topical. Moreover, genes are thought to play a very substantial role in biological evolution and development of the human species, thus making this a topic requiring due consideration. With this commentary, we explore how concepts based in evolutionary biology could contribute to better assess the implications of human germline modifications, assuming they were widely employed. We conclude our brief analysis by summarizing key issues requiring resolution and potential approaches to progress them. Overall, the aim is to contribute to the debate on human genetic enhancement by looking not only at the future, as it is so often done, but also at our evolutionary past.
The noun ‘enhancement’ comes from the verb ‘enhance’, meaning ‘to increase or improve’. The verb enhance can be traced back to the vulgar Latin inaltiare and late Latin inaltare (‘raise, exalt’), from ‘ altare ’ (‘make high’) and altus (‘high’), literally ‘grown tall’. For centuries human enhancement has populated our imagination outlined by stories ranging from the myths of supernormal strengths and eternal life to the superpowers illustrated by the 20th century comic books superheroes. The desire of overcoming normal human capacities and the transformation to an almost ‘perfect’ form has been part of the history of civilization, extending from arts and religion to philosophy. The goal of improving the human condition and health has always been a driver for innovation and biomedical developments.
In the broadest sense, the process of human enhancement can be considered as an improvement of the ‘limitations’ of a ‘natural version’ of the human species with respect to a specific reference in time, and to different environments, which can vary depending on factors such as, for example, climate change. The limitations of the human condition can be physical and/or mental/cognitive (e.g. vision, strength or memory). This poses relevant questions of what a real or perceived human limitation is in the environment and times in which we are living and how it can be shifted over time considering social norms and cultural values of modern societies. Besides, the impact that overcoming these limitations will have on us humans, and the environment, should also be considered. For example, if we boost the immune system of specific people, this may contribute to the development/evolution of more resistant viruses and bacteria or/and lead to new viruses and bacteria to emerge. In environmental terms, enhancing the longevity of humans could contribute to a massive increase in global population, creating additional pressures on ecosystems already under human pressure.
Two decades ago, the practices of human enhancement have been described as ‘biomedical interventions that are used to improve human form or functioning beyond what is necessary to restore or sustain health’ [ 10 ]. The range of these practices has now increased with technological development, and they are ‘any kind of genetic, biomedical, or pharmaceutical intervention aimed at improving human dispositions, capacities, or well-being, even if there is no pathology to be treated’ [ 11 ]. Practices of human enhancement could be visualized as upgrading a ‘system’, where interventions take place for a better performance of the original system. This is far from being a hypothetical situation. The rapid progress within the fields of nanotechnology, biotechnology, information technology and cognitive science has brought back discussions about the evolutionary trajectory of the human species by the promise of new applications which could provide abilities beyond current ones [ 12 , 13 ]. If such a possibility was consciously embraced and actively pursued, technology could be expected to have a revolutionary interference with human life, not just helping humans in achieving general health and capabilities commensurate with our current ones but helping to overcome human limitations far beyond of what is currently possible for human beings. The emergence of new technologies has provided a broader range of potential human interventions and the possibility of transitioning from external changes to our bodies (e.g. external prosthesis) to internal ones, especially when considering genetic manipulation, whose changes can be permanent and transmissible.
The advocates of a far-reaching human enhancement have been referred to as ‘transhumanists’. In their vision, so far, humans have largely worked to control and shape their exterior environments (niche construction) but with new technologies (e.g. biotechnology, information technology and nanotechnology) they will soon be able to control and fundamentally change their own bodies. Supporters of these technologies agree with the possibility of a more radical interference in human life by using technology to overcome human limitations [ 14–16 ], that could allow us to live longer, healthier and even happier lives [ 17 ]. On the other side, and against this position, are the so-called ‘bioconservatives’, arguing for the conservation and protection of some kind of ‘human essence’, with the argument that it exists something intrinsically valuable in human life that should be preserved [ 18 , 19 ].
There is an ongoing debate between transhumanists [ 20–22 ] and bioconservatives [ 18 , 19 , 23 ] on the ethical issues regarding the use of technologies in humans. The focus of this commentary is not centred on this debate, particularly because the discussion of these extreme, divergent positions is already very prominent in the public debate. In fact, it is interesting to notice that the ‘moderate’ discourses around this topic are much less known. In a more moderate view, perhaps one of the crucial questions to consider, independently of the moral views on human enhancement, is whether human enhancement (especially if considering germline heritable genetic interventions) is a necessary development, and represents an appropriate use of time, funding and resources compared to other pressing societal issues. It is crucial to build space for these more moderate, and perhaps less polarized voices, allowing the consideration of other positions and visions beyond those being more strongly projected so far.
Ethical and societal discussions on what constitutes human enhancement will be fundamental to support the development of policy frameworks and regulations on new technological developments. When considering the ethical implications of human enhancement that technology will be available to offer now and in the future, it could be useful to group the different kinds of human enhancements in the phenotypic and genetic categories: (i) strictly phenotypic intervention (e.g. ranging from infrared vision spectacles to exoskeletons and bionic limbs); (ii) somatic, non-heritable genetic intervention (e.g. editing of muscle cells for stronger muscles) and (iii) germline, heritable genetic intervention (e.g. editing of the C–C chemokine receptor type 5 (CCR5) gene in the Chinese baby twins, discussed later on). These categories of enhancement raise different considerations and concerns and currently present different levels of acceptance by our society. The degree of ethical, societal and environmental impacts is likely to be more limited for phenotypic interventions (i) but higher for genetic interventions (ii and iii), especially for the ones which are transmissible to future generations (iii).
The rapid advances in technology seen in the last decades, have raised the possibility of ‘radical enhancement’, defined by Nicholas Agar, ‘as the improvement of human attributes and abilities to levels that greatly exceed what is currently possible for human beings’ [ 24 ]. Genetic engineering offers the possibility of such an enhancement by providing humans a profound control over their own biology. Among other technologies, genetic engineering comprises genome editing (also called gene editing), a group of technologies with the ability to directly modify an organism’s DNA through a targeted intervention in the genome (e.g. insertion, deletion or replacement of specific genetic material) [ 25 ]. Genome editing is considered to achieve much greater precision than pre-existing forms of genetic engineering. It has been argued to be a revolutionary tool due to its efficiency, reducing cost and time. This technology is considered to have many applications for human health, in both preventing and tackling disease. Much of the ethical debate associated with this technology concerns the possible application of genome editing in the human germline, i.e. the genome that can be transmitted to following generations, be it from gametes, a fertilized egg or from first embryo divisions [ 26–28 ]. There has been concern as well as enthusiasm on the potential of the technology to modify human germline genome to provide us with traits considered positive or useful (e.g. muscle strength, memory and intelligence) in the current and future environments.
Genetic engineering: therapy or enhancement and predictability of outcomes
To explore some of the possible implications of heritable interventions we will take as an example the editing (more specifically ‘deletion’ using CRISPR genome editing technology) of several base pairs of the CCR5 gene. Such intervention was practised in 2018 in two non-identical twin girls born in China. Loss of function mutations of the CCR5 had been previously shown to provide resistance to HIV. Therefore, the gene deletion would be expected to protect the twin baby girls from risk of transmission of HIV which could have occurred from their father (HIV-positive). However, the father had the infection kept under control and the titre of HIV virus was undetectable, which means that risk of transmission of HIV infection to the babies was negligible [ 29 ].
From an ethical ground, based on current acceptable practices, this case has been widely criticized by the scientific community beside being considered by many a case of human enhancement intervention rather than therapy [ 29 , 30 ]. One of the questions this example helps illustrate is that the ethical boundary between a therapy that ‘corrects’ a disorder by restoring performance to a ‘normal’ scope, and an intervention that ‘enhances’ human ability outside the accepted ‘normal’ scope, is not always easy to draw. For the sake of argument, it could be assumed that therapy involves attempts to restore a certain condition of health, normality or sanity of the ‘natural’ condition of a specific individual. If we take this approach, the question is how health, normality and sanity, as well as natural per se, are defined, as the meaning of these concepts shift over time to accommodate social norms and cultural values of modern societies. It could be said that the difficulty of developing a conceptual distinction between therapy and enhancement has always been present. However, the potential significance of such distinction is only now, with the acceleration and impact of technological developments, becoming more evident.
Beyond ethical questions, a major problem of this intervention is that we do not (yet?) know exactly the totality of the effects that the artificial mutation of the CCR5 may have, at both the genetic and phenotypic levels. This is because we now know that, contrary to the idea of ‘one gene-one trait’ accepted some decades ago, a gene—or its absence—can affect numerous traits, many of them being apparently unrelated (a phenomenon also known as pleiotropy). That is, due to constrained developmental interactions, mechanisms and genetic networks, a change in a single gene can result in a cascade of multiple effects [ 31 ]. In the case of CCR5, we currently know that the mutation offers protection against HIV infection, and also seems to increase the risk of severe or fatal reactions to some infectious diseases, such as the influenza virus [ 32 ]. It has also been observed that among people with multiple sclerosis, the ones with CCR5 mutation are twice as likely to die early than are people without the mutation [ 33 ]. Some studies have also shown that defective CCR5 can have a positive effect in cognition to enhance learning and memory in mice [ 34 ]. However, it’s not clear if this effect would be translated into humans. The example serves to illustrate that, even if human enhancement with gene editing methods was considered ethically sound, assessing the totality of its implications on solid grounds may be difficult to achieve.
Genetic engineering and human evolution: large-scale impacts
Beyond providing the opportunity of enhancing human capabilities in specific individuals, intervening in the germline is likely to have an impact on the evolutionary processes of the human species raising questions on the scale and type of impacts. In fact, the use of large-scale genetic engineering might exponentially increase the force of ‘niche construction’ in human evolution, and therefore raise ethical and practical questions never faced by our species before. It has been argued that natural selection is a mechanism of lesser importance in the case of current human evolution, as compared to other organisms, because of advances in medicine and healthcare [ 35 ]. According to such a view, among many others advances, natural selection has been conditioned by our ‘niche-construction’ ability to improve healthcare and access to clean water and food, thus changing the landscape of pressures that humans have been facing for survival. An underlying assumption or position of the current debate is that, within our human species, the force of natural selection became minimized and that we are somehow at the ‘end-point’ of our evolution [ 36 ]. If this premise holds true, one could argue that evolution is no longer a force in human history and hence that any human enhancement would not be substituting itself to human evolution as a key driver for future changes.
However, it is useful to remember that, as defined by Darwin in his book ‘On the Origin of the Species’, natural selection is a process in which organisms that happen to be ‘better’ adapted to a certain environment tend to have higher survival and/or reproductive rates than other organisms [ 37 ]. When comparing human evolution to human genetic enhancement, an acceptable position could be to consider ethically sound those interventions that could be replicated naturally by evolution, as in the case of the CCR5 gene. Even if this approach was taken, however, it is important to bear in mind that human evolution acts on human traits sometimes increasing and sometimes decreasing our biological fitness, in a constant evolutionary trade-off and in a contingent and/or neutral—in the sense of not ‘progressive’—process. In other worlds, differently from genetic human enhancement, natural selection does not ‘ aim ’ at improving human traits [ 38 ]. Human evolution and the so-called genetic human enhancement would seem therefore to involve different underlying processes, raising several questions regarding the implications and risks of the latter.
But using genetic engineering to treat humans has been proposed far beyond the therapeutic case or to introduce genetic modifications known to already occur in nature. In particular, when looking into the views expressed on the balance between human evolution and genetic engineering, some argue that it may be appropriate to use genetic interventions to go beyond what natural selection has contributed to our species when it comes to eradicate vulnerabilities [ 17 ]. Furthermore, when considering the environmental, ecological and social issues of contemporary times, some suggest that genetic technologies could be crucial tools to contribute to human survival and well-being [ 20–22 ]. The possible need to ‘engineer’ human traits to ensure our survival could include the ability to allow our species to adapt rapidly to the rate of environmental change caused by human activity, for which Darwinian evolution may be too slow [ 39 ]. Or, for instance, to support long-distance space travel by engineering resistance to radiation and osteoporosis, along with other conditions which would be highly advantageous in space [ 40 ].
When considering the ethical and societal merits of these propositions, it is useful to consider how proto-forms of enhancement has been approached by past human societies. In particular, it can be argued that humans have already employed—as part of our domestication/‘selective breeding’ of other animals—techniques of indirect manipulation of genomes on a relatively large scale over many millennia, albeit not on humans. The large-scale selective breeding of plants and animals over prehistoric and historic periods could be claimed to have already shaped some of our natural environment. Selective breeding has been used to obtain specific characteristics considered useful at a given time in plants and animals. Therefore, their evolutionary processes have been altered with the aim to produce lineages with advantageous traits, which contributed to the evolution of different domesticated species. However, differently from genetic engineering, domestication possesses inherent limitations in its ability to produce major transformations in the created lineages, in contrast with the many open possibilities provided by genetic engineering.
When considering the impact of genetic engineering on human evolution, one of questions to be considered concerns the effects, if any, that genetic technology could have on the genetic pool of the human population and any implication on its resilience to unforeseen circumstances. This underlines a relevant question associated with the difference between ‘health’ and biological fitness. For example, a certain group of animals can be more ‘healthy’—as domesticated dogs—but be less biologically ‘fit’ according to Darwin’s definition. Specifically, if such group of animals are less genetically diverse than their ancestors, they could be less ‘adaptable’ to environmental changes. Assuming that, the human germline modification is undertaken at a global scale, this could be expected to have an effect, on the distribution of genetically heritable traits on the human population over time. Considering that gene and trait distributions have been changing under the processes of evolution for billions of years, the impact on evolution will need to be assessed by analysing which genetic alterations have been eventually associated with specific changes within the recent evolutionary history of humans. On this front, a key study has analysed the implications of genetic engineering on the evolutionary biology of human populations, including the possibility of reducing human genetic diversity, for instance creating a ‘biological monoculture’ [ 41 ]. The study argued that genetic engineering will have an insignificant impact on human diversity, while it would likely safeguard the capacity of human populations to deal with disease and new environmental challenges and therefore, ensure the health and longevity of our species [ 41 ]. If the findings of this study were considered consistent with other knowledge and encompassing, the impact of human genetic enhancements on the human genetic pool and associated impacts could be considered secondary aspects. However, data available from studies on domestication strongly suggests that domestication of both animals and plans might lead to not only decreased genetic diversity per se, but even affect patterns of variation in gene expression throughout the genome and generally decreased gene expression diversity across species [ 42–44 ]. Given that, according to recent studies within the field of biological anthropology recent human evolution has been in fact a process of ‘self-domestication’ [ 45 ], one could argue that studies on domestication could contribute to understanding the impacts of genetic engineering.
Beyond such considerations, it is useful to reflect on the fact that human genetic enhancement could occur on different geographical scales, regardless of the specific environment and geological periods in which humans are living and much more rapidly than in the case of evolution, in which changes are very slow. If this was to occur routinely and on a large scale, the implications of the resulting radical and abrupt changes may be difficult to predict and its impacts difficult to manage. This is currently highlighted by results of epigenetics studies, and also of the microbiome and of the effects of pollutants in the environment and their cumulative effect on the development of human and non-human organisms alike. Increasingly new evidence indicates a greater interdependence between humans and their environments (including other microorganisms), indicating that modifying the environment can have direct and unpredictable consequences on humans as well. This highlight the need of a ‘systems level’ approach. An approach in which the ‘bounded body’ of the individual human as a basic unit of biological or social action would need to be questioned in favour of a more encompassing and holistic unit. In fact, within biology, there is a new field, Systems Biology, which stresses the need to understand the role that pleiotropy, and thus networks at multiple levels—e.g. genetic, cellular, among individuals and among different taxa—play within biological systems and their evolution [ 46 ]. Currently, much still needs to be understood about gene function, its role in human biological systems and the interaction between genes and external factors such as environment, diet and so on. In the future if we do choose to genetically enhance human traits to levels unlikely to be achieved by human evolution, it would be crucial to consider if and how our understanding of human evolution enable us to better understand the implications of genetic interventions.
New forms of human enhancement are increasingly coming to play due to technological development. If phenotypic and somatic interventions for human enhancement pose already significant ethical and societal challenges, germline heritable genetic intervention, require much broader and complex considerations at the level of the individual, society and human species as a whole. Germline interventions associated with modern technologies are capable of much more rapid, large-scale impacts and seem capable of radically altering the balance of humans with the environment. We know now that beside the role genes play on biological evolution and development, genetic interventions can induce multiple effects (pleiotropy) and complex epigenetics interactions among genotype, phenotype and ecology of a certain environment. As a result of the rapidity and scale with which such impact could be realized, it is essential for ethical and societal debates, as well as underlying scientific studies, to consider the unit of impact not only to the human body but also to human populations and their natural environment (systems biology). An important practicable distinction between ‘therapy’ and ‘enhancement’ may need to be drawn and effectively implemented in future regulations, although a distinct line between the two may be difficult to draw.
In the future if we do choose to genetically enhance human traits to levels unlikely to be achieved by human evolution, it would be crucial to consider if and how our understanding of humans and other organisms, including domesticated ones, enable us to better understand the implications of genetic interventions. In particular, effective regulation of genetic engineering may need to be based on a deep knowledge of the exact links between phenotype and genotype, as well the interaction of the human species with the environment and vice versa .
For a broader and consistent debate, it will be essential for technological, philosophical, ethical and policy discussions on human enhancement to consider the empirical evidence provided by evolutionary biology, developmental biology and other disciplines.
This work was supported by Fundação para a Ciência e a Tecnologia (FCT) of Portugal [CFCUL/FIL/00678/2019 to M.A.].
Conflict of interest : None declared.
Pham P , Roux S , Matonti F et al. Post-implantation impedance spectroscopy of subretinal micro-electrode arrays, OCT imaging and numerical simulation: towards a more precise neuroprosthesis monitoring tool . J Neural Eng 2013 ; 10 : 046002 .
Google Scholar
Maghami MH , Sodagar AM , Lashay A et al. Visual prostheses: the enabling technology to give sight to the blind . J Ophthal Vis Res 2014 ; 9 : 494 – 505 .
Weitz AC , Nanduri D , Behrend MR et al. Improving the spatial resolution of epiretinal implants by increasing stimulus pulse duration . Sci Transl Med 2015 ; 7 : 318ra203.
Bouton CE , Shaikhouni A , Annetta NV et al. Restoring cortical control of functional movement in a human with quadriplegia . Nature 2016 ; 533 : 247 – 50 .
Geddes L. First paralysed person to be ‘reanimated’ offers neuroscience insights. Technique moves man’s arm by decoding his thoughts and electrically stimulating his own muscles . Nat News 2016 ; 533 .
Squires JE. Artificial blood . Science 2002 ; 295 : 1002 – 5 .
Lowe KC. Blood substitutes: from chemistry to clinic . J Mater Chem 2006 ; 16 : 4189 – 96 .
Moradi S , Jahanian-Najafabadi A , Roudkenar MH. Artificial blood substitutes: first steps on the long route to clinical utility . Clin Med Insights Blood Disord 2016 ; 9 : 33 – 41 .
Powell R , Kahane G , Savulescu J. Evolution, genetic engineering, and human enhancement . Philos Technol 2012 ; 25 : 439 – 58 .
Parens E (ed.). Enhancing Human Traits: Ethical and Social Implications . Washington, DC : Georgetown University Press , 1998 .
Google Preview
Giubilini A , Sanyal S. Challenging human enhancement. In: Clarke S , Savulescu J , Coady T et al. (eds). The Ethics of Human Enhancement: Understanding the Debate . Oxford : Oxford University Press , 2016 .
Elliott C. Better Than Well: American Medicine Meets the American Dream . New York, NY : WWW Norton & Company, Inc ., 2003 .
Kramer P. Listening to Prozac . London : Fourth Estate , 1994 .
Moravec H. Mind Children: The Future of Robot and Human Intelligence . Cambridge : Harvard University Press , 1990 .
Bostrom N. Human genetic enhancements: a transhumanist perspective . J Value Inq 2003 ; 37 : 493 – 506 .
Kurzweil R. The Singularity is Near: When Humans Transcend Biology . New York, NY : Viking , 2005 .
Harris J. Enhancing Evolution: The Ethical Case for Making Better People . Princeton, NJ : Princeton University Press , 2010 .
Fukuyama F. Our Posthuman Future: Consequences of the Biotechnology Revolution . New York, NY : Picador , 2002 .
Sandel M. The Case Against Perfection: Ethics in the Age of Genetic Engineering . Cambridge : The Belknap Press of Harvard University Press , 2007 .
Savulescu J , Persson I. The perils of cognitive enhancement and the urgent imperative to enhance the moral character of humanity . J Appl Philos 2008 ; 25 : 162 – 77 .
Buchanan A. Beyond Humanity . Oxford : Oxford University Press , 2011 .
Persson I , Savulescu J. Moral enhancement, freedom, and the god machine . Monist 2012 ; 95 : 399 – 421 .
Leon K. Ageless bodies, happy souls: biotechnology and the pursuit of perfection . New Atlantis 2003 ; 1 : 9 – 28 .
Agar N. Humanity’s End: Why We Should Reject Radical Enhancement . Cambridge : MIT Press , 2010 .
Gaj T , Gersbach CA , Barbas CF III ,. ZFN, TALEN, and CRISPR/Cas based methods for genome engineering . Trends Biotechnol 2013 ; 3 : 397 – 405 .
Baltimore D , Berg P , Botchan M et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification . Science 2015 ; 348 : 36 – 8 .
Otieno MO. CRISPR/Cas9 human genome editing: challenges, ethical concerns and implications . J Clin Res Bioeth 2015 ; 6 : 253 .
Ishii T. Germline genome-editing research and its socio-ethical implications . Trends Mol Med 2015 ; 21 : 473 – 81 .
Bionews.org.uk. First Genome-edited Babies: A Very Different Perception of Ethics , 2018 . https://www.bionews.org.uk/page_140060 (27 August 2019, date last accessed).
Cyranoski D. CRISPR-baby scientist fails to satisfy his critics . Nat News 2018 ; 564 : 13 – 4 .
Galis F , Metz JA. Evolutionary novelties: the making and breaking of pleiotropic constraints . Integr Comp Biol 2007 ; 47 : 409 – 19 .
Falcon A , Cuevas MT , Rodriguez-Frandsen A et al. CCR5 deficiency predisposes to fatal outcome in influenza virus infection . J Gen Virol 2015 ; 96 : 2074 – 8 .
Gade-Andavolu R , Comings DE , MacMurray J et al. Association of CCR5 Δ32 deletion with early death in multiple sclerosis . Genet Med 2004 ; 6 : 126 – 31 .
Zhou M , Greenhill S , Huang S et al. CCR5 is a suppressor for cortical plasticity and hippocampal learning and memory . eLife 2016 ; 5 : e20985 .
Tibayrenc M , Ayala FJ (eds). On Human Nature: Biology, Psychology, Ethics, Politics, and Religion . London : Academic Press , 2017 .
Baldi P. The Shattered Self: The End of Natural Evolution . Cambridge : MIT Press , 2001 .
Darwin C. On the Origin of Species by Means of Natural Selection, or, the Preservation of Favoured Races in the Struggle for Life . London : J. Murray , 1859 .
Gould SJ. The Structure of Evolutionary Theory . Belknap, NY : Harvard University Press , 2002 .
Rees M. Our Final Century: Will the Humans Race Survive the Twenty-first Century? Eastbourne : Gardners Books , 2003 .
Nuffield Council on Bioethics. Genome Editing: An Ethical Review . London : Nuffield Council on Bioethics , 2016 .
Powell R. The evolutionary biological implications of human genetic engineering . J Med Philos 2012 ; 37 : 204 – 26 .
Liu W , Chen L , Zhang S et al. Decrease of gene expression diversity during domestication of animals and plants . BMC Evol Biol 2019 ; 19 : 1 – 11 .
Fages A , Hanghøj K , Khan N et al. Tracking five millennia of horse management with extensive ancient genome time series . Cell 2019 ; 177 : 1419 – 35 .
Zhang J , Wang X , Yao J et al. Effect of domestication on the genetic diversity and structure of Saccharina japonica populations in China . Sci Rep 2017 ; 7 : 42158 .
Theofanopoulou C , Gastaldon S , O’Rourke T et al. Self-domestication in Homo sapiens : insights from comparative genomics . PLoS One 2018 ; 13 : e0196700 .
Capra F , Luisi PL. The Systems View of Life . Cambridge : Cambridge University Press , 2014
- genetic engineering
Email alerts
Citing articles via, affiliations.
- Online ISSN 2050-6201
- Copyright © 2024 International Society for Evolution, Medicine, and Public Health
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Genetic Engineering
- First Online: 28 March 2021
Cite this chapter

- David B. Resnik 13
Part of the book series: The International Library of Bioethics ((ILB,volume 86))
221 Accesses
In this chapter I will apply the PP to ethical and policy issues related to genetic engineering of microbes, plants, animals, and human beings. I will argue that the PP can provide some useful insights into these issues, due to the scientific and morally uncertainty surrounding the consequences of genetic engineering for public health, the environment, society, and patients.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
By “genetic engineering” I mean technologies that involve direct modification or alteration of the genomes of cells or organisms. Changes brought about by genetic engineering might or might not be inheritable, depending on the type of change and the organism. Modification of the genomes of somatic cells in humans (discussed below) does not normally result in inheritable genetic changes, but modification of human germ cells, sperm, eggs, or embryos does (Resnik et al. 1999 ). Modification of bacterial genomes always results in inheritable genetic changes because bacteria are unicellular organisms. Ooplasm transfer, nuclear transfer, and reproductive cloning in human beings raise important ethical and social issues, but these procedures are not genetic engineering, according to my definition, because their purposes is not modify genomes, even though they involve the manipulation of genetic material. Synthetic biology uses genetic engineering methods to design cells, organisms, and biological system that do not already exist in the natural world (Biotechnology Innovation Organization 2020b ).
Some viruses encode their genetic information in RNA (ribonucleic acid).
A polymer is a large molecule.
James Watson (1928–) and Francis Crick (1916–2004) won the Nobel Prize in Physiology of Medicine in 1962 for discovering the structure of DNA. Their model was confirmed by Rosalind Franklin’s x-ray crystallography data, Watson and Crick did not name Franklin as an author on the paper that described their model of the structure of DNA. Franklin (1920–1958) was also not awarded the Nobel Prize for her contribution, because she died of ovarian cancer in 1958, and the Nobel Prize is not awarded posthumously (Maddox 2003 ).
Because mitochondria have their own DNA, scientists have speculated that mitochondria were at one time independent organisms that became incorporated into primordial, unicellular organisms (Alberts et al. 2015 ).
Prokaryotes are single-celled organisms with no distinct cell nucleus or organelles.
Mitochondria replicate independently of the cell.
Most higher life forms, including most plants, mammals, and human beings, are diploid (Alberts et al. 2015 ).
Many species of plants and animals that reproduce sexually can also propagate asexually. Growing a new plant from a cutting is a form of asexual propagation.
Plant stem cells can also generate different tissue types.
Berg, Gilbert, and Sanger won the Nobel Prize in chemistry in 1980 for their development of recombinant DNA techniques (Nobel Prize.org 2021 ).
Doudna and Charpentier won the Nobel Prize in Chemistry in 2000 for the discovery of CRISPR (Ledford and Callaway 2020 ).
Laboratory animals are used to produce monoclonal antibodies. An antigen is introduced into the animal, which produces antibodies in its lymphocyte cells. These cells are cultured and then antibodies are isolated. Since these antibodies would be rejected by the human immune system, the cells are genetically modified so that they produce antibodies with a human protein component, or humanized antibodies. The genetically modified cells are then cultured and humanized antibodies are isolated for production (GenScript 2020 ).
Somatic cells are cells other than the reproductive or germ cells, such as skin, nerve, muscle, liver or bone marrow cells.
Monsanto has developed GM crops (known as Bt crops) that produce Bacillus thuringiensis toxins, which are deadly to insects. Farmers were already using these toxins as pesticides were Bt crops were developed (Resnik 2012 ).
Monsanto has developed GM crops (known as “Roundup Ready” crops) that are immune to the effects of glyphosate, the active ingredient in the widely-used herbicide Roundup ™. Farmers can control weeds with damaging their crops by spraying their crops with Roundup (Resnik 2012 ).
Golden rice, for example, contains more beta carotene than normal rice (McDivitt 2019 ).
In 2018, 228 million people worldwide contracted malaria and 405,000 people died from the disease (World Health Organization 2020a ). About 390 million people contract the dengue virus each year and about 4000 die from the disease (World Health Organization 2020b ).
Oxitec has also genetically engineered diamondback moths (Plutella xylostella) to control these populations. Diamondback moths are a destructive pests that feed on cauliflower, cabbage, broccoli and canola (Campbell 2020a ).
E.g. Bt crops. See Footnote 12.
These are the sorts of problems encountered by the natural law approaches to morality, discussed in Chapter 3 .
Most defenders of the slippery slope argument in genetic only apply it to using genome editing in humans, but it could be applied to other applications of genetic engineering.
I am assuming that GM microbes will not be intentionally released into the environment, which would create risks not discussed here. Scientists have developed GM microbes to clean up oil spills but have not deployed them yet, mostly due to regulatory issues. In nature, microbes already play an important role in cleaning up oil spills (Ezezika and Singer 2010 ).
The reproduction rate is how many people infected persons infect. R 0 = 1 means that an infected person infects one more person on average; R 0 = 2 means an infected person infects two people on average.
It is worth noting, however, that a voluntary moratorium was a reasonable option when this technology was emerging in the 1970s.
As noted in Chapter 6 , a black market for alcohol emerged during Prohibition era in the US (1919–1933). The desire to avoid creating a black market for any product is an relevant to regulatory actions that involve prohibitions.
As a side note, members of Greenpeace broke into a research farm in Australia in 2011 and destroyed an entire crop of GM wheat. Members of another environmental damaged a crop of golden rice in the Philippines (Zhang et al. 2016 ).
To date, 156 Nobelists have signed the petition (Nobel Prize Winners 2016 ).
For a review of the GM food safety literature, also see Domingo ( 2016 ).
It is worth noting the long-term animal studies pose some scientific and technical challenges because most of the rodent species used in these types of experiments have a lifespan of about three years and normally develop tumors and other health problems as they age. So, it can be difficult to determine whether an adverse effect in a laboratory animal is due to an exposure to a GM food or the natural aging process. A two-year study published by Séralini et al. ( 2012 ) claiming that mice fed a diet of Roundup Ready GM corn had more tumors than mice fed the normal diet (the control group) was later retracted by the journal due to serious methodological flaws that undermined the validity of the data (Resnik 2015a ).
See Footnote 12.
Davidson ( 2001 ) defends a principle of charity for interpreting language. The basic idea here is that one should interpret a speaker’s statements as being rational, other things being equal. Interpreting disagreements about GM foods/crops as based on differing value priorities portrays these disagreements as rational, rather than based on irrational fear or ignorance.
It is also worth noting that bans on GM plants can create black markets because of the high demand for these products.
As of the writing of this book, Kenya is currently rethinking its ban on GM crops (Meeme 2019 ).
Most of the debate about chimeras so far has focused on inserting human cells into early animal embryos (or blastocysts), not on inserting human genes into animals.
It is also worth noting that a ban would probably create a black market because demand for GM animals and animal products it high.
There is a potential regulatory gap in the genetic engineering of animals for meat or animal products. Although regulations and ethical guidelines require IACUCs to review and oversee genetic engineering of animals for research conducted at academic institutions, there are no such requirements for genetic engineering of animals for non-research purposes, such as meat production. One could argue that companies that genetically engineer animals for non-research purposes should form ethics committees similar to IACUCs to oversee these activities.
Anderson led the research team that conducted the world’s first human gene therapy clinical trial. The experiment used an adenovirus vector to insert the adenosine deaminase gene into the T-cells of two young children with combined immunodeficiency. The trial showed that the procedure was safe and effective even if did not cure the patients (Blaese et al. 1995 ). In 2006, Anderson was convicted of molesting and sexually abusing a girl over a four-year period, beginning when she was 10 years old, and he served 12 years in prison. Anderson maintains that he is innocent and that his conviction was based on falsified evidence (Begley 2018 ).
See Footnote 29.
An example of somatic genetic enhancement would be a transferring a gene to an adult male to stimulate production of testosterone to enhance athletic and sexual performance.
It is worth noting that not everyone regards genetic enhancement immoral or morally questionable. The transhumanist movement embraces various forms of enhancement to benefit mankind and allow people to express creative freedom (Harris 2007 ; Bostrom 2008 , 2010 ; More and Vita-More 2013 ; Porter 2017 ; Rana and Samples 2019 ).
Some have attempted to define health in terms of a normal range of variation for an organism. In medicine, a normal physiological trait is a trait that falls within a range of variation for healthy functioning of the organism (Boorse 1977 ; Schaffner 1993 ). For example, normal fasting blood sugar levels range from 60 mg/dL to 100 mg/dL (WebMD 2020 ). Fasting blood sugar levels that are too high cause diabetes and levels that are too low cause hypoglycemia, both of which are unhealthy conditions. However, normality cannot be equated with the statistical norm for a population, since the statistical norm might be unhealthy. If most people in a population have a fasting blood sugar greater than 100 mg/dL, we would not say that a fasting blood sugar greater than 100 mg/dL is normal, even though it would be the statistical norm for that population. Thus, the concept of a normal range of variation cannot be defined statistically and depends on a broader concept of health, which may be influenced by moral, social, and cultural factors.
Some argue that “gene therapy” is a misleading term because it implies that the genetic interventions are likely to benefit the patient or human subject, when often they do not (Henderson et al. 2006 ).
See Resnik ( 2018a ) for discussion of additional safety protections for subjects enrolled in clinical research.
In 1996, the US Congress passed a ban, known as the Dickey-Wicker amendment, on the use of federal funds to create human embryos for research (Green 2001 ). Though the ban has been interpreted differently by different administrations, it is still in effect.
For further discussion of creating embryos for research, see Green ( 2001 ).
I will assume that parents who are willing to use medical technology to prevent the birth of children with genetic diseases view abortion as morally acceptable, at least for this purpose.
Prenatal genetic testing can also be used to avoid giving birth to children with chromosomal abnormalities, such as Trisomy 21 (Down Syndrome).
Embryos that are not implanted would be destroyed. I am assuming that parents would view this as morally acceptable.
See Resnik et al. ( 1999 ) and National Academies of Sciences, Engineering, and Medicine ( 2017 ) for additional examples of monogenic disorders that GGE might be used to prevent.
The concept of a parent can be confusing here, because people who related to the child genetically might not be related socially. The concept of a parent can be even more confusing when surrogate pregnancy is used to produce children, since woman who gestates and gives birth to the child might not be genetically related to the child, if she is carrying a fetus created by another couple in vitro.
This is one of the themes of the science fiction movie GATTACA.
This cost estimate is based on dividing the total cost of the Human Genome Project--$3 billion—by three. The Human Genome Project was a US-funded research project that took place from 1990 to 2003. Although sequencing the human genome was the primary goal of the project, it also included other activities, such as studies of human diseases, model organisms, genetic technologies, computational methods, and ethical issues (Human Genome Project 2020 ).
Interestingly, two of the scientists who called for the moratorium, David Baltimore and Paul Berg, participated in the Asilomar conference on recombinant DNA (discussed earlier).
These studies could include the creation of human embryos to study the safety and efficacy of GGE methods and techniques (Liang et al. 2015 ).
This is an example of the problem of incoherence discussed in Chapter 4 .
Alopecia areata is a condition that leads to hair loss. It is thought to have a genetic basis (McIntosh 2017 ).
The moratorium would not apply to GGE for research purposes.
The moratorium would not apply to research on embryos created by GGE, which would be necessary to obtain the knowledge needed to better understand the safety and efficacy of using GGE to produce children (Liang et al. 2015 ; Baltimore et al. 2015 ).
Agar, N. 2014. Truly Human Enhancement: A Philosophical Defense of Limits . Cambridge, MA: MIT Press.
Book Google Scholar
Alberts, B., A.D. Johnson, J. Lewis, D. Morgan, M. Raff, K. Roberts, and P. Walter. 2015. Molecular Biology of the Cell , 6th ed. New York, NY: W. W. Norton.
Google Scholar
American Association for the Advancement of Science. 2000. Human Inheritable Genetic Modifications: Assessing Scientific, Ethical, Religious, and Policy Issues . Washington, DC: American Association for the Advancement of Science.
American Association for the Advancement of Science. 2012. Statement by the AAAS Board of Directors on labeling of genetically modified foods, October 2012. Available at: http://www.aaas.org/sites/default/files/AAAS_GM_statement.pdf . Accessed 18 Jan 2021.
American College of Obstetricians and Gynecologists. 2019. Prenatal genetic screening tests. Available at: https://www.acog.org/Patients/FAQs/Prenatal-Genetic-Screening-Tests?IsMobileSet=false . Accessed 18 Jan 2021.
Anderson, W.F. 1985. Human Gene Therapy: Scientific and Ethical Considerations. Journal of Medicine and Philosophy 10 (3): 275–291.
Article Google Scholar
Anderson, W.F. 1989. Human Gene Therapy: Why Draw a Line? Journal of Medicine and Philosophy 14 (6): 81–93.
Annas, G.J., L.B. Andrews, and R.M. Isasi. 2002. Protecting the Endangered Human: Toward an International Treaty Prohibiting Cloning and Inheritable Alterations. American Journal of Law and Medicine 28: 151–178.
Araki, A., and T. Ishii. 2016. Providing Appropriate Risk Information on Genome Editing for Patients. Trends in Biotechnology 34 (2): 86–90.
Arms Control Association. 2018. The Biological Weapons Convention (BWS) at a Glance. Available at: https://www.armscontrol.org/factsheets/bwc . Accessed 18 Jan 2021.
Baeshen, N.A., M.N. Baeshen, A. Sheikh, R.S. Bora, M.M. Ahmed, H.A. Ramadan, K.S. Saini, and E.M. Redwan. 2014. Cell Factories for Insulin Production. Microbial Cell Factories 13: 141.
Baltimore, D., P. Berg, M. Botchan, D. Carroll, R.A. Charo, G. Church, J.E. Corn, G.Q. Daley, J.A. Doudna, M. Fenner, H.T. Greely, M. Jinek, G.S. Martin, E. Penhoet, J. Puck, S.H. Sternberg, J.S. Weissman, and K.R. Yamamoto. 2015. A Prudent Path Forward for Genomic Engineering and Germline Gene Modification. Science 348 (6230): 36–38.
Bates, K.G. 2014. A Chosen Exile: Black People Passing in White America. NRP, October 7. Available at: https://www.npr.org/sections/codeswitch/2014/10/07/354310370/a-chosen-exile-black-people-passing-in-white-america . Accessed 18 Jan 2021.
Baylis, F. 2019. Altered Inheritance: CRISPR and the Ethics of Human Genome Editing . Cambridge, MA: Harvard University Press.
BBC News. 2015. Is Opposition to Genetically Modified Food Irrational? BBC News , June 3. Available at: https://www.bbc.com/news/science-environment-32901834 . Accessed 18 Jan 2021.
Beauchamp, T.L., and D. DeGrazia. 2020. Principles of Animal Research Ethics . New York, NY: Oxford University Press.
Begley S. 2018. Out of Prison, the ‘Father of Gene Therapy’ Faces a Harsh Reality: A Tarnished Legacy and an Ankle Monitor. STAT , July 23. Available at: https://www.statnews.com/2018/07/23/w-french-anderson-father-of-gene-therapy/ . Accessed 18 Jan 2021.
Berger, E., and B. Gert. 1991. Genetic Disorders and the Ethical Status of Germ-Line Gene Therapy. Journal of Medicine and Philosophy 16 (6): 667–683.
Beriain, I. 2018. Human Dignity and Gene Editing: Using Human Dignity as an Argument Against Modifying the Human Genome and Germline Is a Logical Fallacy. EMBO Reports 19 (10): e46789.
Berry, R. 2013. The Ethics of Genetic Engineering . New York, NY: Routledge.
Biello, D. 2010. Genetically Modified Crops on the Loose and Evolving in the U.S. Midwest. Scientific American , August 6. Available at: https://www.scientificamerican.com/article/genetically-modified-crop/ . Accessed 18 Jan 2021.
Billings, L.K., and J.C. Florez. 2010. The Genetics of Type 2 Diabetes: What Have We Learned from GWAS? Annals of New York Academy of Science 1212: 59–77.
Biofuels International. 2018. GM Yeast Could Fix Food vs. Fuel Debate Around Bioethanol. Biofuels International , April 4. Available at: https://biofuels-news.com/news/gm-yeast-could-fix-food-vs-fuel-debate-around-bioethanol/ . Accessed 26 Feb 2020.
Biotechnology Innovation Organization. 2020b. Genetically Engineered Animals: Frequently Asked Questions. Available at: https://archive.bio.org/articles/genetically-engineered-animals-frequently-asked-questions . Accessed 18 Jan 2021.
Blackford, R. 2014. Humanity Enhanced: Genetic Choice and the Challenge for Liberal Democracies . Cambridge, MA: MIT Press.
Blaese, R.M., K.W. Culver, A.D. Miller, C.S. Carter, T. Fleisher, M. Clerici, G. Shearer, L. Chang, Y. Chiang, P. Tolstoshev, J.J. Greenblatt, S.A. Rosenberg, H. Klein, M. Berger, C.A. Mullen, W.J. Ramsey, L. Muul, R.A. Morgan, and W.F. Anderson. 1995. T Lymphocyte-Directed Gene Therapy for ADA-SCID: Initial Trial Results After 4 Years. Science 270 (5235): 475–480.
Blancke, S. 2015. Is Opposition to Genetically Modified Food Irrational? Scientific American , August 18. Available at: https://www.scientificamerican.com/article/why-people-oppose-gmos-even-though-science-says-they-are-safe/ . Accessed 18 Jan 2021.
Blendon, R.J., M.T. Gorski, and J.M. Benson. 2016. The Public and the Gene-Editing Revolution. New England Journal of Medicine 374 (15): 1406–1411.
Boone, C.K. 1988. Bad axioms in Genetic Engineering. Hastings Center Report 18 (4): 9–13.
Bodner, A. 2015. Preventing Escape of GMO Salmon. Biology Fortified , November 20. Available at: https://biofortified.org/2015/11/gmo-salmon/ . Accessed 18 Jan 2021.
Boorse, C. 1977. Health as a Theoretical Concept. Philosophy of Science 44: 542–573.
Borges, B.J., O.M. Arantes, A.A. Fernandes, J.R. Broach, and P.M. Fernandes. 2018. Genetically Modified Labeling Policies: Moving Forward or Backward? Frontiers in Bioengineering and Biotechnology 6: 181.
Bostrom, N. 2010. Letter from Utopia (Version 1.9). Studies in Ethics, Law, and Technology 2: 1–7.
Bostrom, N. 2008. Why I Want to Be a Posthuman When I Grow Up. In Medical Enhancement and Posthumanity , ed. B. Gordijn and R. Chadwick, 107–137. Dordrecht, Netherlands: Springer.
Buchanan, A., D.W. Brock, N. Daniels, and D. Wikler. 2000. From Chance to Choice: Genetics and Justice . Cambridge, UK: Cambridge University Press.
Callahan, D. 1995. Setting Limits: Medical Goals in an Aging Society with “A Response to My Critics” . Washington, DC: Georgetown University Press.
Campbell, M. 2020a. World’s First Genetically Engineered Moth Is Released into an Open Field. Technology Networks , January 29. Available at: https://www.technologynetworks.com/genomics/news/world-first-genetically-engineered-moth-is-released-into-an-open-field-329960 . Accessed 18 Jan 2021.
Campbell, M. 2020b. Genetically Engineered Bacteria Protect Honey Bees Against Parasites. Technology Networks , February 24. Available at: https://www.technologynetworks.com/genomics/news/genetically-engineered-bacteria-protect-honey-bees-against-parasites-331209 . Accessed 18 Jan 2021.
Caplan, A. 1995. Moral Matters . New York, NY: Wiley.
Caplan, A. 1997. The Concepts of Health, Illness, and Disease. In Medical Ethics , 2nd ed, ed. R. Veatch, 57–74. Sudbury, MA: Jones and Bartlett.
Carlson, E.A. 2001. The Unfit: A History of a Bad Idea . Cold Spring Harbor, NY: Cold Spring Harbor Press.
Centers for Disease Control and Prevention. 2019. Heart Disease Facts. Available at: https://www.cdc.gov/heartdisease/facts.htm . Accessed 18 Jan 2021.
Centers for Disease Control and Prevention and National Institutes of Health. 2009. Biosafety in Microbiological and Biomedical Laboratories, 5th ed. Available at: https://www.cdc.gov/labs/pdf/CDC-BiosafetyMicrobiologicalBiomedicalLaboratories-2009-P.PDF . Accessed 18 Jan 2021.
Christensen J. 2018. The Five Most Expensive Drugs in the United States. CNN , May 11. Available at: https://www.cnn.com/2018/05/11/health/most-expensive-prescription-drugs/index.html . Accessed 18 Jan 2021.
Cilluffo, A., and N.G. Ruiz. 2019. World’s Population Is Projected to Nearly Stop Growing by the End of the Century. Pew Research Center , June 17. Available at: https://www.pewresearch.org/fact-tank/2019/06/17/worlds-population-is-projected-to-nearly-stop-growing-by-the-end-of-the-century/ . Accessed 18 Jan 2021.
Coelho, A.C., and J.D. García. 2015. Biological Risks and Laboratory-Acquired Infections: A Reality That Cannot Be Ignored in Health Biotechnology. Frontiers in Bioengineering and Biotechnology 3: 56.
Cohen J. 2019a. China’s CRISPR Push in Animals Promises Better Meat, Novel Therapies, and Pig Organs for People. Science , July 31. Available at: https://www.sciencemag.org/news/2019/07/china-s-crispr-push-animals-promises-better-meat-novel-therapies-and-pig-organs-people . Accessed 18 Jan 2021.
Cohen, J. 2019b. Deaf Couple May Edit Embryo’s DNA to Correct Hearing Mutation. Science , October 21. Available at: https://www.sciencemag.org/news/2019/10/deaf-couple-may-edit-embryo-s-dna-correct-hearing-mutation . Accessed 18 Jan 2021.
Cole-Turner, R. 1997. Genes, Religion and Society: The Developing Views of the Churches. Science and Engineering Ethics 3: 273–288.
Collins, M., and A. Thrasher. 2015. Gene Therapy: Progress and Predictions. Proceedings of Biological Sciences 282: 1821.
Conrow, J. 2018. Developing Nations Lead the Growth of GMO Crops. Alliance for Science , June 29. Available at: https://allianceforscience.cornell.edu/blog/2018/06/developing-nations-lead-growth-gmo-crops/ . Accessed 18 Jan 2021.
Convention on Biological Diversity. 2020. Available at: https://www.cbd.int/ . Accessed 18 Jan 2021.
Cornish, L. 2018. Understanding the Continued Opposition to GMOs. Devex , January 22. Available at: https://www.devex.com/news/understanding-the-continued-opposition-to-gmos-91888 . Accessed 18 Jan 2021.
Cossins, D. 2015. Will We Ever See GM Meat? BBC Future . March 9. Available at: https://www.bbc.com/future/article/20150309-will-we-ever-eat-gm-meat . Accessed 18 Jan 2021.
Costa, J.R., B.E. Bejcek, J.E. McGee, A.I. Fogel, K.R. Brimacombe, and R. Ketteler. 2017. Genome Editing Using Engineered Nucleases and Their Use in Genomic Screening. In Assay Guidance Manual , ed. S. Sittampalam et al. Bethesda, MD: Eli Lilly and Company and the National Center for Advancing Translational Sciences. Available at: https://www.ncbi.nlm.nih.gov/books/NBK464635/ . Accessed 18 Jan 2021.
Cummings, J.P. 2018. The Lifetime Economic Burden of Monogenic Diseases and the Social Motivations for Their Treatment with Genetic Therapy. Thesis. Rochester Institute of Technology. Available at: https://scholarworks.rit.edu/cgi/viewcontent.cgi?article=10984&context=theses . Accessed 18 Jan 2021.
Cyranoski, D. 2020. What CRISPR-Baby Prison Sentences Mean for Research. Nature 577: 154–155.
Daniell, H. 2002. Molecular Strategies for Gene Containment in Transgenic Crops. Nature Biotechnology 20 (6): 581–586.
Darwin, C. 1859. The Origin of Species by Means of Natural Selection . London, UK: John Murray.
Davidson, D. 2001. Inquiries into Truth and Interpretation , 2nd ed. Oxford, UK: Clarendon Press.
Davis, D.S. 2001. Genetic Dilemmas: Reproductive Technology, Parental Choices, and Children’s Futures . New York, NY: Routledge.
De Wert, G., B. Heindryckx, G. Pennings, A. Clarke, U. Eichenlaub-Ritter, C.G. van El, F. Forzano, M. Goddijn, H.C. Howard, D. Radojkovic, E. Rial-Sebbag, W. Dondorp, B.C. Tarlatzis, M.C. Cornel, and European Society of Human Genetics and the European Society of Human Reproduction and Embryology. 2018. Responsible Innovation in Human Germline Gene Editing: Background Document to the Recommendations of ESHG and ESHRE. European Journal of Human Genetics 26 (4): 450–470.
Domingo, J.L. 2016. Safety Assessment of GM Plants: An Updated Review of the Scientific Literature. Food and Chemical Toxicology 95: 12–18.
Doyle, A., M.P. McGarry, N.A. Lee, and J.J. Lee. 2012. The Construction of Transgenic and Gene Knockout/Knockin Mouse Models of Human Disease. Transgenic Research 21 (2): 327–349.
Duan, J.J., M. Marvier, J. Huesing, G. Dively, and Z.Y. Huang. 2008. A Meta-Analysis of Effects of Bt Crops on Honey Bees (Hymenoptera: Apidae). PLoS One 3 (1): e1415.
Dubljević, V. 2019. Neuroethics, Justice and Autonomy: Public Reason in the Cognitive Enhancement Debate . Cham, Switzerland: Springer.
Dunn, S.E., J.L. Vicini, K.C. Glenn, D.M. Fleischer, and M.J. Greenhawt. 2017. The Allergenicity of Genetically Modified Foods from Genetically Engineered Crops: A Narrative and Systematic Review. Annals of Allergy, Asthma and Immunology 119 (3): 214–222.
Environmental Protection Agency. 2020b. EPA’s Regulation of Biotechnology for Use in Pest Management. Available at: https://www.epa.gov/regulation-biotechnology-under-tsca-and-fifra/epas-regulation-biotechnology-use-pest-management . Accessed 18 Jan 2021.
European Commission. 2020. GMO Legislation. Available at: https://ec.europa.eu/food/plant/gmo/legislation_en . Accessed 18 Jan 2021.
Ezezika, O.C., and P.A. Singer. 2010. Genetically Engineered Oil-Eating Microbes for Bioremediation: Prospects and Regulatory Challenges. Technology in Society 32 (4): 331–335.
Fagan, J., M. Antoniou, and C. Robinson. 2014. GMO Myths and Truths , 2nd ed. London, UK: Earth Open Source.
Fernandez-Cornejo, J., S. Wechsler, M. Livingston, and L. Mitchell. 2014. Genetically Engineered Crops in the United States. U.S. Department of Agriculture, Economic Research Report 162, February. Available at: https://www.ers.usda.gov/webdocs/publications/45179/43668_err162.pdf . Accessed 18 Jan 2021.
Food and Drug Administration. 2020a. Animals with Intentional Genomic Alterations: Consumer Q & A. Available at: https://www.fda.gov/animal-veterinary/animals-intentional-genomic-alterations/consumer-qa . Accessed 19 Jan 2021.
Food and Drug Administration. 2020b. Oxitec Mosquito. Available at: https://www.fda.gov/animal-veterinary/animals-intentional-genomic-alterations/oxitec-mosquito . Accessed 19 Jan 2021.
Food and Drug Administration. 2020c. Therapeutic Cloning and Genome Modification. Available at: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/therapeutic-cloning-and-genome-modification . Accessed 19 Jan 2021.
Food and Drug Administration. 2020d. What Is the Approval Process for Generic Drugs? Available at: https://www.fda.gov/drugs/generic-drugs/what-approval-process-generic-drugs . Accessed 19 Jan 2021.
Forabosco, F., M. Löhmus, L. Rydhmer, and L.F. Sundström. 2013. Genetically Modified Farm Animals and Fish in Agriculture: A Review. Livestock Science 153 (1–3): 1–9.
Frank, S.A. 2014. Somatic Mosaicism and Disease. Current Biology 24 (2): R577–R581.
Fukuyama, F. 2002. Our Posthuman Future: Consequences of the Biotechnology Revolution . New York: Picador.
Funk, C., and M. Hefferon. 2018. Public Views of Gene Editing for Babies Depend on How It Would Be Used. Pew Research Center , July 26. Available at: https://www.pewresearch.org/science/2018/07/26/public-views-of-gene-editing-for-babies-depend-on-how-it-would-be-used/ . Accessed 19 Jan 2021.
Gallo, A.M., D. Wilkie, M. Suarez, R. Labotka, R. Molokie, A. Thompson, P. Hershberger, and B. Johnson. 2010. Reproductive Decisions in People with Sickle Cell Disease or Sickle Cell Trait. Western Journal of Nursing Research 32 (8): 1073–1090.
Geib, C. 2018. Changing Regulations Mean Genetically Modified Meat Could Soon Be on Your Plate. Futurism , March 14. Available at: https://futurism.com/genetically-modified-meat-fda-usda . Accessed 19 Jan 2021.
Genetic Literacy Project. 2018. New Generation of GMO Crops Could Dramatically Boost Biofuel Production. Available at: https://geneticliteracyproject.org/2018/01/15/new-generation-gmo-crops-dramatically-boost-biofuel-production/ . Accessed 19 Jan 2021.
Genetic Literacy Project. 2020. GMO FAQs. Available at: https://gmo.geneticliteracyproject.org/FAQ/where-are-gmos-grown-and-banned/ . Accessed 19 Jan 2021.
GenScript. 2020. What Are Monoclonal Antibodies? Available at: https://www.genscript.com/how-to-make-monoclonal-antibodies.html . Accessed 19 Jan 2021.
GM Watch. 2019. International Scientists Urge Precaution with Gene Drives: New Study. GM Watch , May 21. https://www.gmwatch.org/en/news/latest-news/18951-international-scientists-urge-precaution-with-gene-drives-new-study . Accessed 19 Jan 2021.
GMO Answers. 2020a. What GMO Crops Are Currently Available on the Market? Available at: https://gmoanswers.com/current-gmo-crops?gclid=CjwKCAiAhc7yBRAdEiwAplGxX265z5GBxlV4Y4pqKVOfiooF2qfFs91eOW8InUo3yuJGH_B39BkoDxoCY2gQAvD_BwE . Accessed 19 Jan 2021.
GMO Answers. 2020b. Nine Things You Need to Know About GMO Salmon. Available at: https://gmoanswers.com/nine-9-things-you-need-know-about-gmo-salmon . Accessed January.
Gonzaludo, N., J.W. Belmont, V.G. Gainullin, and R.J. Taft. 2019. Estimating the Burden and Economic Impact of Pediatric Genetic Disease. Genetics in Medicine 21: 1781–1789.
Green, R.M. 2001. The Human Embryo Research Debates: Bioethics in the Vortex of Controversy . New York, NY: Oxford University Press.
Guillemaud, T., E. Lombaert, and D. Bourguet. 2016. Conflicts of Interest in GM Bt Crop Efficacy and Durability Studies. PLoS One 11 (12): e0167777.
Gurevich, R. 2020. How Much Does IVF Really Cost? Very Well Family, March 5. Available at: https://www.verywellfamily.com/how-much-does-ivf-cost-1960212 . Accessed 19 Jan 2021.
Harmon, A. 2016. Fighting Lyme Disease in the Genes of Nantucket’s Mice. New York Times , June 7, A15.
Harris, J. 1992. Wonderwoman and Superman: The Ethics of Human Biotechnology . Oxford, UK: Oxford University Press.
Harris, J. 2007. Enhancing Evolution: The Ethical Case for Making Better People . Princeton, NJ: Princeton University Press.
He, K., L.R. Wilkens, D.O. Stram, L.N. Kolonel, B.E. Henderson, A.H. Wu, L. Le Marchand, and C.A. Haiman. 2011. Generalizability and Epidemiologic Characterization of Eleven Colorectal Cancer GWAS Hits in Multiple Populations. Cancer Epidemiology and Biomarkers and Prevention 20 (1): 70–81.
Henderson, G.E., M.M. Easter, C. Zimmer, N.M. King, A.M. Davis, B.B. Rothschild, L.R. Churchill, B. Wilfond, and D.K. Nelson. 2006. Therapeutic Misconception in Early Phase Gene Transfer Trials. Social Science and Medicine 62 (1): 239–253.
Henkel, R.D., T. Miller, and R.S. Weyant. 2012. Monitoring Select Agent Theft, Loss and Release Reports in the United States—2004–2010. Applied Biosafety 18: 171–180.
Hjältén, J., and E.P. Axelsson. 2015. GM Trees with Increased Resistance to Herbivores: Trait Efficiency and Their Potential to Promote Tree Growth. Frontiers in Plant Science , May 1. Available at: https://doi.org/10.3389/fpls.2015.00279 . Accessed 19 Jan 2021.
Holdrege, C. 2008. Understanding the Unintended Effects of Genetic Manipulation. The Nature Institute . Available at: https://natureinstitute.org/txt/ch/nontarget.php . Accessed 19 Jan 2021.
Horgan, J. 2017. Has the Era of Gene Therapy Finally Arrived? Scientific American , September 1. Available at: https://blogs.scientificamerican.com/cross-check/has-the-era-of-gene-therapy-finally-arrived/ . Accessed 19 Jan 2021.
Hou, Z., and Z. Zhang. 2019. Inserting DNA with CRISPR. Science 365 (6448): 25–26.
House, K. 2019. China Quietly Confirms Birth of Third Gene-Edited Baby. Futurism , December 30. Available at: https://futurism.com/neoscope/china-confirms-birth-third-gene-edited-baby . Accessed 19 Jan 2021.
Hryhorowicz, M., J. Zeyland, R. Słomski, and D. Lipiński. 2017. Genetically Modified Pigs as Organ Donors for Xenotransplantation. Molecular Biotechnology 59 (9–10): 435–444.
Hübner, D. 2018. Human-animal Chimeras and Hybrids: An Ethical Paradox Behind Moral Confusion? The Journal of Medicine and Philosophy 43 (2): 187–210.
Human Fertilisation and Embryology Authority. 2020. About Us. Available at: https://www.hfea.gov.uk/about-us/ . Accessed 19 Jan 2021.
Human Genome Project. 2020. Human Genome Project Budget. Available at: https://web.ornl.gov/sci/techresources/Human_Genome/project/budget.shtml . Accessed 19 Jan 2021.
International Service for the Acquisition of Agri-biotech Applications. 2018. Gm Crops and the Environment. Available at: https://www.isaaa.org/resources/publications/pocketk/4/default.asp . Accessed: 19 Jan 2021.
Johnston, T. 2005. In One’s Own Image: Ethics and the Reproduction of Deafness. Journal of Deaf Studies and Deaf Education 10 (4): 426–441.
Juengst, E. 1997. Can Enhancement Be Distinguished from Prevention in Genetic Medicine? Journal of Medicine and Philosophy 22 (2): 125–142.
Justlabelit.org. 2020. Labelling Around the World. Available at: http://www.justlabelit.org/right-to-know-center/labeling-around-the-world/ . Accessed 19 Jan 2021.
Kaebnick, G.E., E. Heitman, J.P. Collins, J.A. Delborne, W.G. Landis, K. Sawyer, L.A. Taneyhill, and D.E. Winickoff. 2016. Precaution and Governance of Emerging Technologies. Science 354 (6313): 710–711.
Kaemmerer, W.F. 2018. How Will the Field of Gene Therapy Survive Its Success? Bioengineering and Translational Medicine 3 (2): 166–177.
Kaiser Family Foundation. 2016. Medicaid Coverage of Family Planning Benefits: Results from a State Survey. Available at: https://www.kff.org/report-section/medicaid-coverage-of-family-planning-benefits-results-from-a-state-survey-fertility-services/ . Accessed 19 Jan 2021.
Kelle, A. 2013. Beyond Patchwork Precaution in the Dual-Use Governance of Synthetic Biology. Science and Engineering Ethics 19 (3): 1121–1139.
Kevles, D.J. 1985. In the Name of Eugenics: Genetics and the Uses of Human Heredity . Cambridge, MA: Harvard University Press.
Kids Health. 2018. Osteogenesis Imperfecta (Brittle Bone Disease). Available at: https://kidshealth.org/en/parents/osteogenesis-imperfecta.html . Accessed 19 Jan 2021.
Kimman, T.G., E. Smit, and M.R. Klein. 2008. Evidence-Based Biosafety: A Review of the Principles and Effectiveness of Microbiological Containment Measures. Clinical Microbiology Reviews 21 (3): 403–425.
Kimmelman, J. 2010. Gene Transfer and the Ethics of First-in-Human Research: Lost in Translation . Cambridge, UK: Cambridge University Press.
Kitcher, P. 1996. The Lives to Come: the Genetic Revolution and Human Possibilities . New York, NY: Simon and Schuster.
Koch, T. 2020. Transhumanism, Moral Perfection, and Those 76 Trombones. Journal of Medicine and Philosophy 45 (2): 179–192.
Koplin, J.J., C. Gyngell, and J. Savulescu. 2020. Germline Gene Editing and the Precautionary Principle. Bioethics 34 (1): 49–59.
Koplin, J.J., and D. Wilkinson. 2019. Moral Uncertainty and the Farming of Human-Pig Chimeras. Journal of Medical Ethics 45 (7): 440–446.
Kriebel, D., J. Tickner, P. Epstein, J. Lemons, R. Levins, E.L. Loechler, M. Quinn, R. Rudel, T. Schettler, and M. Stoto. 2001. The Precautionary Principle in Environmental Science. Environmental Health Perspectives 109 (9): 871–876.
Kumar, P., J. Radhakrishnan, M.A. Chowdhary, and P.F. Giampietro. 2001. Prevalence and Patterns of Presentation of Genetic Disorders in a Pediatric Emergency Department. Mayo Clinic Proceedings 76 (8): 777–783.
Kumar, S.R.P., D.M. Markusic, M. Biswas, K.A. High, and R.W. Herzog. 2016. Clinical Development of Gene Therapy: Results and Lessons from Recent Successes. Molecular Therapy—Methods and Clinical Development 3: 16034.
Kuzma, J. 2016. A Missed Opportunity for U.S. Biotechnology Regulation. Science 353 (6305): 1211–1213.
Lander, E.S., F. Baylis, F. Zhang, E. Charpentier, P. Berg, C. Bourgain, B. Friedrich, J.K. Joung, J. Li, D. Liu, L. Naldini, J.B. Nie, R. Qiu, B. Schoene-Seifert, F. Shao, S. Terry, W. Wei, and E.L. Winnacker. 2019. Adopt a Moratorium on Heritable Genome Editing. Nature 567 (7747): 165–168.
Lanphier, E., F. Urnov, S.E. Haecker, M. Werner, and J. Smolenski. 2015. Don’t Edit the Human Germ Line. Nature 519: 410–411.
Ledford, H., and E. Callaway. 2020. Pioneers of CRISPR Gene Editing Win Nobel in Chemistry. Nature 586: 346–347.
Lee, B. 2018. What Are Biologics? 5 Examples of Biological Drugs You May Already Be Taking. Good RX , June 13. Available at: https://www.goodrx.com/blog/biologics-biological-drugs-examples/ . Accessed 19 Jan 2021.
Le Page, M. 2020. Human Genes Have Been Added to Pigs to Create Skin for Transplants. New Scientist , January 29. Available at: https://www.newscientist.com/article/2231579-human-genes-have-been-added-to-pigs-to-create-skin-for-transplants/#ixzz6GPggXYEP . Accessed 19 Jan 2021.
Liang, P., Y. Xu, X. Zhang, C. Ding, R. Huang, Z. Zhang, J. Lv, X. Xie, Y. Chen, Y. Li, Y. Sun, Y. Bai, Z. Songyang, W. Ma, C. Zhou, and J. Huang. 2015. CRISPR/Cas9-Mediated Gene Editing in Human Tripronuclear Zygotes. Protein and Cell 6 (5): 363–372.
Losey, J.E., L.S. Rayor, and M.E. Carter. 1999. Transgenic Pollen Harms Monarch Larvae. Nature 399: 214.
Lucht, J.M. 2015. Public Acceptance of Plant Biotechnology and GM Crops. Viruses 7 (8): 4254–4281.
Maddox, B. 2003. Rosalind Franklin: The Dark Lady of DNA . New York, NY: HarperCollins.
Main, D. 2017. USDA Agrees to Not Regulate Genetically Modified GRASS on the Loose in Oregon. Newsweek , January 31. Available at: https://www.newsweek.com/usda-agrees-not-regulate-gmo-grass-loose-oregon-550942 . Accessed 19 Jan 2021.
Mamcarz, E., S. Zhou, T. Lockey, H. Abdelsamed, S.J. Cross, G. Kang, Z. Ma, J. Condori, J. Dowdy, B. Triplett, C. Li, G. Maron, J.C. Aldave Becerra, J.A. Church, E. Dokmeci, J.T. Love, A.C. da Matta Ain, H. van der Watt, X. Tang, W. Janssen, B.Y. Ryu, S.S. De Ravin, M.J. Weiss, B. Youngblood, J.R. Long-Boyle, S. Gottschalk, M.M. Meagher, H.L. Malech, J.M. Puck, M.J. Cowan, and B.P. Sorrentino. 2019. Lentiviral Gene Therapy Combined with Low-Dose Busulfan in Infants with SCID-X1. New England Journal of Medicine 380 (16): 1525–1534.
Marshall, D.A., E.I. Benchimol, A. MacKenzie, D.D. Duque, K.V. MacDonald, T. Hartley, H. Howley, A. Hamilton, M. Gillespie, F. Malam, and K. Boycott. 2019. Direct Health-Care Costs for Children Diagnosed with Genetic Diseases Are Significantly Higher Than for Children with Other Chronic Diseases. Genetics in Medicine 21: 1049–1057.
Maslen, H., N. Faulmüller, and J. Savulescu. 2014. Pharmacological Cognitive Enhancement-How Neuroscientific Research Could Advance Ethical Debate. Frontiers in Systems Neuroscience 8: 107.
Maziarz, R.T. 2019. CAR T-Cell Therapy Total Cost Can Exceed $1.5 Million Per Treatment. Healio , May 29. Available at: https://www.healio.com/hematology-oncology/cell-therapy/news/online/%7B124396e7-1b60-4cff-a404-0a2baeaf1413%7D/car-t-cell-therapy-total-cost-can-exceed-15-million-per-treatment . Accessed 19 Jan 2021.
McDivitt, P. 2019. Golden Rice: The GMO Crop Loved by Humanitarians, Opposed by Greenpeace. Genetic Literacy Project , November 8. Available at: https://geneticliteracyproject.org/2019/11/08/golden-rice-the-gmo-crop-loved-by-humanitarians-opposed-by-greenpeace/ . Accessed 19 Jan 2021.
McDonald, J. 2007. Could Genetically Modified Crops Be Killing Honeybees? SF Gate , March 10. Available at: https://www.sfgate.com/homeandgarden/article/Could-genetically-modified-crops-be-killing-bees-2611496.php . Accessed 19 Jan 2021.
McGee, G. 2000. The Perfect Baby: Parenthood in the New World of Cloning and Genetics , 2nd ed. Lanham, MD: Rowman and Littlefield.
McIntosh, J. 2017. What’s to Know About Alopecia Areata? Medical News Today , December 22. Available at: https://www.medicalnewstoday.com/articles/70956#home-remedies . Accessed 19 Jan 2021.
Meeme, V. 2019. Kenya Reconsidering GMO Crop Ban for Food Security. Alliance for Science , April 30. Available at: https://allianceforscience.cornell.edu/blog/2019/04/kenya-reconsidering-gmo-crop-ban-support-food-security/ . Accessed 19 Jan 2021.
Mehlman, M.J. 2009. The Price of Perfection: Individualism and Society in the Era of Biomedical Enhancement . Baltimore, MD: Johns Hopkins University Press.
Merler, S., M. Ajelli, L. Fumanelli, and A. Vespignani. 2013. Containing the Accidental Laboratory Escape of Potential Pandemic Influenza Viruses. BMC Medicine 11: 252.
Messer, K.D., S. Bligh, M. Costanigro, and H.M. Kaiser. 2015. Process Labeling of Food: Consumer Behavior, the Agricultural Sector, and Policy Recommendations. Council for Agricultural Science and Technology 10: 1–16.
Miller, F.G., and S. Joffe. 2009. Benefit in Phase 1 Oncology Trials: Therapeutic Misconception or Reasonable Treatment Option? Clinical Trials 5 (6): 617–623.
Miliotou, A.N., and L.C. Papadopoulou. 2018. CAR T-Cell Therapy: A New Era in Cancer Immunotherapy. Current Pharmaceutical Biotechnology 19 (1): 5–18.
Mitchell, C.B., E.D. Pellegrino, J.B. Elshtain, J.F. Kilner, and S.B. Rae. 2007. Biotechnology and the Human Good . Washington, DC: Georgetown University Press.
Molteni, M. 2018. Now You Can Sequence Your Whole Genome for Just $200. Wired , November 11. Available at: https://www.wired.com/story/whole-genome-sequencing-cost-200-dollars/ . Accessed 19 Jan 2021.
More, M., and N. Vita-More (eds.). 2013. The Transhumanist Reader: Classical and Contemporary Essays on the Science, Technology, and Philosophy of the Human Future . New York, NY: Wiley-Blackwell.
Moritz, R. 2020. Community Engagement on Pathogen Research. Presentation to the National Science Advisory Board for Biosecurity, January 24. Bethesda, MD.
Murphy, D. 2020. Concepts of Health and Disease. Stanford Encyclopedia of Philosophy . Available at: https://plato.stanford.edu/entries/health-disease/ . Accessed 19 Jan 2021.
National Academies of Sciences, Engineering, and Medicine. 2016a. Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values . Washington, DC: National Academies Press.
National Academies of Sciences, Engineering, and Medicine. 2016b. Genetically Engineered Crops: Experiences and Prospects . Washington, DC: National Academies Press.
National Academies of Sciences, Engineering, and Medicine. 2017. Human Genome Editing: Science, Ethics, and Governance . Washington, DC: National Academies Press.
National Conference of State Legislatures. 2019. State Laws Related to Insurance Coverage for Infertility Treatment . Available at: https://www.ncsl.org/research/health/insurance-coverage-for-infertility-laws.aspx . Accessed 19 Jan 2021.
National Heart, Lung, and Blood Institute. 2020. Cell Sickle Disease. Available at: https://www.nhlbi.nih.gov/health-topics/sickle-cell-disease . Accessed 19 Jan 2021.
National Human Genome Research Institute. 2017. How Does Genome Editing Work? Available at: https://www.genome.gov/about-genomics/policy-issues/Genome-Editing/How-genome-editing-works . Accessed 19 Jan 2021.
National Institutes of Health. 2020a. Stem Cell Information. Available at: https://stemcells.nih.gov/ . Accessed 19 Jan 2021.
National Research Council. 2004. Biotechnology in the Age of Terrorism . Washington, DC: National Academies Press.
National Research Council. 2011. Guide for the Care and Use of Laboratory Animals , 8th ed. Washington, DC: National Academies Press.
Neuhaus, C.P. 2018. Community Engagement and Field Trials of Genetically Modified Insects and Animals. Hastings Center Report 48 (1): 25–36.
Nobel Prize.org. 2021. The Nobel Prize in Chemistry 1980. Available at: https://www.nobelprize.org/prizes/chemistry/1980/berg/lecture/ . Accessed 10 Jan 2021.
Nobel Prize Winners. 2016. Letter to Greenpeace, June 26. Available at: https://www.supportprecisionagriculture.org/nobel-laureate-gmo-letter_rjr.html . Accessed 19 Jan 2021.
Nogrady, B. 2020. What the Data Say About Asymptomatic COVID Infections. Nature 587: 534–535.
Norero, D. 2016. Genetically Modified Crops and the Exaggeration of “Interest Conflict.” Cornell Alliance for Science , November 3. Available at: https://allianceforscience.cornell.edu/blog/2016/11/genetically-modified-crops-and-the-exaggeration-of-interest-conflict/ . Accessed 19 Jan 2021.
Normile, D. 2004. Infectious Diseases: Mounting Lab Accidents Raise SARS Fears. Science 304: 659–661.
Normile, D. 2018. Shock Greets Claim of CRISPR-Edited Babies. Science 362 (6418): 978–979.
Normile, D. 2019. China Tightens Rules on Gene Editing. Science 363 (6431): 1023.
Nozick, R. 1974. Anarchy, State, Utopia . New York, NY: Basic Books.
Nuffield Council on Bioethics. 2016. Genome Editing: An Ethical Review. Available at: https://www.nuffieldbioethics.org/publications/genome-editing-an-ethical-review . Accessed 13 Mar 2020.
Organizing Committee of the Second International Summit on Human Genome Editing. 2018. Concluding Statement. Available at: http://www8.nationalacademies.org/onpinews/newsitem.aspx?RecordID=11282018b . Accessed 19 Jan 2021.
Ormandy, E.H., J. Dale, and G. Griffin. 2011. Genetic Engineering of Animals: Ethical Issues, Including Welfare Concerns. The Canadian Veterinary Journal 52 (5): 544–550.
Parens, E. (ed.). 1998. Enhancing Human Traits: Ethical and Social Implications . Washington, DC: Georgetown University Press.
Parens, E., and A. Asch. 1999. The Disability Rights Critique of Prenatal Genetic Testing: Reflections and Recommendations. Hastings Center Report 29 (5): S1–22.
Park, A. 2019. Experts Are Calling for a Ban on Gene Editing of Human Embryos. Time Magazine , March 13. Available at: https://time.com/5550654/crispr-gene-editing-human-embryos-ban/ . Accessed 19 Jan 2021.
Pew Research Center. 2016. Public Opinion About Genetically Modified Foods and Trust in Scientists Connected with These Foods. Pew Research Center , December 1. Available at: https://www.pewresearch.org/science/2016/12/01/public-opinion-about-genetically-modified-foods-and-trust-in-scientists-connected-with-these-foods/ . Accessed 19 Jan 2021.
Poppy, G. 2000. GM Crops: Environmental Risks and Non-target Effects. Trends in Plant Science 5 (1): 4–6.
Porter, A. 2017. Bioethics and Transhumanism. Journal of Medicine and Philosophy 42 (3): 237–260.
Porterfield, A., and J. Entine. 2018. ‘Substantial Equivalence’: Are GMOs as Safe as Other Conventional and Organic Foods? Genetic Literacy Project , May 11. Available at: https://geneticliteracyproject.org/2018/05/11/substantial-equivalence-are-gmos-as-safe-as-other-conventional-organic-foods/ . Accessed 19 Jan 2021.
President’s Commission for the Study of Ethical Problems in Medicine and Biomedical and Behavioral Research. 1982. Washington, DC: President’s Commission.
President’s Council on Bioethics. 2002. Human Cloning and Human Dignity: An Ethical Inquiry . Washington, DC: President’s Council on Bioethics.
President’s Council on Bioethics. 2003. Beyond Therapy: Biotechnology and the Pursuit of Happiness . New York, NY: Harper Perennial.
Proctor, R. 1988. Racial Hygiene: Medicine Under the Nazis . Cambridge, MA: Harvard University Press.
Public Health Emergency. 2015. Biosafety Levels. Available at: https://www.phe.gov/s3/BioriskManagement/biosafety/Pages/Biosafety-Levels.aspx . Accessed 19 Jan 2021.
Ragnedda, M., and G.W. Muschert (eds.). 2015. The Digital Divide . New York, NY: Routledge.
Rana, F.R., and K.R.Samples. 2019. Humans 2.0: Scientific, Philosophical, and Theological Perspectives on Transhumanism . Covina, CA: Reasons to Believe.
Rasko, J.E., G.M. O’Sullivan, and R.A. Ankeny (eds.). 2006. The Ethics of Inheritable Genetic Modification: a Dividing Line? Cambridge, UK: Cambridge University Press.
Rawls, J. 2005. Political Liberalism , 2nd ed. New York: Columbia University Press.
Regan, T. 1983. The Case for Animal Rights . Berkeley, CA: University of California Press.
Reiss, M.J., and R. Straughan. 1996. Improving Nature? The Science and Ethics of Genetic Engineering . Cambridge, UK: Cambridge University Press.
Resnik, D.B. 1993. Debunking the Slippery Slope Argument Against Human Germ-Line Gene Therapy. Journal of Medicine and Philosophy 19 (1): 23–40.
Resnik, D.B. 2000a. The Moral Significance of the Therapy/Enhancement Distinction in Human Genetics. Cambridge Quarterly of Healthcare Ethics 9 (3): 365–377.
Resnik, D.B. 2000b. Of Maize and Men: Reproductive Control and the Threat to Genetic Diversity. Journal of Medicine and Philosophy 25 (4): 451–467.
Resnik, D.B. 2001. DNA Patents and Human Dignity. Journal of Law, Medicine, and Ethics 29 (2): 153–165.
Resnik, D.B. 2007. Embryonic Stem Cell Patents and Human Dignity. Health Care Analysis 15 (3): 211–222.
Resnik, D.B. 2011. Ethical Issues Concerning Transgenic Animals in Biomedical Research. In The Ethics of Animal Research: Exploring the Controversy , ed. J. Garrett, 169–179. Cambridge, MA: MIT Press.
Resnik, D.B. 2012. Environmental Health Ethics . Cambridge, UK: Cambridge University Press.
Resnik, D.B. 2015a. Retracting Inconclusive Research: Lessons from the Séralini GM Maize Feeding Study. Journal of Agricultural and Environmental Ethics 28 (4): 621–633.
Resnik, D.B. 2015b. Food and Beverage Policies and Public Health Ethics. Health Care Analysis 23 (2): 122–133.
Resnik, D.B. 2018a. The Ethics of Research with Human Subjects: Protecting People, Advancing Science, Promoting Trust . Cham, Switzerland: Springer.
Resnik, D.B. 2018b. Ethics of Community Engagement in Field Trials of Genetically Modified Mosquitoes. Developing World Bioethics 18 (2): 135–143.
Resnik, D.B. 2019a. Two Unresolved Issues in Community Engagement for Field Trials of Genetically Modified Mosquitoes. Pathogens and Global Health 113 (5): 238–245.
Resnik, D.B. 2019b. How Should Engineered Nanomaterials Be Regulated for Public and Environmental Health? AMA Journal of Ethics 21 (4): E363–369.
Resnik, D.B., and D. Vorhaus. 2006. Genetic Modification and Genetic Determinism. Philosophy, Ethics, and Humanities in Medicine 1: 9.
Resnik, D.B., H. Steinkraus, and P. Langer. 1999. Human Germ-Line Gene Therapy: Scientific, Moral and Political Issues . Georgetown, TX: RG Landes.
Resnik, D.B., and P. Langer. 2001. Human Germline Gene Therapy Reconsidered. Human Gene Therapy 12 (11): 1449–1458.
Ridley, M. 2000. Genome: The Autobiography of a Species in 23 Chapters . New York, NY: Harper Collins.
Rifkin, J. 1983. Algeny . New York, NY: Viking Press.
Rigby, B. 2017. Growth Hormones in Meat: Myths and Reality. Climbing Nutrition , February 24. Available at: https://www.climbingnutrition.com/diet/growth-hormones-meat-myths-reality/ . Accessed 19 Jan 2021.
Robert, J.S., and F. Baylis. 2003. Crossing Species Boundaries. American Journal of Bioethics 3 (3): 1–13.
Robertson, J.A. 1994. Children of Choice: Freedom and the New Reproductive Technologies . Princeton, NJ: Princeton University Press.
Rollin, B. 1995. The Frankenstein Syndrome: Ethical and Social Issues in the Genetic Engineering of Animals . Cambridge, UK: Cambridge University Press.
Russell, W., and R. Birch. 1959. Principles of Humane Animal Experimentation . Springfield, IL: Charles C. Thomas.
Sandel, M.J. 2009. The Case Against Perfection: Ethics in the Age of Genetic Engineering . Cambridge, MA: Harvard University Press.
Savulescu, J. 2002. Education and Debate: Deaf Lesbians, “Designer Disability,” and the Future of Medicine. British Medical Journal 325 (7367): 771–773.
Schaffner, K.F. 1993. Discovery and Explanation in Biology and Medicine . Chicago, IL: University of Chicago Press.
Schuppli, C., D. Fraser, and M. McDonald. 2004. Expanding the Three Rs to Meet New Challenges in Humane Animal Experimentation. Alternative to Laboratory Animals 32: 515–532.
Science and Environmental Health Network. 1998. Wingspread Statement on the Precautionary Principle. Available at: http://www.who.int/ifcs/documents/forums/forum5/wingspread.doc . Accessed: 19 Jan 2021.
Sears, M.K., R.L. Hellmich, D.E. Stanley-Horn, K.S. Oberhauser, J.M. Pleasants, H.R. Mattila, B.D. Siegfried, and G.P. Dively. 2001. Impact of Bt Corn Pollen on Monarch Butterfly Populations: A Risk Assessment. Proceedings of the National Academy of Sciences of the United States of America 98 (21): 11937–11942.
Séralini, G.E., E. Clair, R. Mesnage, S. Gress, N. Defarge, M. Malatesta, D. Hennequin, and J.S. de Vendômois. 2012. Long Term Toxicity of a Roundup Herbicide and a Roundup-Tolerant Genetically Modified Maize. Food and Chemical Toxicology 50 (11): 4221–4231. Retraction in: Food and Chemical Toxicology 63: 244.
Shamoo, A.E., and D.B. Resnik. 2015. Responsible Conduct of Research , 3rd ed. New York, NY: Oxford University Press.
Shendure, J., G.M. Findlay, and M.W. Snyder. 2019. Genomic Medicine–Progress, Pitfalls, and Promise. Cell 177 (1): 45–57.
Simmons, D. 2008. The Use of Animal Models in Studying Genetic Disease: Transgenesis and Induced Mutation. Nature Education 1 (1): 70.
Singer, P. 2009. Animal Liberation , reissue ed. New York, NY: Harper Perennial.
Spinello, R.A. 2016. Cyberethics: Morality and Law in Cyberspace , 6th ed. Boston: MA: Jones and Bartlett.
Stöppler, M.C. 2019. Genetic Diseases. Medicine.net . Available at: https://www.medicinenet.com/genetic_disease/article.htm . Accessed 19 Jan 2021.
Streiffer, R. 2005. At the Edge of Humanity: Human Stem Cells, Chimeras, and Moral Status. Kennedy Institute of Ethics Journal 15 (4): 347–370.
Szasz, T. 1961. The Myth of Mental Illness . New York, NY: Harper.
Tait, J. 2001. More Faust Than Frankenstein: The European Debate About the Precautionary Principle and Risk Regulation for Genetically Modified Crops. Journal of Risk Research 4 (2): 175–189.
The Business Research Company. 2019. Global Biologic Market Size and Segments, March 20. Available at: https://www.globenewswire.com/news-release/2019/03/27/1774114/0/en/Global-Biologics-Market-Size-and-Segments.html . Accessed 20 Jan 2021.
Thompson, P.B. 1993. Genetically Modified Animals: Ethical Issues. Journal of Animal Science 71 (Suppl. 3): 51–56.
Tratar, U.L., S. Horvat, and M. Cemazar. 2018. Transgenic Mouse Models in Cancer Research. Frontiers in Oncology 8 (July 20): 268.
Treatment Solutions. 2017. Are GMO Bacteria Safe for Wastewater Treatment? Available at: https://aosts.com/gmo-bacteria-safe-wastewater-treatment/ . Accessed 26 Feb 2020.
United Nations Educational, Scientific, and Cultural Organization. 2020. UNESCO Panel of Experts Calls for Ban on “Editing” of Human DNA to Avoid Unethical Tampering with Hereditary Traits. Available at: https://en.unesco.org/news/unesco-panel-experts-calls-ban-editing-human-dna-avoid-unethical-tampering-hereditary-traits . Accessed 20 Jan 2021.
United States Department of Agriculture. 2018. Establishing the National Bioengineered Food Disclosure Standard. Available at: https://www.usda.gov/media/press-releases/2018/12/20/establishing-national-bioengineered-food-disclosure-standard . Accessed 20 Jan 2021.
United States Department of Agriculture. 2020. Biotechnology Frequently Asked Questions. Available at: https://www.usda.gov/topics/biotechnology/biotechnology-frequently-asked-questions-faqs . Accessed 20 Jan 2021.
United States Department of Homeland Security. 2008. National Bio and Agro-Defense Facility Final Environmental Impact Statement, Appendix B . Washington, DC: US Department of Homeland Security.
Urry, L.A., M.L. Cain, S.A. Wasserman, P.V. Minorsky, and J.B. Reece. 2016. Campbell Biology , 11th ed. New York, NY: Pearson.
Walters, L., and J.G. Palmer. 1997. The Ethics of Human Gene Therapy . New York, NY: Oxford University Press.
Walton, D. 2017. The Slippery Slope Argument in the Ethical Debate on Genetic Engineering of Humans. Science and Engineering Ethics 23 (6): 1507–1528.
Wang, H., and H. Yang. 2019. Gene-Edited Babies: What Went Wrong and What Could Go Wrong. PLoS Biology 17 (4): e3000224.
Wareham, C., and C. Nardini. 2015. Policy on Synthetic Biology: Deliberation, Probability, and the Precautionary Paradox. Bioethics 29 (2): 118–125.
Warwick, S.I., H.J. Beckie, and L.M. Hall. 2009. Gene Flow, Invasiveness, and Ecological Impact of Genetically Modified Crops. Annals of the New York Academy of Sciences 1168 (1): 72–99.
WebMD. 2020. What Are Normal Blood Sugar Levels? Available at: https://www.webmd.com/diabetes/qa/what-are-normal-blood-sugar-levels . Accessed 20 Jan 2021.
Werth, J., L. Boucher, D. Thornby, S. Walker, and G. Charles. 2013. Changes in Weed Species Since the Introduction of Glyphosate-Resistant Cotton. Crop and Pasture Science 64 (8): 791–798.
Whiteside, K. 2006. Precautionary Politics: Principle and Practice in Confronting Environmental Risk . Cambridge, MA: MIT Press.
Whitlock J. 2019. Gender Reassignment Surgery. Very Well Health, November 8. Available at: https://www.verywellhealth.com/sex-reassignment-surgery-srs-3157235 . Accessed 20 Jan 2021.
Wolinetz, C.D., and F.S. Collins. 2019. NIH Pro Germline Editing Moratorium. Nature 567: 175.
World Health Organization. 2020a. Malaria. Available at: https://www.who.int/malaria/en/ . Accessed 20 Jan 2021.
World Health Organization. 2020b. Dengue and Severe Dengue. Available at: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue . Accessed 20 Jan 2021.
World Health Organization. 2020c. Determinants of Health. Available at: https://www.who.int/hia/evidence/doh/en/ . Accessed 20 Jan 2021.
Yabroff, K.R., J. Lund, D. Kepka, and A. Mariotto. 2011. Economic Burden of Cancer in the United States: Estimates, Projections, and Future Research. Cancer Epidemiology, Biomarkers and Prevention 20 (10): 2006–2014.
Yourgenome.org. 2020. What Are Single Gene Disorders? Available at: https://www.yourgenome.org/facts/what-are-single-gene-disorders . Accessed 20 Jan 2021.
Zhang, C., R. Wohlhueter, and H. Zhang. 2016. Genetically Modified Foods: A Critical Review of Their Promise and Problems. Food Science and Human Wellness 5 (3): 116–123.
Zhang, X.H., L.Y. Tee, X.G. Wang, Q.S. Huang, and S.H. Yang. 2015. Off-Target Effects in CRISPR/Cas9-Mediated Genome Engineering. Molecular Therapy—Nucleic Acids 4: e264.
Download references
Author information
Authors and affiliations.
National Institutes of Health, National Institute of Environmental Health Sciences, Research Triangle Park, NC, USA
David B. Resnik
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to David B. Resnik .
Rights and permissions
Reprints and permissions
Copyright information
© 2021 This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply
About this chapter
Resnik, D.B. (2021). Genetic Engineering. In: Precautionary Reasoning in Environmental and Public Health Policy. The International Library of Bioethics, vol 86. Springer, Cham. https://doi.org/10.1007/978-3-030-70791-0_7
Download citation
DOI : https://doi.org/10.1007/978-3-030-70791-0_7
Published : 28 March 2021
Publisher Name : Springer, Cham
Print ISBN : 978-3-030-70790-3
Online ISBN : 978-3-030-70791-0
eBook Packages : Religion and Philosophy Philosophy and Religion (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Help | Advanced Search
Computer Science > Artificial Intelligence
Title: crispr-gpt: an llm agent for automated design of gene-editing experiments.
Abstract: The introduction of genome engineering technology has transformed biomedical research, making it possible to make precise changes to genetic information. However, creating an efficient gene-editing system requires a deep understanding of CRISPR technology, and the complex experimental systems under investigation. While Large Language Models (LLMs) have shown promise in various tasks, they often lack specific knowledge and struggle to accurately solve biological design problems. In this work, we introduce CRISPR-GPT, an LLM agent augmented with domain knowledge and external tools to automate and enhance the design process of CRISPR-based gene-editing experiments. CRISPR-GPT leverages the reasoning ability of LLMs to facilitate the process of selecting CRISPR systems, designing guide RNAs, recommending cellular delivery methods, drafting protocols, and designing validation experiments to confirm editing outcomes. We showcase the potential of CRISPR-GPT for assisting non-expert researchers with gene-editing experiments from scratch and validate the agent's effectiveness in a real-world use case. Furthermore, we explore the ethical and regulatory considerations associated with automated gene-editing design, highlighting the need for responsible and transparent use of these tools. Our work aims to bridge the gap between beginner biological researchers and CRISPR genome engineering techniques, and demonstrate the potential of LLM agents in facilitating complex biological discovery tasks.
Submission history
Access paper:.
- HTML (experimental)
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .
An official website of the United States government
The .gov means it's official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you're on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- Browse Titles
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Board on Agriculture and Natural Resources; Committee on Genetically Engineered Crops: Past Experience and Future Prospects. Genetically Engineered Crops: Experiences and Prospects. Washington (DC): National Academies Press (US); 2016 May 17.

Genetically Engineered Crops: Experiences and Prospects.
- Hardcopy Version at National Academies Press
5 Human Health Effects of Genetically Engineered Crops
In this chapter, the committee examines the evidence that substantiates or negates specific hypotheses and claims about the health risks and benefits associated with foods derived from genetically engineered (GE) crops. There are many reviews and official statements about the safety of foods from GE crops (for example, see Box 5-1 ), but to conduct a fresh examination of the evidence, the committee read through a large number of articles with original data so that the rigor of the evidence could be assessed.
Sample of Statements About the Safety of Genetically Engineered Crops and Food Derived from Genetically Engineered Crops.
Some of the evidence available to the committee came from documents that were part of the U.S. regulatory process for GE crops conducted by the U.S. Environmental Protection Agency (EPA), the U.S. Department of Agriculture (USDA), and the U.S. Food and Drug Administration (FDA). Other evidence came from studies published by regulatory agencies in other countries or by companies, nongovernmental organizations (NGOs), and academic institutions. The committee also sought evidence from the public and from the speakers at its public meetings and webinars. 1
The committee thinks that it is important to make clear that there are limits to what can be known about the health effects of any food, whether non-GE or GE. If the question asked is “Is it likely that eating this food today will make me sick tomorrow?” researchers have methods of getting quantitative answers. However, if the question is “Is it likely that eating this food for many years will make me live one or a few years less than if I never eat it?” the answer will be much less definitive. Researchers can provide probabilistic predictions that are based on the available information about the chemical composition of the food, epidemiological data, genetic variability across populations, and studies conducted with animals, but absolute answers are rarely available. Furthermore, most current toxicity studies are based on testing individual chemicals rather than chemical mixtures or whole foods because testing of the diverse mixtures of chemicals experienced by humans is so challenging ( Feron and Groten, 2002 ; NRC, 2007 ; Boobis et al., 2008 ; Hernández et al., 2013 ).
With regard to the issue of uncertainty, it is useful to note that many of the favorable institutional statements about safety of foods from GE crops in Box 5-1 contain caveats, for example: “no overt consequences,” “no effects on human health have been shown,” “are not per se more risky,” and “are not likely to present risks for human health.” Scientific research can answer many questions, but absolute safety of eating specific foods and the safety of other human activities is uncertain.
The review in this chapter begins with an examination of what is known about the safety of foods from non-GE plants and how they are used as counterparts to those from GE crops in food-safety testing. U.S. food-safety regulatory testing for GE products and GE food-safety studies conducted outside the agency structure are then assessed. A variety of hypothesized health risks posed by and benefits of GE crops are examined, and the chapter concludes with a short discussion of the challenges that society will face in assessing the safety of GE foods that are likely to be developed with emerging genetic-engineering technologies.
- COMPARING GENETICALLY ENGINEERED CROPS WITH THEIR COUNTERPARTS
An oft-cited risk of GE crops is that the genetic-engineering process could cause “unnatural” changes in a plant's own naturally occurring proteins or metabolic pathways and result in the unexpected production of toxins or allergens in food ( Fagan et al., 2014 ). Because analysis of risks of the product of the introduced transgene itself is required during risk assessment, the argument for unpredicted toxic chemicals in GE foods is based on the assumption that a plant's endogenous metabolism is more likely to be disrupted through introduction of new genetic elements via genetic engineering than via conventional breeding or normal environmental stresses on the plant. The review below begins by discussing natural chemical constituents of plants in the context of food safety to provide a background on what the natural plant toxins are and how they vary in non-GE plants. The review then goes on to explain the premise used by regulatory agencies to compare GE crops with their non-GE counterparts.
Endogenous Toxins in Plants
Most chemicals of primary metabolism (for example, those involved in the formation of carbohydrates, proteins, fats, and nucleic acids) are shared between animals and plants and are therefore unlikely to be toxic. Perceived risks associated with alterations of plant compounds arise mainly from alterations of plant-specific molecules, popularly known as plant natural products and technically named secondary metabolites. Collectively, there are more than 200,000 secondary metabolites in the plant kingdom ( Springob and Kutchan, 2009 ). Crop species vary in the number of secondary metabolites that they produce. For example, potato ( Solanum tuberosum ) is known for its high diversity of secondary metabolites and can have more than 20 sesquiterpenes (a single group of related compounds), some of which are thought to confer resistance to diseases ( Kuc, 1982 ). The concentrations of these secondary metabolites within some tissues in a particular plant species may vary from high—for example, chlorogenic acids alone make up about 12 percent of the dry matter of green coffee beans ( Ferruzzi, 2010 )—to trace amounts (many minor saponins in legumes) and may be associated with particular stages of plant development (some found only in seeds) or may increase in response to external stimuli, such as pathogen or herbivore attack, drought, or altered mineral nutrition ( Small, 1996 ; Pecetti et al., 2006 ; Nakabayashi et al., 2014 ). Many secondary metabolites function as protective agents, for example, by absorbing damaging ultraviolet radiation ( Treutter, 2006 ), acting as antinutrients ( Small, 1996 ), or killing or halting insects and pathogens that damage crops ( Dixon, 2001 ). Plant secondary metabolites that protect against pathogen attack have been classified as either phytoanticipins (if they exist in a preformed state in a plant before exposure to a pathogen) or phytoalexins (if their synthesis and accumulation are triggered by pathogen attack) ( VanEtten et al., 1994 ; Ahuja et al., 2012 ). The toxic properties of some plant compounds are understood, but most of these compounds have not been studied. Some secondary metabolites and other products (such as proteins and peptides) in commonly consumed plant materials can be toxic to humans when consumed in large amounts, and examples are listed below:
- Steroidal glycoalkaloids in green potato skin, which can cause gastrointestinal discomfort or, more severely, vomiting and diarrhea.
- Oxalic acid in rhubarb, which can cause symptoms ranging from breathing difficulty to coma.
- Gossypol in cottonseed oil and cake, which can cause respiratory distress, anorexia, impairment of reproductive systems, and interference with immune function in monogastric animals.
- Nonprotein amino acid canavanine in alfalfa sprouts, which can be neurotoxic.
- Hemolytic triterpene saponins in many legume species, which can increase the permeability of red blood cell membranes.
- Cyanogenic glycosides in almonds and cassava, which can cause cyanide poisoning.
- Phototoxic psoralens in celery, which are activated by ultraviolet sunlight and can cause dermatitis and sunburn and increase the risk of skin cancer.
Friedman (2006) provided information that demonstrated that some glycoalkaloids in potato can have both harmful and beneficial effects. The Food and Agriculture Organization has recognized that foods often contain naturally occurring food toxins or antinutrients but that at naturally occurring concentrations in common diets they can be safely consumed by humans ( Novak and Haslberger, 2000 ; OECD, 2000 ). The health risks associated with some secondary metabolites in common foodstuffs are generally well understood, and the plants are either harvested at times when the concentrations of the compounds are low, the tissues with the highest concentrations of toxins are discarded, or, as in the case of cassava ( Manihot esculenta ), the food is prepared with special methods to remove the toxic compounds. In other cases, food preparation may be the cause of the presence of a toxic compound (for example, the formation of the probable carcinogen acrylamide when potatoes are fried at high temperatures or when bread is toasted). Plant breeders have generally screened for toxins that are typical of the plant group from which a crop was domesticated and have excluded plants that have high concentrations of the compounds.
Unintended changes in the concentrations of secondary metabolites can result from conventional breeding ( Sinden and Webb, 1972 ; Hellenas et al., 1995 ). In some cases, conventionally bred varieties have been taken off the market because of unusually high concentrations of a toxic compound, as in the case of a Swedish potato variety that was banned from sale in the 1980s because of high concentrations of glycoalkaloids ( Hellenas et al., 1995 ).
Rather than being a cause of worry, many secondary metabolites are perceived as having potential health benefits for humans and are consumed in increasingly large quantities ( Murthy et al., 2015 ). Examples include the isoflavone phytoestrogens found in a number of leguminous plants, such as soybean ( Glycine max ) and clover ( Trifolium spp.), which have been ascribed beneficial activities, including chemoprevention of breast and prostate cancers, cardiovascular disease, and post-menopausal ailments ( Dixon, 2004 ; Patisaul and Jefferson, 2010 ). Also, various perceived antioxidants, such as anthocyanins ( Martin et al., 2013 ), and some saponins may have anticancer activity ( Joshi et al., 2002 ). There is, however, disagreement as to whether many of the compounds are beneficial or toxic at the concentrations consumed in herbal medicines or dietary supplements (see, for example, Patisaul and Jefferson, 2010 ).
FINDING: Crop plants naturally produce an array of chemicals that protect against herbivores and pathogens. Some of these chemicals can be toxic to humans when consumed in large amounts.
Substantial Equivalence of Genetically Engineered and Non–Genetically Engineered Crops
A major question addressed in the regulation of GE crops is whether the concentrations of the toxic secondary metabolites are affected by genetic engineering. In addition to the plant toxins, nutrients, introduced genes, and proteins and their metabolic products in specific GE crops are assessed with a comparative approach that is generally encompassed by the concept of substantial equivalence.
The concept of substantial equivalence has a long history in safety testing of GE foods. The term and concept were “borrowed from the [U.S. FDA's] definition of a class of new medical devices that do not differ materially from their predecessors, and thus, do not raise new regulatory concerns” ( Miller, 1999:1042 ). No simple definition of substantial equivalence is found in the regulatory literature on GE foods. In 1993, the Organisation for Economic Co-operation and Development (OECD) explained that the “concept of substantial equivalence embodies the idea that existing organisms used as food, or as a source of food, can be used as the basis for comparison when assessing the safety of human consumption of a food or food component that has been modified or is new” ( OECD, 1993:14 ).
The Codex Alimentarius Commission's Guideline for the Conduct of Food Safety Assessment of Foods Derived from Recombinant-DNA Plants is careful to state that “the concept of substantial equivalence is a key step in the safety assessment process. However, it is not a safety assessment in itself; rather it represents the starting point which is used to structure the safety assessment of a new food relative to its conventional counterpart” ( CAC, 2003:2 ). The Codex guideline also makes clear that a safety assessment of a new food based on the concept of substantial equivalence “does not imply absolute safety of the new product; rather, it focuses on assessing the safety of any identified differences so that the safety of the new product can be considered relative to its conventional counterpart” ( CAC, 2003:2 ). The OECD (2006) came to a similar conclusion. Conflict among stakeholders often comes into play during the determination of what constitutes evidence of differences from substantial equivalence sufficient to justify a detailed food-safety assessment.
The Codex Alimentarius Commission concluded that the concept of substantial equivalence “aids in the identification of potential safety and nutritional issues and is considered the most appropriate strategy to date for safety assessment of foods derived from recombinant-DNA plants” ( CAC, 2003:2 ). Despite some criticism of the substantial-equivalence concept itself (for example, Millstone et al., 1999 ) and operational problems (for example, Novak and Haslberger, 2000 ), it remains the cornerstone for GE food-safety assessment by regulatory agencies. The present committee examined its use in practice and its empirical limitations.
The precautionary principle, which is described in more detail in Chapter 9 (see Box 9-2 ) is a deliberative principle related to the regulation of health, safety, and the environment and typically involves taking measures to avoid uncertain risks. The precautionary principle has been interpreted in a number of ways, but it is not necessarily incompatible with use of the concept of substantial equivalence. In the case of foods, including GE foods, it can be reasonably argued that even a small adverse chronic effect should be guarded against, given that billions of people could be consuming the foods. However, the degree of precaution taken in the face of uncertainty is a policy decision that varies among countries and according to the specific uncertainty being considered. For example, many European countries and the European Union (EU) as a whole generally take a more precautionary approach with GE foods and climate change whereas the United States has historically taken a more precautionary approach with tobacco products and ozone depletion ( Wiener et al., 2011 ). The reader is directed to Chapter 9 for further discussion of how different regulatory frameworks address uncertainty in the safety of GE foods.
Some differences between a GE food and its non-GE counterpart are intentional and identifiable (for example, the presence of a Bt toxin in maize kernels) or are due to practices directly associated with the use of the GE crops (for example, increased use of glyphosate). Some of the risks posed by the intended changes can be anticipated on the basis of the physiological and biochemical characteristics of the engineered change. There are often established protocols for assessing such risks, especially when a change involves exposure to a known toxin. However, other risks have been hypothesized for GE crops because previous uses of a trait (for example, Bt as an insecticidal spray) did not have consumption of the GE plant products as the route of exposure. New routes of exposure could result in unanticipated effects.
In contrast with such intended differences, some potential differences between GE crops and their non-GE counterparts are unintentional and can be difficult to anticipate and discern ( NRC, 2004 ). Two general sources of unintended differences could affect food safety:
- Unintended effects of the targeted genetic changes on other characteristics of the food (for example, the intended presence of or increase in one compound in plant cells could result in changes in plant metabolism that affect the abundance of other compounds).
- Unintended effects associated with the genetic-engineering process (for example, DNA changes resulting from plant tissue culture).
Much of the concern voiced by some citizens and scientists about the safety of GE foods is focused on potential risks posed by unintended differences. Some of the biochemical and animal testing done by or for government agencies is aimed at assessing the toxicity of such unintended differences, but what is adequate and appropriate testing for assessing specific toxicities is often difficult to determine. In some cases, the unintended effects are somewhat predictable or can be determined; in such cases, tests can be designed. In other cases, the change or risk could be something that has not even been considered, so the only effective testing is of the whole food itself. As discussed in Chapter 6 , there is a tradeoff between costs of such testing and societal benefits of reduction in risks.
The approach of comparing new varieties to existing varieties is just as applicable to crops developed by conventional plant breeding as it is to GE crops (see Chapter 9 ). The discussion above on endogenous toxins (see section “Endogenous Toxins in Plants”) shows that such crops pose some risks. The 2000 National Research Council report Genetically Modified Pest-Protected Plants found that “there appears to be no strict dichotomy between the risks to health and the environment that might be posed by conventional and transgenic pest-protected plants” ( NRC, 2000:4 ). Similarly, the 2004 National Research Council report Safety of Genetically Engineered Foods found that all forms of conventional breeding and genetic engineering may have unintended effects and that the probability of unintended effects of genetic engineering falls within the range of unintended effects of diverse conventional-breeding methods. The 2002 National Research Council report Environmental Effects of Transgenic Plants found that “the transgenic process presents no new categories of risk compared to conventional methods of crop improvement but that specific traits introduced by both approaches can pose unique risks” ( NRC, 2002:5 ). That finding remains valid with respect to food safety and supports the conclusion that novel varieties derived from conventional-breeding methods could be assessed with the substantial-equivalence concept.
FINDING: The concept of substantial equivalence can aid in the identification of potential safety and nutritional issues related to intended and unintended changes in GE crops and conventionally bred crops.
FINDING: Conventional breeding and genetic engineering can cause unintended changes in the presence and concentrations of secondary metabolites.
- OVERVIEW OF U.S. REGULATORY TESTING OF RISKS TO HUMAN HEALTH
Although the committee agrees that crops developed through conventional breeding could result in food-safety risks, its statement of task focuses on GE crops. Furthermore, there have been claims and counterclaims about the relative safety of GE crops and their associated technologies compared with conventionally bred crops and their associated technologies. Therefore, the remainder of this chapter examines possible risks and benefits associated with GE crops and assesses the methods used to test them in and beyond government regulatory systems.
Whether testing is done for regulatory purposes or beyond the regulatory realm, it typically involves three categories of testing: acute or chronic animal toxicity tests, chemical compositional analysis, and allergenicity testing or prediction. Although the precision, transparency, specific procedures, and interpretation of results vary among countries, criticisms about the adequacy of testing are not so much country-specific as they are method- and category-specific. For example, there may be arguments about whether a 90-day whole-food animal test is more appropriate than a 28-day test, but the bigger issue is about whether whole-food testing is appropriate. The committee uses a description of the U.S. testing methods as an example, but it mostly examines the criticism of food-safety testing more broadly.
The structure of the U.S. regulatory process for GE crops based on the Coordinated Framework for the Regulation of Biotechnology is briefly reviewed in Chapter 3 and is examined in more detail in Chapter 9 . The focus in this chapter is on the testing itself. The present section provides insight into U.S. procedures by describing the risk-testing methods used for two examples of traits in commercialized GE crops: Bt toxins and crop resistance to the herbicides glyphosate and 2,4-D.
Regulatory Testing of Crops Containing Bt Toxins
EPA considers plant-produced Bt toxins to be “plant-incorporated protectants,” a class of products generally defined as “a pesticidal substance that is intended to be produced and used in a living plant, or in the produce thereof, and the genetic material necessary for the production of that pesticidal substance” (40 CFR §174.3). EPA specifically exempts plant-incorporated protectants whose genetic material codes for a pesticidal substance that is derived from plants that are sexually compatible. Bt toxin genes are not exempted because they come from bacteria (see Chapter 9 for regulatory details).
For Bt toxins produced by GE crops, EPA took into consideration that there was already toxicity testing of Bt toxins in microbial pesticides and that the toxins were proteins that, if toxic, typically show almost immediate toxicity at low doses ( EPA, 2001a ; also see Box 5-2 ). The pesticidal safety tests mostly involved acute toxicity testing in mice and digestibility studies in simulated gastric fluids because one characteristic of food allergens is that they are not rapidly digested by such fluids.
Cry1F Testing by the U.S. Environmental Protection Agency.
Box 5-2 provides a verbatim example of the procedures used for testing as reported in EPA fact sheets for the Cry1F Bt toxin so that readers can see what is involved in the testing. The actual research is not typically done by EPA itself. The registrant is usually responsible for testing. Results of the tests of Cry1F show no clinical signs of any toxicity even when Cry1F protein was fed at 576 mg/kg body weight, which would be the equivalent of about ¼ cup of pure Cry1F for a 90.7-kilogram (200-pound) person. Another part of the testing described in Box 5-2 is allergenicity testing. Concerns about the EPA testing methods are discussed in sections below on each category of testing.
Regulatory Testing of Crops Resistant to Glyphosate and 2,4-D and of the New Uses of the Herbicides Themselves
The regulatory actions taken for herbicide-resistant (HR) crops are different from regulatory actions taken to assess Bt crops. With Bt crops, regulatory actions are related to the crop itself. With HR crops, there are regulatory processes for the plant itself and separate regulatory processes for the new kind of exposure that can accompany spraying of a herbicide on a crop or on a growth stage of a crop that has never been sprayed prior to availability of the GE variety.
EPA governs the registration of herbicides such as glyphosate and 2,4-D. Both glyphosate and 2,4-D were registered well before the commercialization of GE crops. However, EPA has authority to re-examine herbicides if their uses or exposure characteristics change.
A good example of such re-examination was the 2014 EPA registration of the Dow AgroSciences Enlist Duo® herbicide, which contains both glyphosate and 2,4-D for use on GE maize ( Zea mays ) and soybean. Because the glyphosate component of Enlist Duo had already been in use on GE maize and soybean, EPA did not conduct further testing of glyphosate alone. However, 2,4-D was registered previously only for applications to maize up to 20 centimeters tall and for preplant applications to soybean. The proposed use of 2,4-D on GE crops was expected to change use patterns and exposure and thereby triggered a safety assessment of the new use 2,4-D. Additionally, EPA compared the toxicity of the formulation that contained both herbicides to the toxicity of the individual herbicides and concluded the formulation did not show greater toxicity or risk compared to either herbicide alone.
In the human health risk assessment portion of the EPA Enlist Duo registration document, the following tests and results with 2,4-D were considered ( EPA, 2014a ):
- An acute dietary test in rats that found a lowest observed-adverse-effect level (LOAEL) of 225 mg/kg (about 1 ounce per 200-pound person).
- A chronic-dietary-endpoint, extended one-generation reproduction toxicity study in rats that found a LOAEL of 46.7 mg/kg-day in females and higher in males.
- Inhalation tests involving data from a 28-day inhalation toxicity study in rats that found a LOAEL of 0.05 mg/L-day.
- Dermal tests that showed no dermal or systemic toxicity after repeated applications to rabbits at the limit dose of 1000 mg/kg-day.
- Reviews of epidemiological and animal studies, which did not support a linkage between human cancer and 2,4-D exposure.
Analysis of the results of those tests and agronomic and environmental assessments resulted in the product's registration.
EPA received over 400,000 comments in response to the initial proposal to register the new use of 2,4-D. Some of the concerns submitted to EPA were similar to ones some members of the public expressed in public comments to the committee, including questions about whether EPA had considered toxicity of only the active ingredient or of the formulated herbicide and whether it had tested for synergistic effects of 2,4-D and glyphosate. EPA (2014b:7) responded that
acute oral, dermal, and inhalation data, skin and eye irritation data, and skin sensitization data are available for the 2,4-D choline salt and glyphosate formulation for comparison with the 2,4-D parent compound and glyphosate parent compound data, and these test results show similar profiles. The mixture does not show a greater toxicity compared to either parent compound alone. Although no longer duration toxicity studies are available, toxic effects would not be expected as the maximum allowed 2,4-D exposure is at least 100-fold below levels where toxicity to individual chemicals might occur, and exposure to people is far below even that level.
The committee did not have access to the actual data from the registrant. 2
EPA does not regulate the commercialization of the GE herbicide-resistant crops themselves. That is the role of USDA's Animal and Plant Health Inspection Service (APHIS) under the Plant Protection Act. Under its statutory authority, APHIS controls and prevents the spread of plant pests (see Box 3-5 ). On the basis of a plant-pest risk assessment (USDA–APHIS, 2014a), APHIS concluded that Enlist™ GE herbicide-resistant maize and soybean engineered to be treated with the Enlist Duo herbicide (containing glyphosate and 2,4-D) were unlikely to become plant pests and deregulated them on September 18, 2014 (USDA–APHIS, 2014b). In its document on the decision to deregulate Enlist GE herbicide-resistant maize and soybean (USDA–APHIS, 2014a:ii), APHIS states a general policy that “if APHIS concludes that the GE organism is unlikely to pose a plant pest risk, APHIS must then issue a regulatory determination of nonregulated status, since the agency does not have regulatory authority to regulate organisms that are not plant pests. When a determination of nonregulated status has been issued, the GE organism may be introduced into the environment without APHIS' regulatory oversight.”
FDA did not identify any safety or regulatory issues in its consultation with Dow AgroSciences on the Enlist maize and soybean varieties ( FDA, 2013 ). FDA also explained the basis of Dow's conclusion that Enlist soybean is not “materially different in composition” from other soybean varieties ( FDA, 2013 ):
Dow reports the results of compositional analysis for 62 components in soybean grain, including crude protein, crude fat, ash, moisture, carbohydrates, [acid detergent fiber] ADF, [neutral detergent fiber] NDF, total dietary fiber (TDF), lectin, phytic acid, raffinose, stachyose, trypsin inhibitor, soy isoflavones (i.e., total daidzein, total genistein, total glycitein), minerals, amino acids, fatty acids, and vitamins. No statistically significant differences in the overall treatment effect and the paired contrasts between each of the DAS-44406-6 soybean treatment groups and the control were observed for 29 of the components. A statistically significant difference in the overall treatment effect was observed for 16 components (crude protein, carbohydrates (by difference), NDF, calcium, potassium, cystine, palmitic acid, oleic acid, linoleic acid, linolenic acid, behenic acid, folic acid, γ-tocopherol, total tocopherol, lectin, and trypsin inhibitor). However, differences between the control and the DAS-44406-6 treatment groups were small in magnitude. Differences between DAS-44406-6 soybean and the control were considered not biologically relevant because the mean values were either within the ranges generated using the reference lines, consistent with the ranges of values in the published literature, or both.
FINDING: U.S. regulatory assessment of GE herbicide-resistant crops is conducted by USDA, and by FDA when the crop can be consumed, while the herbicides are assessed by EPA when there are new potential exposures.
FINDING: When mixtures of herbicides are used on a new GE crop, EPA assesses the interaction of the mixture as compared to the individual herbicidal compounds.
Technical Assessment of Human Health Risks Posed by Genetically Engineered Crops
As explained in Chapter 2 , the development and use of GE crops is governed by more than national and regional regulatory standards. In the cases of the GE crops commercially available in the United States and some other countries in 2015, inputs from many public and private institutions regarding their specific concerns have influenced the type and extent of GE crop food-safety tests conducted by companies, agencies, and other researchers. Many stakeholders have criticized the testing used by U.S. and other national regulatory agencies for lacking rigor (for example, Hilbeck et al., 2015 ). Researchers in companies, NGOs, and universities have sometimes conducted more extensive safety tests than are required by national agencies or have reanalyzed existing data, as described below. All testing as of 2015 fell into three categories: animal testing, compositional analysis, and allergenicity testing and prediction.
Animal Testing
Short-term and long-term rodent testing with compounds and whole foods.
One common criticism of the animal testing conducted by or for regulatory agencies in the United States and elsewhere is related to its short duration (for example, Séralini et al., 2014 ; Smith, 2014 ). Indeed, there is a range in the duration and doses within the test protocols used by regulatory agencies that depends in part on the product. Doses for subchronic and chronic toxicity studies are such that the lowest dose (exposure level), which is many times higher than expected for human exposure, is set to ensure that it does not elicit acute adverse effects that would interfere with examining the potential chronic-effect endpoints. As can be seen in the discussion above, EPA conducted an extended one-generation reproduction toxicity study in male and female rats in its assessment of 2,4-D, and it relied on previous long-term studies for the assessment of cancer risk associated with it. For assessment of the Bt toxin Cry1F and for the bacterially derived proteins in 2,4-D-resistant maize and soybean, company testing submitted to EPA, FDA, and USDA relied on acute toxicity testing. In all the cases above, the experiments were conducted by adding large amounts of a single test chemical to an animal's diet. Tests with high concentrations of a chemical are typical of EPA testing protocols for pesticides.
What is different between GE crop evaluation and that of general agricultural chemicals is the use of “whole food” tests. These tests are aimed at assessing potential hazards due to the combined intentional and unintentional changes that might have been caused by the genetic engineering of the crop. In such tests, it is not possible to use concentrations higher than what is in the crop itself because potential unintended effects are not typically known. Thus, it is impossible for a researcher to know what compounds should be increased in concentration in a fabricated diet, and the only way to assess such unintended effects is to feed the actual GE crop to test animals. For testing GE maize, soybean, and rice ( Oryza sativa ), 3 flour from kernels or seed is added to an animal's diet and constitutes between about 10–60 percent of the diet. The high percentages can be used because the crop products are nutritious for the animal. In the case of whole foods that are not typically part of a rodent's diet, whether GE or non-GE, it is impossible to achieve very high concentrations of the test food because it would cause nutritional imbalance. The whole-food tests done for regulatory agencies are generally conducted for 28 or 90 days with rats, but some researchers have run tests for multiple generations.
The utility of the whole-food tests has been questioned by a number of government agencies and by industry and academic researchers (for example, Ricroch et al., 2014 ), and they are not an automatic part of the regulatory requirements of most countries that have specific GE food-testing requirements ( CAC, 2008 ; Bartholomaeus et al., 2013 ). However, in its 2010 report A Decade of EU-Funded GMO Research (2001–2010) , the European Directorate-General for Research and Innovation concluded that “the data from a well-designed 90-day rodent feeding study, together with data covering the gene insert, the compositional analysis, and the toxicity of the novel gene product, form the optimal basis for a comparative assessment of the safety of [genetically engineered] food and its conventional counterpart in the pre-market situation” ( EC, 2010a:157 ). The European Food Safety Authority (EFSA) developed principles and guidance for establishing protocols for 90-day whole-food studies in rodents at the European Commission's request ( EFSA, 2011b ), and 90-day, whole-food studies were made mandatory by the European Commission ( EC, 2013 ). Most studies reported in the peer-reviewed literature have concluded that there was a lack of adverse effects of biological or toxicological significance (see, for example, Knudsen and Poulsen, 2007 ; MacKenzie et al., 2007 ; He et al., 2008 , 2009 ; Onose et al., 2008 ; Liu et al., 2012 ), even though some of the studies found statistically significant differences between the GE and non-GE comparator in toxicity.
The criticisms of whole-food tests come from two perspectives. One perspective is that whole-food studies cannot provide useful tests of food safety because they are not sensitive enough to detect differences (see, for example, Bartholomaeus et al., 2013 ; Kuiper et al., 2013 ; Ricroch et al., 2013a , 2014 ) and that animal testing is not needed because other types of required testing ensure safety ( Bartholomaeus et al., 2013 ; Ricroch et al., 2014 ). Ricroch et al. (2014) pointed to the costs of the 90-day tests, which they reported as being €250,000 (in 2013 money). The second perspective is that whole-food tests could be useful, but there is concern about their design and conduct or about the parties who conduct them (the companies commercializing the GE crops). That perspective is evident in Séralini et al. (2007) , Domingo and Bordonaba (2011) , Hilbeck et al. (2015) , and Krimsky (2015) . Boxes 5-3 and 5-4 describe some of the specific procedures and practices involved in doing these tests.
Common Procedures for Rodent Toxicity Studies for Safety Evaluation.
Laboratory Practices for Consistency among Studies.
The committee heard from invited speakers ( Entine, 2014 ; Jaffe, 2014 ) and members of the public who provided comments at meetings and it received a number of written public comments highlighting the work of one research group ( Séralini et al., 2012 , 2014 ) that has conducted a number of whole-food studies of GE herbicide-resistant and insect-resistant crops and of direct consumption of glyphosate. Some comments made to the committee pointed to the publications of that research group as evidence that GE crops and foods derived from GE crops were deleterious to human health; other comments questioned the robustness and accuracy of the research. The committee also heard from the lead researcher himself at one of its meetings ( Séralini, 2014 ). Because of the attention garnered by this specific research group, the committee examined the primary research paper from the group and many articles related to it ( Box 5-5 ).
Controversial Results of an Animal Feeding Study of Genetically Engineered Crops and Glyphosate.
A general question that remains for all whole-food studies using animals is, How many animals, tested for how long, are needed to assess food safety when a whole food is tested? That question is related to the question of how large an effect the tested food would have to have on the animal for it to be detected with the experiment. The statistical procedure called power analysis can answer the first question, but the committee did not find such analyses in articles related to GE crop whole-food studies. The EFSA scientific committee ( EFSA, 2011b ) provided general guidance on power analysis. Figure 5-2 , from the EFSA report, shows the relationship between the number of experimental units (cages with two animals) per treatment group and the power of an experiment in standard-deviation units. Standard deviations quantify how much the measurement of a trait or effect varies among animals that have been given the same diet. The report concluded that, if researchers follow OECD Test No. 408 of 10 males and 10 females per treatment ( OECD, 1998a ), a test should be able to detect a difference equal to about 1 standard deviation (with 90-percent confidence) unless the food has a different effect on males and females, in which case, the smallest difference that could be detected would be about 1.5 standard deviations from the experimental mean.
General statistical information on the number of experimental units needed per treatment group as a function of standardized effect size for 80-percent and 90-percent power and 5-percent significance level using a two-sided t test. SOURCE: EFSA (2011b). (more...)
Because the relationship is quite abstract for the nonstatistician, the committee examined the size of the standard deviations in a number of whole-food safety articles. It found that the sizes of the standard deviations compared with the mean value of a measured trait depended heavily on the trait being measured and on the specific research article. For example, in the Hammond et al. (2004) study, the average white blood cell count for the four treatments, each with 9 or 10 female Sprague-Dawley rats, is 6.84 10 3 /µl, and the average standard deviation is 1.89 10 3 /µl. On the basis of rough calculations, this test would have the power to discern statistically whether the GE food caused an increase in white blood cell count of about 35 percent with about 90-percent confidence. If the male white blood cell count effects and standard deviations were similar to those in females, the test could have found about a 25-percent increase.
OECD (1998a) made general recommendations, such as those used in Hammond et al. (2004) , for the number of units (cages with two animals) per treatment. Following these guidelines leads to the assumption that less than a 25-percent change in the white blood cell count was not biologically relevant. The EU Standing Committee on the Food Chain and Animal Health adopted the mandatory use of 90-day whole-food testing of GE crops, and its protocols generally follow OECD guidelines for the testing of chemicals ( EC, 2013 ).
EFSA also published a document ( EFSA, 2011c ) that focused specifically on the questions, What is statistical significance? and What is biological relevance? The accessibly written document makes clear that the two are very different and that it is important to decide how large a difference is biologically relevant before designing an experiment to test a null hypothesis of no difference. The problem in most whole-food animal studies is in determining how large a biological difference is relevant. Most of the statistically significant differences observed in the literature on the animal-testing data were around a 10- to 30-percent change, but the authors do not give detailed explanations of why they conclude that a statistically significant difference is not biologically relevant. A general statement is sometimes made that the difference is within the range for the species, but because the range of values for the species typically come from multiple laboratories, such a statement is not useful unless the laboratories, instrumentation, and health of the animals were known to be comparable.
Clearly, the European Commission relied on both expert judgment and citizen concerns in making its assessment of biological relevance of the effects of GE foods in requiring 90-day testing. It is reasonable to ask what balance of the two is the basis for this judgment. As pointed out by the 2002 National Research Council report, “risk analysis of transgenic plants must continue to fulfill two distinct roles: (1) technical support for regulatory decision making and (2) establishment and maintenance of regulatory legitimacy” ( NRC, 2002:6 ). Fulfilling the two roles can lead to different country-specific and region-specific decisions. This issue is discussed further in Chapter 9 .
One specific criticism of the 90-day whole-food studies revolves around an EU-funded project conducted by Poulsen et al. (2007) in which rice was genetically engineered to produce the kidney bean lectin, agglutinin E-form, which is known to have toxic properties. In a 90-day test, rats were fed diets of 60-percent rice with the lectin gene or 60-percent rice without the lectin gene. The researchers concluded that they did not find any meaningful differences between the two treatments. However, in a treatment in which the diets were spiked with 0.1-percent recombinant lectin (a high dose), biological effects including significant differences in weight of small intestines, stomach, and pancreas and in plasma biochemistry were found. Poulsen et al. included results from a preceding 28-day feeding study and compositional analyses of the rice diets. The criticism involves the question, If a whole-food study with a known toxin does not demonstrate effects, how can the test be considered useful? ( Bartholomaeus et al., 2013 ). If a whole-food study with an animal finds statistically significant effects, there is obviously a need for further safety testing, but when there is a negative result, there is uncertainty as to whether there is an adverse effect on health. In the specific case of lectin gene in rice, one could argue that the statistical power of the whole-food test was insufficient or that, when the toxin is in the structure of the food, it is no longer toxic so the food is safe.
Other Long-Term Studies with Rodents
In addition to the work of Séralini et al. (2012 , 2014 ), there have been other long-term rodent studies, some of which included multiple generations. Magana-Gomez and de la Barca (2009) , Domingo and Bordonaba (2011) , Snell et al. (2012) , and Ricroch et al. (2013b) reviewed the studies. Some found no statistically significant differences, but quite a few found statistically significant differences that the authors generally did not consider biologically relevant, typically without providing data on what was the normal range. In the multigeneration studies, the sire and dam are dosed via the diet before conception, and the parent generation and pups are dosed via the diet throughout the duration of the study to determine multiple generational outcomes, including growth, behavior, and phenotypic characteristics. Some studies have looked at three or four generations. For example, Kiliç and Akay (2008) conducted a three-generation rat study in which 20 percent of the diet was Bt maize or a non- Bt maize that otherwise was genetically similar. All generations of female and male rats were fed the assigned diets, and the third-generation offspring that were fed the diets were sacrificed after 3.5 months for analysis. The authors found statistical differences in kidney and liver weights and long kidney glomerular diameter between the GE and non-GE treatments but considered them not biologically relevant. Similarly, statistically significant differences were observed in amounts of globulin and total protein between the two groups. There was no presentation of standards used for judging what would be a biologically relevant difference or for what the normal range was in the measurements.
The standard deviations in measurements of the traits (that is, effects) of individual animals in a treatment in the long-term studies were similar to those of studies of shorter duration. Therefore, the power of the tests to detect statistically significant differences was in the range of 10–30 percent. The committee could not find justification for considering this statistical power sufficient. It can be argued that the number of replicates (number of units of two animals per treatment) in the studies should be substantially increased, but one argument against an increase in numbers is related to the ethics of subjecting more animals to testing ( EC, 2010b ). One could also argue that it is unethical to conduct an underpowered study. However, most if not all of the rodent studies are based on widely accepted safety evaluation protocols with fixed numbers of animals per treatment. Cultural values regarding precaution for human safety and those regarding the number of animals subjected to testing are in conflict in this case. As pointed out by Snell et al. (2012) , a close examination of the long-term and multigenerational studies reveals that some have problems with experimental design, the most common being that the GE and non-GE sources were not isogenic and were grown in different locations (or unknown locations). Those problems in design make it difficult to determine whether differences are due to the genetic-engineering process or GE trait or to other sources of variation in the nutritional quality of the crops.
In cases in which testing produces equivocal results or tests are found to lack rigor, follow-up experimentation with trusted research protocols, personnel, and publication outlets is needed to decrease uncertainty and increase the legitimacy of regulatory decisions. There is a precedent of such follow-up studies in the literature on GE crop environmental effects that could serve as a general model for follow-up food-safety testing (see Chapter 4 section “Genetically Engineered Crops, Milkweed, and Monarch Butterflies”). The USDA Biotechnology Risk Assessment Research Grants Program has enabled this approach in a few cases.
Beyond Rodent Studies
Mice and rats are typically used in toxicity studies because of their general physiological similarities to humans and their small size, but some farm animals are considered to be better models of human physiology than rodents. The best example is the pig, which is considered to be better than rodents as a model, especially with respect to nutritional evaluations ( Miller and Ullrey, 1987 ; Patterson et al., 2008 ; Litten-Brown et al., 2010 ). Porcine insulin has been used for decades to control blood sugar in patients who have childhood-onset diabetes mellitus (type I diabetes). Pig heart valves are used for human mitral valve replacement, and pig skin has been investigated as a possible donor tissue. The pig is monogastric as is the human, and its gastrointestinal tract absorbs and metabolizes nutrients (lipids and micronutrients) in the same manner as in humans.
Reviews of studies with animals fed GE foods have included studies using both rodents and farm animals ( Bartholomaeus et al., 2013 ; DeFrancesco, 2013 ; Ricroch et al., 2013a , b , 2014 ; Swiatkiewicz et al., 2014 ; Van Eenennaam and Young, 2014 ). Those animal studies have taken advantage of the fact that maize and soybean are major components of the diets of many farm animals. Some of the reported studies that used farm animals have designs similar to those of rodent studies and have variation in duration and replicates similar to that of the rodent experiments. Some of the tests were run for 28 days (for example, Brouk et al., 2011 ; Singhal et al., 2011 ), others for a long term ( Steinke et al., 2010 ) or in multiple generations ( Trabalza-Marinucci et al., 2008 ; Buzoianu et al., 2013b ).
The experiments with pigs are especially relevant. Most of them were conducted in one prolific laboratory ( Walsh et al., 2011 , 2012a , b , 2013 ; Buzoianu et al., 2012a , b , c , d , 2013a , b ). The studies range from examination of short-term growth of piglets to multigenerational studies of sows and piglets, with mixed designs having either generation or both exposed to Bt maize and non- Bt maize. Characteristics measured included food consumption and growth, assessment of organ size and health, immunological markers, and microbial communities. The authors of the studies generally concluded that Bt maize does not affect health of the pigs, but they reported a number of statistically significant differences between Bt maize treatment and control maize treatment. In one experiment ( Walsh et al., 2012a ), the weaned piglets that were fed Bt maize had lower feed-conversion efficiency during days 14–30 (P > 0.007) but no significant effect over the full span of the experiment. In another experiment ( Buzoianu et al., 2013b ), there was lower efficiency in the Bt treatment during days 71–100 (P > 0.01) but again no effect over the full span of the experiment.
In those experiments with pigs and experiments with other farm animals and rodents, there was apparently one source of the GE food and one source of the non-GE food per study, and it is generally not clear that the food sources were isogenic or grown in the same location. That makes it difficult to determine whether any statistical differences found were due to the engineered trait or to the batches of food used, which in at least some experiments varied in nutrient content and may have differed in bioactive compounds (produced in response to plant stressors), which may have a profound effect on outcomes of nutritional studies. Another issue is that many statistical tests were performed in most studies. That could result in accumulation of false-positive results ( Panchin and Tuzhikov, 2016 ). Although this is not a situation in which a stringent correction for doing multiple tests is called for ( Dunn, 1961 ), there is reason to be cautious in interpretation of statistical significance of individual results because multiple tests can lead to artifactual positive results. The issue of multiple test results is common in many fields, and one approach used in genetics is to use the initial tests for hypothesis generation with follow-up experiments that test an a priori hypothesis (for example, Belknap et al., 1996 ). If a straightforward application of Bonferonni correction is used, each animal study that measures multiple outcomes, whether for GE crops or any other potential toxicant, could require over 1,000 animals to obtain reasonable statistical power ( Dunn, 1961 ).
In addition to the literature on controlled experiments with livestock, Van Eenennaam and Young (2014) reviewed the history of livestock health and feed-conversion ratios as the U.S. livestock industry shifted from non-GE to GE feed. Producers of cattle, milk cows, pigs, chickens, and other livestock are concerned about the efficiency of conversion of animal feed into animal biomass because it affects profit margins. The data examined start as early as 1983 and run through 2011. Therefore, livestock diets shifted from all non-GE feed to mostly GE feed within the duration of the study. Van Eenennaam and Young found that, if anything, the health and feed-conversion efficiencies of livestock had increased since the introduction of GE crops but that the increase was a steady rise, most likely because of more efficient practices not associated with use of GE feed. In the studies that they reviewed, the number of animals examined was large (thousands). Of course, most livestock are slaughtered at a young age, so that data cannot address the issue of longevity directly. However, given the general relationship between general health and longevity, the data are useful.
FINDING: The current animal-testing protocols based on OECD guidelines for the testing of chemicals use small samples and have limited statistical power; therefore, they may not detect existing differences between GE and non-GE crops or may produce statistically significant results that are not biologically meaningful.
FINDING: In addition to experimental data, long-term data on the health and feed-conversion efficiency of livestock that span a period before and after introduction of GE crops show no adverse effects on these measures associated with introduction of GE feed. Such data test for correlations that are relevant to assessment of human health effects, but they do not examine cause and effect.
RECOMMENDATION: Before an animal test is conducted, it is important to justify the size of a difference between treatments in each measurement that will be considered biologically relevant.
RECOMMENDATION: A power analysis for each characteristic based on standard deviations in treatments in previous tests with the animal species should be done whenever possible to increase the probability of detecting differences that would be considered biologically relevant.
RECOMMENDATION: In cases in which early published studies produced equivocal results regarding health effects of a GE crop, followup experimentation using trusted research protocols, personnel, and publication outlets should be used to decrease uncertainty and increase the legitimacy of regulatory decisions.
RECOMMENDATION: Public funding in the United States should be provided for independent follow-up studies when equivocal results are found in reasonably designed initial or preliminary experimental tests.
Compositional Analysis
Compositional analysis of genetically engineered crops.
As part of the regulatory process of establishing substantial equivalence, GE crop developers submit data comparing the nutrient and chemical composition of their GE plant with a similar (isoline) variety of the crop. In the United States, submitting such data to FDA is voluntary, although as of 2015 this seems to always be done by developers. Developers and regulators compare key components of the GE variety with published reference guides that list the concentrations and variabilities of nutrients, antinutrients, and toxicants that occur in crops already in the food supply. 4 The section “Regulatory Testing of Crops with Resistance to Glyphosate and 2,4-D and the New Uses of the Herbicides Themselves” earlier in this chapter gives an example of the types of nutrients and chemicals that are generally measured. In the specific case of the soybean resistant to 2,4-D and glyphosate, measurements of 62 components in the soybean were submitted by Dow AgroSciences. There were statistically significant differences between the GE and comparison varieties in 16 of the 62. The differences were considered to be small and within the range of published values for other soybean varieties. They were therefore “considered not biologically relevant.” In compositional analysis, as in some of the whole-food animal testing, it is difficult to know how much of the variance and range in values for the components is due to the crop variety, the growing conditions, and the specific laboratory experimental equipment. In the United States, regulatory agencies require that the comparison be between the GE crop and its isogenic conventionally bred counterpart grown in side-by-side plots. In those cases, it is hard to attribute differences to anything but the genetic-engineering process.
FINDING: Statistically significant differences in nutrient and chemical composition have been found between GE and non-GE plants by using traditional methods of compositional analysis, but the differences have been considered to fall within the range of naturally occurring variation found in currently available non-GE crops.
Composition of Processed Genetically Engineered Foods
General compositional analysis and the specific content of the introduced proteins are typically conducted on raw products, such as maize kernels or soybean seed. However, much of the human consumption of these products occurs after substantial exposure to heat or other processing. If in processing of foods the amounts of GE proteins substantially increase, consumers are potentially exposed to a risk that is different from that anticipated from testing the raw material. In the production of oil, for example, the goal is to separate the oil from other compounds in the raw crop, such as proteins and carbohydrates. Crude oils can contain plant proteins ( Martín-Hernández et al., 2008 ), but in highly purified oils even sophisticated approaches have failed to find any nondegraded proteins ( Hidalgo and Zamora, 2006 ; Martín-Hernández et al., 2008 ). Those results are reflected in the fact that people who are allergic to soybean are not affected by purified oils ( Bush et al., 1985 ; Verhoeckx et al., 2015 ).
A few studies have searched for a means of finding DNA in plant-derived oils to identify the origin of the oil as GE or non-GE for labeling purposes ( Costa et al., 2010a , b ) or to identify the origin of olive oil ( Muzzalupo et al., 2015 ). It is possible to detect DNA, but the amounts are typically diminished in purified oils to 1 percent or less of the original content. Similarly, Oguchi et al. (2009) were not able to find any DNA in purified beet sugar. Some countries exempt products from labeling if GE protein or DNA is not detectable. For example, in Japan, where foods with GE ingredients typically require labeling, oil, soy sauce, and beet sugar are excluded because of degradation of GE proteins and DNA ( Oguchi et al., 2009 ). Australia and New Zealand have similar exemptions from labeling for such highly refined foods as sugars and oils ( FSANZ, 2013 ).
The detection of GE protein and DNA in other processed foods depends on the type of processing. For example, the amount of the Bt protein Cry1Ab detected by immunoassay in tortillas depends on cooking time ( de Luis et al., 2009 ). The detected amount of Cry9C protein remaining in samples of corn bread, muffins, and polenta was about 13, 5, and 3 percent of the amount in the whole-grain maize ( Diaz et al., 2002 ). For Cry1Ab in rice, Wang et al. (2015) found that baking was more effective in lowering the detection using polyclonal antibodies of the Cry1Ab protein than microwaving, but 20 minutes of baking at 180°C left almost 40 percent of the protein intact. Heat denaturation of proteins can lower antibody binding to epitopes and cause lower detection of GE proteins.
FINDING: The amount of GE protein and DNA in food ingredients can depend on the specific type of processing; some foods contain no detectable protein and little DNA. In a few countries that have manda tory labeling of GE foods, that is taken into account, and food without detectable GE DNA or GE protein is not labeled.
Newer Methods for Assessing Substantial Equivalence
As explained in Chapter 2 , governance of GE crops includes regulatory governance. Although not required to by governing bodies, companies and academic researchers have moved beyond the typical measurements of food composition to newer technologies that involve transcriptomics, proteomics, and metabolomics. The new methods provide a broad, nontargeted assessment of thousands of plant characteristics, including the concentrations of most of the messenger RNAs, proteins, and small molecules in a plant or food. These methods are more likely to detect changes in a GE crop than the current regulatory approaches. If a GE crop has been changed only as intended, any changes observed in these -omics measurements theoretically should be predictable in a given environment. The science behind the methods, including the current limitations of their interpretation, is discussed in Chapter 7 . The discussion here focuses on how the methods have already been applied in the assessment of risk of health effects of currently commercialized GE crops.
Ricroch et al. (2011) reviewed -omics data from 44 studies of crops and detailed studies of the model plant Arabidopsis thaliana . Of those studies, 17 used transcriptomics, 12 used proteomics, and 26 used metabolomic methods. Ricroch (2013) updated the number of studies to 60. The committee found that many more studies had been done since those reviews were published, and many of them have used multiple -omics approaches. The sophistication of the studies has increased ( Ibáñez et al., 2015 ) and is likely to increase further. As recommended in Chapter 7 , there is a need to develop further and share databases that contain detailed -omics data ( Fukushima et al., 2014 ; Simó et al., 2014 ).
In some studies of GE plants in which simple marker genes were added, there were almost no changes in the transcriptome ( El Ouakfaoui and Miki, 2005 ), but use of other -omics methods has revealed changes ( Ren et al., 2009 ). For example, in a comparison of glyphosate-resistant soybean and non-GE soybean, García-Villalba et al. (2008) found that three free amino acids, an amino acid precursor, and flavonoid-derived secondary metabolites (liquiritigenin, naringenin, and taxifolin) had greater amounts in the GE soybean and 4-hydroxy-l-threonine was present in the non-GE soybean, but not in the GE variety. They hypothesized that the change in the flavonoids may have been because the modified EPSPS enzyme (a key enzyme of the shikimate pathway leading to aromatic amino acids) introduced to achieve glyphosate resistance could have different enzymatic properties that influenced the amounts of aromatic amino acids. The committee was not aware of such a hypothesis before this metabolomic study. (A concern was expressed in a comment submitted to the committee that the EPSPS transgene would cause endocrine disruption. The committee found no evidence to suggest that the changes found by García-Villalba et al. would have such an effect.)
On the basis of previous experimentation, it is predicted that, when a gene for a nonenzymatic protein (such as a Bt toxin gene) is added to a plant, there will be very few changes in the plant's metabolism ( Herman and Price, 2013 ). However, when a gene has been added specifically to alter one metabolic pathway of a plant, a number of predicted and unpredicted changes have been found. For example, Shepherd et al. (2015) found that, when they downregulated enzymes (that is, decreased expression or activity) involved in the production of either of two toxic glycoalkaloids (alpha-chaconine and alpha-solanine) in a GE potato with RNA-interfering transgenes that regulated synthesis of one toxic glycoalkaloid, the other compound usually increased. When they downregulated production of both compounds, beta-sitosterol and fucosterol increased. Neither of these compounds has the degree of toxicity associated with alpha-chaconine and alpha-solanine. Other compounds also differed from controls in concentration, but some of the changes may have been due to products generated during the tissue-culture process used in these experiments and not to the transgenes.
Many of the studies have found differences between the GE plants and the isogenic conventionally bred counterparts, but for many components there is more variation among the diverse conventionally bred varieties than between the GE and non-GE lines ( Ricroch et al., 2011 , Ricroch, 2013 ). Furthermore, the environmental conditions and the stage of the fruit or seed affect the finding. Chapter 7 addresses the future utility of the -omics approaches in assessing the biological effects of genetic engineering.
FINDING: In most cases examined, the differences found in comparisons of transcriptomes, proteomes, and metabolomes in GE and non-GE plants have been small relative to the naturally occurring variation found in conventionally bred crop varieties due to genetics and environment.
FINDING: If an unexpected change in composition beyond the natural range of variation in conventionally bred crop varieties were present in a GE crop, -omics approaches would be more likely to find the difference than current methods.
FINDING: Differences in composition found by using -omics methods do not, on their own, indicate a safety problem.
Food Allergenicity Testing and Prediction
Allergenicity is a widespread adverse effect of foods, several plants, tree and grass pollens, industrial chemicals, cosmetics, and drugs. Self-reporting of lifetime allergic responses to each of the most common food allergens (milk, egg, wheat, soy, peanut, tree nuts, fish, and shellfish) ranges from 1 to 6 percent of the population ( Nwaru et al., 2014 ). Allergies are induced in a two-step process: sensitization from an initial exposure to a foreign protein or peptide followed by elicitation of the allergic response on a second exposure to the same or similar agent. Sensitization and elicitation are generally mediated by immunoglobulins, primarily IgE, and the responses may range from minor palatal or skin itching and rhinitis to severe bronchial spasms and wheezing, anaphylaxis, and death. In addition to IgE responses to food allergens, IgA has been identified as an inducible immune mediator primarily in the gastrointestinal mucosa in response to foods, foreign proteins, pathogenic microorganisms, and toxins. The role of IgA in classical allergy has been investigated ( Macpherson et al., 2008 ).
Assessment of the potential allergenicity of a food or food product from a GE crop is a special case of food-toxicity testing and is based on two scenarios: transfer of any protein from a plant known to have food-allergy properties and transfer of a protein that could be a de novo allergen. Predictive animal testing for allergens in foods (GE and non-GE) is not sufficient for allergy assessment ( Wal, 2015 ). Research efforts are ongoing to discover or develop an animal model that predicts sensitization to allergy ( Ladics and Selgrade, 2009 ), but so far none has proved predictive ( Goodman, 2015 ). Therefore, researchers have relied on multiple indirect methods for predicting whether an allergic response could be caused by a protein that is either added to a food by genetic engineering or appears in the food as an unintended effect of genetic engineering. Endogenous protein concentrations with known allergic properties also have to be monitored because it is possible that their concentration could increase due to genetic engineering.
A flow diagram of the interactive approach to allergen testing recommended by the Codex Alimentarius Commission ( CAC, 2009 ) and EFSA (2010 , 2011a ) is presented in Figure 5-3 ( Wal, 2015 ); Box 5-2 describes the EPA testing of the Bt toxin Cry1F that generally follows this approach. The logic behind the approach starts with the fact that any gene for a protein that comes from a plant that is known to cause food allergies has a higher likelihood of causing allergenicity than any gene from a plant that does not cause an allergic response. If the introduced protein is similar to a protein already known to be an allergen, it becomes suspect and should be tested in people who have an allergy to the related protein. Finally, if a protein fits none of the above characteristics but is not digested by simulated gastric fluid, it could be a novel food allergen. The latter factor comes from research demonstrating that some, but not all, proteins already known to be food allergens are resistant to digestion by gut fluid.
Flow chart summarizing the weight-of-evidence approach for assessment of allergenicity of a newly expressed protein in genetically engineered (GE) organisms. SOURCE: CAC (2009) and EFSA (2010, 2011a) in Wal (2015). NOTE: This approach starts with questions (more...)
There is one case in which that approach was used and a GE crop with allergenicity issues was detected early and prevented from being commercialized, and a second case in which a GE crop was withdrawn from the market based on the possibly that it included a food allergen. In the first case, research was conducted on a soybean line genetically engineered to produce a Brazil nut ( Bertholletia excelsa ) protein, which was a known allergen. Sera from patients allergic to Brazil nut protein were available and tested positive for activity against the GE soybean protein. Because the segregation from the human food supply of GE soybean with that protein could not be guaranteed, the project was halted ( Nordlee et al., 1996 ). The soybean variety was never commercialized.
In the second case, EPA allowed a Bt maize variety developed by Aventis CropScience with a potential for allergenicity (due to decreased digestion of the protein Cry9c in simulated gastric fluid) to be sold as cattle feed under the name StarLink™; because of the potential for allergenicity, the variety was not approved for direct human consumption. However, the Bt protein was found in human food, so the maize variety was removed from all markets. After that incident, EPA no longer distinguished between Bt proteins in human food versus in animal feed ( EPA, 2001b ). Bt crop varieties are approved in the United States for all markets or none.
The interactive approach for testing should work for GE crops when the testing is for a transgene that is expressed by the plant as a protein that does not affect its metabolism (for example, Bt toxins). The testing does not cover endogenous allergens whose concentrations have been increased by unintended effects of genetic engineering. In 2013, the European Commission set a requirement for assessing endogenous allergens in GE crops ( EC, 2013 ). A number of articles since then have supported the approach ( Fernandez et al., 2013 ) or have found it unnecessary and impractical ( Goodman et al., 2013 ; Graf et al., 2014 ). Soybean is an example of a crop that has endogenous allergens. A paper on endogenous soybean allergens concluded that there is enough knowledge of only some soybean allergens for proper testing ( Ladics et al., 2014 ). As emphasized by Wal (2015) , there is considerable variation among conventionally bred varieties in the concentrations of endogenous allergens, especially when they are grown under different conditions. Therefore, the existing variation must be taken into consideration in assessing a GE variety. Of course, the issue is not only the magnitude of variation but the potential change in the overall exposure of the global human population to the allergen.
One example of an existing potential allergen of concern is gamma-zein, one of the storage proteins produced in the maize kernel that is a comparably hard-to-digest protein ( Lee and Hamaker, 2006 ). Concern was expressed to the committee that GE maize may have higher amounts of gamma-zein, which could be allergenic ( Smith, 2014 ). Krishnan et al. (2010) found that young pigs consuming maize generate antibodies against gamma-zein. That observation and the fact that the protein withstands pepsin digestion suggest that gamma-zein could be an allergen. In a comparison of the Bt maize line MON810 with non- Bt maize, known maize allergens, including the 27-kDa and 50-kDa gamma-zein proteins, were not found to be in significantly different amounts ( Fonseca et al., 2012 ). On the other hand, conventionally bred Quality Protein Maize is reported to have a 2 to 3 fold higher concentration of the 27-kDa gamma-zein protein ( Wu et al., 2010 ). There is one patent for decreasing gamma-zein through genetic engineering. 5
There can be a connection between immune response and allergenicity. One well-cited study brought up in the public comment period was that by Finamore et al. (2008) , who assessed the effect of Bt maize ingestion on the mouse gut and peripheral immune system. They found that Bt maize produced small but statistically significant changes in percentage of T and B cells and of CD4+, CD8+, γδT, and αβT subpopulations at gut and peripheral sites and alterations of serum cytokines in weanlings fed for 30 days and in aged mice. However, there was no significant response in weaning mice that were fed for 90 days, which they related to further maturation of the immune system. They concluded that there was no evidence that the Bt toxin in maize caused substantial immune dysfunction. Similarly, Walsh et al. (2012a) did not find immune function changes in a long-term pig feeding study (80 or 110 days) on Bt MON810 maize compared with non-GE maize. Overall, no changes of concern regarding Bt maize feeding and altered immune response have been found.
At a public meeting that the committee held on health effects of GE foods, a question was raised about whether current testing for allergenicity is insufficient because some people do not have acidic conditions in their stomachs. Regarding that issue, digestibility of the proteins is assessed with simulated gastric fluid (0.32 percent pepsin, pH 1.2, 37°C), under the premise that an undigested protein may lead to the absorption of a novel allergenic fragment ( Astwood et al., 1996 ; Herman et al., 2006 ). Stomach fluid is typically acidic, with a pH of 1.5–3.5, which is the range at which pepsin (the digestive enzyme of the stomach) is active, and the volume of stomach fluid is 20–200 mL (about 1–3 ounces). Simulated gastric fluid was developed to represent human gastric conditions in the stomach and is used in bioavailability studies of drugs and foods ( U.S. Pharmacopeia, 2000 ).
In general, if the pH of the stomach is greater than 5, pepsin will not be active, and less breakdown of large proteins will take place. Hence, the usefulness of simulated gastric fluid in the case of a less acidic (higher pH) stomach is questionable, whether used for non-GE foods or GE foods. Untersmayr and Jensen-Jarolim (2008:1301) concluded that “alterations in the gastric milieu are frequently experienced during a lifetime either physiologically in the very young and the elderly or as a result of gastrointestinal pathologies. Additionally, acid-suppression medications are frequently used for treatment of dyspeptic disorders.” Trikha et al. (2013) used a group of 4,724 children (under 18 years old) who had received a diagnosis of gastroesophageal reflux disease (GERD) and who were treated with gastric acid-suppressive medication and matched with 4,724 children who had GERD but were not so treated. Those treated with acid-reducing medicine were more than 1.5 times as likely to have a diagnosis of food allergy as those who were not so treated. The difference between the two GERD groups was statistically significant (hazard ratio, 1.68; 95-percent confidence interval, 1.15–2.46).
The National Research Council report Safety of Genetically Engineered Foods pointed out that there were important limitations in allergenicity predictions that could be done before commercialization ( NRC, 2004 ). Since that report was published, there have been improvements in the allergen database, and research has been funded to improve precommercialization prediction. However, as the committee heard from an invited speaker, “no new methods have been demonstrated to predict sensitization and allergy in the absence of proven exposure” ( Goodman, 2015 ). Before commercialization, the general population will probably not have been exposed to an allergen similar enough to an allergen in a GE plant to cause cross-reactivity, so it would be useful to use the precommercialization tests only as a rough predictor. To ensure that allergens did not remain in the food system, the Safety of Genetically Engineered Foods report called for a two-step process of precommercialization testing and post-commercialization testing. Even though progress has been made on allergenicity prediction since that report was published in 2004, the committee found that post-commercialization testing would be useful in ensuring that no new allergens are introduced. There have been no steps toward post-commercialization testing since 2004. The committee recognized that such testing would be logistically challenging, as described in a scientific report to EFSA ( ADAS, 2015 ). Post-commercialization surveillance of such specific agents as drugs and medical devices is difficult, but there is generally a well-defined endpoint to look for in patients. In the case of food, the detection of an allergic response to a particular protein would be confounded by multiple exposures in the diet. However, several region-wide human populations have been exposed to GE foods for many years whereas others have not; this could enable an a priori hypothesis to be tested that populations that have been exposed to foods from specific GE crops will not show a higher rate of allergic response to such foods.
FINDING: For crops with endogenous allergens, knowing the range of allergen concentrations in a broad set of crop varieties grown in a variety of environments is helpful, but it is most important to know whether adding a GE crop to the food supply will change the general exposure of humans to the allergens.
FINDING: Because testing for allergenicity before commercialization could miss allergens to which the population had not previously been exposed, post-commercialization allergen testing would be useful in ensuring that consumers are not exposed to allergens, but such testing would be difficult to conduct.
FINDING: There is a substantial population of persons who have higher than usual stomach pH, so tests of digestibility of proteins in simulated acidic gastric fluid may not be relevant to this population.
- GENETICALLY ENGINEERED CROPS AND OCCURRENCE OF DISEASES AND CHRONIC CONDITIONS
The overall results of short-term and long-term animal studies with rodents and other animals and other data on GE-food nutrient and secondary compound composition convinces many (for example, Bartholomaeus et al., 2013 ; Ricroch et al., 2013a , b ; Van Eenennaam and Young, 2014 ) but not all involved researchers (for example, Dona and Arvanitoyannis, 2009 ; Domingo and Bordonaba, 2011 ; Hilbeck et al., 2015 ; also see DeFrancesco, 2013 ) that currently marketed GE foods are as safe as foods from conventionally bred crops. The committee received comments from an invited speaker ( Smith, 2014 ) and from the public regarding the possible relationship between increases in the incidence of specific chronic diseases and the introduction of GE foods into human diets. Appendix F includes a representative list of the comments about GE food safety that were sent to the committee through the study's website. The comments mentioned concerns about such chronic diseases as cancers, diabetes, and Parkinson's; possible organ-specific injuries (liver and kidney toxicity); and such disorders as autism and allergies. Smith (2003:39) made the claim that “diabetes rose by 33 percent from 1990 to 1998, lymphatic cancers are up, and many other illnesses are on the rise. Is there a connection to [genetically modified] foods? We have no way of knowing because no one has looked for one.”
As part of the committee's effort to respond to its task to “assess the evidence for purported negative effects of GE crops and their accompanying technologies,” it used available peer-reviewed data and government reports to assess whether any health problems may have increased in frequency in association with commercialization of GE crops or were expected to do so on the basis of the results of toxicity studies. The committee presents additional biochemical data from animal experiments but relies mostly on epidemiological studies that used time-series data. The epidemiological data for some specific health problems are generally robust over time (for example, cancers) but are less reliable for others. The committee presents the available data knowing that they include a number of sources of bias, including changes over time in survey methods and in the tools for detection of specific chronic diseases. As imperfect as the data may be, they are in some cases the only information available beyond animal experiments for formulating or testing hypotheses about possible connections between a GE food and a specific disease. The committee points out that the lack of rigorous data on incidence of disease is not only a problem for assessing effects of GE foods on health. More rigorous data on time, location, and sociocultural trends in disease would enable better assessment of potential health problems caused by environmental factors and other products from new technologies.
Cancer Incidence
A review of the American Cancer Society's database indicates that mortality from cancers in the United States and Canada has continued to decrease or stabilized in all categories except cancers of the lung and bronchus attributable to smoking. The decreases in mortality are due in part to early detection and improved treatment, so mortality data can mask the rate at which cancers occur. For that reason, the committee sought data on cancer incidence rather than cancer mortality. Figures 5-4 and 5-5 show changes in cancer incidence in U.S. women and men, respectively, from 1975 to 2011 ( NCI, 2014 ). If GE foods were causing a substantial number of specific cancers, the incidence of those cancers would be expected to show a change in slope in the time source after 1996, when GE traits were first available in commercial varieties of soybean and maize. The figures show that some cancers have increased and others decreased, but there is no obvious change in the patterns since GE crops were introduced into the U.S. food system. Figures 5-6 and 5-7 show cancer incidence in women and men in the United Kingdom, where GE foods are not generally being consumed. For the specific types of cancers that are reported in both the United States and the United Kingdom, there is no obvious difference in the patterns that could be attributed to the increase in consumption of GE foods in the United States. (The absolute numbers cannot be compared because of differences in methodology.)
Trends in cancer incidence in women in the United States, 1975–2011. SOURCE: NCI (2014). NOTE: Age-adjusted to the 2000 U.S. standard population and adjusted for delays in reporting. Dashed line at 1996 indicates year GE soybean and maize were (more...)
Trends in cancer incidence in men in the United States, 1975–2011. SOURCE: NCI (2014). NOTE: Age-adjusted to the 2000 U.S. standard population and adjusted for delays in reporting. Dashed line at 1996 indicates year GE soybean and maize were first (more...)
Cancer incidence in women in the United Kingdom, 1975–2011. DATA SOURCE: Cancer Research UK. Available at http://www.cancerresearchuk.org/health-professional/cancer-statistics. Accessed October 30, 2015. NOTE: Dashed line at 1996 indicates year (more...)
Cancer incidence in men in the United Kingdom, 1975–2011. DATA SOURCE: Cancer Research UK. Available at http://www.cancerresearchuk.org/health-professional/cancer-statistics. Accessed October 30, 2015. NOTE: Dashed line at 1996 indicates year (more...)
Forouzanfar et al. (2011) published data on breast and cervical cancer incidence worldwide from 1980 to 2010. As can be seen in Figure 5-8 , the global incidence of those two cancers has increased. An examination of the plots for North America (high income) (Canada and the United States), where GE foods are eaten, compared with the plots for western Europe, where GE foods generally are not eaten, shows similar increases in incidence of breast cancer and no increase in cervical cancer. The data do not support the hypothesis that GE-food consumption has substantially increased breast and cervical cancer. (The data for North America [high income] and western Europe are different from those in the studies above on the incidence of cancer in the United States and the United Kingdom.)
Global incidence of breast (A) and cervical (B) cancer. SOURCE: Forouzanfar et al. (2011). NOTE: North America (high income): Canada, United States; Western Europe: Andorra, Austria, Belgium, Cyprus, Denmark, Finland, France, Germany, Greece, Iceland, (more...)
Taken together, Figure 5 through Figure 8 do not support the hypothesis that GE foods have resulted in a substantial increase in the incidence of cancer. However, they do not establish that there is no relationship between cancer and GE foods because there can be a delay in the onset of cancer that would obscure a trend, and one could hypothesize that something else has occurred with GE foods in the United States that has lowered cancer incidence and thus obscured a relationship. The committee had limited evidence on which to make its judgments, but the evidence does not support claims that the incidence of cancers has increased because of consumption of GE foods.
There is ongoing debate about potential carcinogenicity of glyphosate in humans. Assessment of glyphosate is relevant to the committee's report because it is the principal herbicide used on HR crops (Livingston, et al. 2015), and it has been shown that there are higher residues of glyphosate in HR soybean treated with glyphosate than in non-GE soybean ( Duke et al., 2003 ; Bøhn et al., 2014 ). Box 5-5 provides details about a study by Séralini et al. (2012 , 2014 ) that concluded that glyphosate causes tumors in rats. The committee found that this study was not conclusive and used incorrect statistical analysis. The most detailed epidemiological study that tested for a relationship between cancer and glyphosate as well as other agricultural chemicals found “no consistent pattern of positive associations indicating a causal relationship between total cancer (in adults or children) or any site-specific cancer and exposure to glyphosate” ( Mink et al., 2012:440 ; also see section below “Health Effects of Farmer Exposure to Insecticides and Herbicides”).
In 1985, EPA classified glyphosate as Group C (possibly carcinogenic to humans) on the basis of tumor formation in mice. However, in 1991, after reassessment of the mouse data, EPA changed the classification to Group E (evidence of noncarcinogenicity in humans) and in 2013 reaffirmed that “based on the lack of evidence of carcinogenicity in two adequate rodent carcinogenicity studies, glyphosate is not expected to pose a cancer risk to humans” ( EPA, 2013:25399 ).
In 2015, the International Agency for Research on Cancer (IARC) of the World Health Organization (WHO) issued a monograph on glyphosate as part of its volume on some organophosphate insecticides and herbicides ( IARC, 2015 ). In the monograph, IARC classified glyphosate in Group 2A (probably carcinogenic to humans). A summary and reasons for the classification were published in Lancet Oncology ( Guyton et al., 2015 ).
The 2015 IARC Working Group found that, although there is “ limited evidence in humans for the carcinogenicity of glyphosate,” there is “ sufficient evidence in experimental animals for the carcinogenicity of glyphosate” ( IARC, 2015:78 ). Furthermore, IARC noted that there is mechanistic support in that glyphosate induces oxidative stress, which could cause DNA damage, and some epidemiological data that support the classification.
EFSA (2015) evaluated glyphosate after the IARC report was released and concluded that glyphosate is unlikely to pose a carcinogenic risk to humans. Canada's health agency concluded that “the level of human exposure, which determines the actual risk, was not taken into account by WHO (IARC)” ( Health Canada, 2015 ). The Canadian agency found that current food and dermal exposure to glyphosate even by those who work directly with glyphosate is not a health concern as long as it is used as directed on product labels ( Health Canada, 2015 ). EPA (2015) found that glyphosate does not interact with estrogen, androgen, or thyroid systems.
A comment to the committee expressed concern that glyphosate breaks down to formaldehyde, which was classified as a known human carcinogen by IARC (2006) . However, this hypothesis was not supported; Franz et al. (1997) used radiolabeled glyphosate and failed to show formation of formaldehyde in the normal environmental degradation of glyphosate.
FINDING: The incidence of a variety of cancer types in the United States has changed over time, but the changes do not appear to be associated with the switch to consumption of GE foods. Furthermore, patterns of change in cancer incidence in the United States are generally similar to those in the United Kingdom and Europe, where diets contain much lower amounts of food derived from GE crops. The data do not support the assertion that cancer rates have increased because of consumption of products of GE crops.
FINDING: There is significant disagreement among expert committees on the potential harm that could be caused by the use of glyphosate on GE crops and in other applications. In determining the risk from glyphosate and formulations that include glyphosate, analyses must take into account both marginal exposure and potential harm.
Kidney Disease
It has been hypothesized that kidney disease may have increased because GE proteins reached the kidney. The committee examined epidemiological data to determine whether there was a correlation between the consumption of GE foods and the prevalence of chronic kidney disease (CKD).
The total prevalence of all stages of CKD in the United States increased 2 percent from about 12 percent in 1988–1994 to 14 percent in 1999–2004, but the total prevalence has not increased significantly since then. Figure 5-9 presents prevalence data on the five progressively more serious, recognized stages of CKD ( USRDS, 2014 ). The greatest percent increase is seen in Stage 3, and based on the study ( USRDS, 2014 ), a large amount of the increase occurred in people with comorbidity of cardiovascular disease. Prevalence of CKD increases substantially with age ( Coresh et al., 2003 ), so the aging of the U.S. population may contribute to the overall increase ( U.S. Census Bureau, 2014 ), as does the increase in diabetes and hypertension ( Coresh et al., 2007 ).
Prevalence of chronic kidney disease by stage among National Health and Nutrition Examination Survey (NHANES) participants, 1988–2012. SOURCE: NHANES 1988–1994, 1999–2004, and 2005–2012; participants 20 years old and older; (more...)
FINDING: The available data on prevalence of chronic kidney disease in the United States show a 2 percent increase from 1988 to 2004, but the increase does not appear to be attributable to consumption of GE foods.
Obesity in humans is a complex condition associated with several genetic and environmental factors—including geography, ethnicity, socioeconomic status, lack of exercise, availability of fresh fruits and vegetables, and less nutritional meals ( Thayer et al., 2012 )—and an altered functioning microbiome ( Turnbaugh et al., 2009 ).
Studies of various species examined body-weight gain when animals were fed a GE crop, a non-GE isogenic comparator, or a non-GE, nonisogenic control. The authors concluded that there were no biologically relevant differences in body-weight gain regardless of the length of the studies ( Rhee et al. 2005 ; Hammond et al., 2006 ; Arjó et al., 2012 ; Buzoianu et al., 2012b ; Ricroch et al., 2013a , b ; Halle and Flachowsky, 2014 ; Nicolia et al. 2014) .
Human population studies have shown that obesity has become more prevalent in the United States (for example, Fryar et al., 2014 ). An (2015) provided a graphic of the change in U.S. adults (sorted by education level) from 1984 to 2013 ( Figure 5-10 ). As can be seen in the figure, the percentage of obese U.S. adults increased until about 2009, at which time it appears to level off. Because there is no increase in the slope after commercialization of GE crops, these data do not support the hypothesis that GE crops have increased obesity. These time-series data do not prove that there is no association, but if one is present, it is not strong.
FIGURE 5-10
Annual trend for adjusted prevalence of obesity in U.S. adults by education level, 1984–2013. SOURCE: An (2015). NOTE: Prevalence of obesity was adjusted to account for gender, age group, and race or ethnicity. Dashed line at 1996 indicates year (more...)
Those statistics on obesity coincide with those on the incidence of type II diabetes in the United States ( Abraham et al., 2015 ) and therefore do not support a relationship between GE crops and type II diabetes.
FINDING: The committee found no published evidence to support the hypothesis that the consumption of GE foods has caused higher U.S. rates of obesity or type II diabetes.
Gastrointestinal Tract Diseases
Although the gastrointestinal tract has evolved to digest dietary proteins in the stomach and small intestine effectively for absorption and use of amino acids, it is normal for some full proteins or their fragments to cross the gut barrier through a paracellular route (between cells) or damaged mucosa and for the immune system, which has a high presence at the interface of the gut wall and the internal circulation, to respond accordingly. It is also not unusual, given the high sensitivity of today's analytical equipment, for proteins or fragments to be detected in minute amounts in different body fluids. Detection methods are not specific to transgene-produced proteins but can find any dietary protein or fragment that is able to pass from the gastrointestinal tract into the bloodstream and tissues. The presence of a dietary protein or its fragment in the bloodstream or in tissues is not unusual or a cause for health concerns.
About 60–70 percent of the body's immune system is in the gastrointestinal tract's gut-associated lymphoid tissue, which has an interface with the gut luminal contents, including toxins, allergens, and the associated microbiota. For GE crops, a public concern has been that the immune system is compromised through ingested transgenic proteins. That possibility has been investigated in animal studies that examined immune system bio-markers and epithelial cell integrity (see section “Beyond Rodent Studies” above and Walsh et al., 2011 ).
It was suggested to the committee in presentations and public comments that fragments of transgenes may have some special properties that would result in human diseases if they were absorbed into the body through the digestive tract. The mechanism by which such genes or proteins would affect the body is not clear, although Smith (2013) hypothesized that consuming GE foods increased gut permeability.
FINDING: The committee could find no published evidence supporting the hypothesis that GE foods generate unique gene or protein fragments that would affect the body.
Celiac Disease
Celiac disease is an autoimmune disorder that affects about 1 percent of the population of western countries. It is triggered in susceptible people by consumption of gluten-containing cereal grains ( Fasano et al., 2003 ; Catassi et al., 2010 ). Symptoms of celiac disease are the result of an immune reaction that causes marked gastrointestinal inflammation in persons susceptible to gliadin, a component of gluten protein found in wheat, rye ( Secale cereale ), and barley ( Hordeum vulgare ) ( Green and Cellier, 2007 ). In addition to exposure to gluten, the etiology of celiac disease is multifactorial and includes genetic predisposition, microbial infection of the gastrointestinal tract, antibiotic exposure, and gastrointestinal erosion ( Riddle et al., 2012 ). Diagnosis is based on detection of serum concentrations (serotypes) of IgA tissue transglutaminase and endomysial antibody IgA, the relief of symptoms upon gluten avoidance, and tissue biopsy. The genetic changes related to the serotyped IgAs are found in about 30 percent of the Caucasian population, but susceptibility to celiac disease is found in only 1 percent of this population ( Riddle et al., 2012 ).
The committee was able to find data on the incidence of celiac disease in the United Kingdom ( West et al., 2014 ; Figure 5-11 ) and a detailed study conducted by the Mayo Clinic in one county in Minnesota ( Murray et al., 2003 ; Ludvigsson et al., 2013 ). In the Minnesota and UK studies, there is a clear pattern of increase in celiac-disease incidence (or at least its detection or the extent of self-reports) that started before 1996 ( Catassi et al., 2010 ), when U.S. citizens began to consume more GE foods and the use of glyphosate increased in the United States but not in the United Kingdom. The increases are similar in magnitude to that found in U.S. military personnel, in whom prevalence increased from 1.3 per 100,000 in 1999 to 6.5 per 100,000 in 2008 ( Riddle et al., 2012 ). The authors cautioned that most cases of celiac disease are undiagnosed. Some of the observed increase may be related to improvements in diagnostic criteria, greater awareness of the disease in physicians and patients, better blood tests, and increases in the number of biopsies. However, recent observations point to an increase in incidence beyond those factors (J. A. Murray, Mayo Clinic, personal communication, February 1, 2016).
FIGURE 5-11
Three-year rolling average incidence of celiac disease in 1990–2011, by age group, in the United Kingdom. SOURCE: West et al., 2014. NOTE: Dashed line at 1996 indicates year genetically engineered soybean and maize were first grown in the United (more...)
On the basis of data collected in the 2009–2010 National Health and Nutrition Examination Survey, Rubio-Tapia et al. (2012) reported a prevalence of celiac disease of 0.71 percent with 1.01 percent in non-Hispanic whites in a sample of 7,798 subjects. It should be noted that there has not been any commercial production of GE wheat, rye, or barley in the world. The committee found no evidence that the introduction of GE foods affected the incidence or prevalence of celiac disease worldwide.
FINDING: Celiac-disease detection began increasing in the United States before the introduction of GE crops and the increased use of glyphosate. It appears to have increased similarly in the United Kingdom, where GE foods are not typically consumed and glyphosate use did not increase. The data are not robust, but they do not show a major difference in the rate of increase in incidence of celiac disease between the two countries.
Food Allergies
Speakers and some members of the public suggested that the prevalence of food allergies has increased because of GE crops. The committee examined records on the prevalence of food allergies in the United States over time. As is clear from Figure 5-12 and Jackson et al. (2013) , the prevalence of food allergies in the United States is rising. For a rough comparator, the committee examined data on hospital admissions for food allergies in the United Kingdom over time ( Figure 5-13 ). UK citizens eat far less food derived from GE crops. The data ( Gupta et al., 2007 ) suggest that food allergies are increasing in the United Kingdom at about the same rate as in the United States (but the types of measurement are different).
FIGURE 5-12
Percentage of children 0–17 years old in the United States with a reported allergic condition in the preceding 12 months, 1997–2011. a Significantly increasing linear trend for food and skin allergy from 1997–1999 to 2009–2011. (more...)
FIGURE 5-13
Trends in hospital admission rates for anaphylaxis related to food allergy by age in the United Kingdom during 1990–2004. SOURCE: Gupta et al. (2007). NOTES: ICD = International Classification of Diseases. Green = ages 0–14 years; blue (more...)
FINDING: The committee did not find a relationship between consumption of GE foods and the increase in prevalence of food allergies.
Autism Spectrum Disorder
Autism is often described by such symptoms as difficulty in communicating, forming personal relationships, and using language and abstract concepts. According to the American Psychiatric Association (2013) , autism spectrum disorder (ASD) encompasses the previous diagnoses of autism, Asperger syndrome, pervasive developmental disorder not otherwise specified, and childhood disintegrative disorder. Accurate diagnosis of ASD can be difficult, but efforts to identify children with ASD have increased in the United States over the last three decades ( CDC, 2014 ).
In the 2010 Centers for Disease Control and Prevention (CDC) survey of ASD in 11 regions of the United States ( CDC, 2014 ), the overall prevalence in children 8 years old was about 1 in 68 (1.47 percent), but there was wide variation among regions and sociocultural groupings of children. The CDC report stated that “the extent to which this variation might be attributable to diagnostic practices, under-recognition of ASD symptoms in some racial/ethnic groups, socioeconomic disparities in access to services, and regional differences in clinical or school-based practices that might influence the findings in this report is unclear” ( CDC, 2014:1 ). The degree to which the increase in ASD prevalence since 1990 is due to improved diagnosis is also unclear.
Before 1990, few children in the United States or the United Kingdom had diagnoses of ASD ( Taylor et al., 2013 ), but the prevalence has increased dramatically in both countries. Researchers in the United States and United Kingdom wrote a report that examined prevalence of ASD in the United Kingdom over time and compared it with that in the United States ( Taylor et al., 2013 ). They concluded that “a continuous simultaneous extraordinary rise in the number of children diagnosed as autistic began in both countries in the early 1990s and lasted for a decade. The distribution of first time diagnosis according to age and gender was the same. These similarities between countries as well as within different locations in each country point to a common etiology for this extraordinary medical case” ( Taylor et al., 2013:5 ). There is a higher prevalence in the United States, but it is difficult to evaluate whether it is because of differences in efforts in and approaches to diagnosis and in sociocultural factors that seem to influence prevalence. The overall similarities in prevalence of ASD in the United Kingdom, where GE foods are rarely eaten, and in the United States, where GE foods are commonly eaten, suggest that the major rise in ASD is not associated with consumption of GE foods.
FINDING: The similarity in patterns of increase in autism spectrum disorder in children in the United States, where GE foods are commonly eaten, and the United Kingdom, where GE foods are rarely eaten, does not support the hypothesis of a link between eating GE foods and prevalence of autism spectrum disorder.
- OTHER HUMAN HEALTH CONCERNS RELATED TO GENETICALLY ENGINEERED CROPS
The committee heard from some members of the public and some invited speakers that ailments of gastrointestinal origin could be caused by GE crops or their associated technologies or by foods derived from GE crops. The committee investigated the evidence available for that hypothesis.
Gastrointestinal Tract Microbiota
The committee received comments from the public that foods derived from GE crops could change the gut microbiota in an adverse way. Three scenarios can be considered as related to the potential effects of GE crops on the gut microbiota: the effect of the transgene product (for example, Bt toxin), unintended alteration of profiles of GE plant secondary metabolites, and herbicide (and adjuvant) residue (for example, glyphosate) and its metabolites in HR crops.
Research on the human gut microbiota (the community of microorganisms that live in the digestive tract) is rapidly evolving with recent reports ( Dethlefsen and Relman, 2011 ; David et al., 2014 ) that suggest that microbiota perturbations occur fairly quickly owing to dietary components or antibiotic treatment. Microbiota composition and state are now well recognized to be linked to noncommunicable chronic diseases and other health problems, so factors that cause either beneficial or adverse changes in the microbiota are of interest to researchers and clinicians. However, the science has not reached the point of understanding how specific changes in microbiota composition affect health and what represents a “healthy” microbiota. The effect of different dietary patterns (for example, high-fat versus high-carbohydrate diets) on the gut microbiota has been linked to metabolic syndrome ( Ley, 2010 ; Zhang et al., 2015 ).
As discussed above, most proteins, including those in GE and conventionally bred crops, are at least partially digested in the stomach by the action of pepsin that is maintained by the acidic pH of the stomach in most people. Further digestion and absorption are a function of the small intestine, where amino acids and dipeptides and tripeptides are absorbed. Therefore, an effect of a dietary protein on the microbiota, whether from GE or non-GE foods, is unlikely. However, there is some evidence that Bt proteins can be toxic to microorganisms ( Yudina et al., 2007 ), and some nondegraded Bt protein is found within the lumen of the gut but not in the general circulation of pigs ( Walsh et al., 2011 ). Buzoianu et al. (2012c , 2013a ) studied the effect of Bt maize feeding on microbiota composition in pigs. In their 2012 study, 110-day feeding of Bt maize (variety MON810) and of isogenic non-GE maize diets led to no differences in cultured Enterobacteriaceae, Lactobacillus , and total anaerobes from the gut; 16S rRNA sequencing showed no differences in bacterial taxa, except the genus Holdemania with which no health effects are associated ( Buzoianu et al., 2012c ). In the follow-up study in which intestinal content of sows and their offspring were examined with 16S rRNA gene sequencing, the only observed difference for major bacterial phyla was that Proteobacteria were less abundant in sows fed Bt maize before farrowing and in offspring at weaning compared with the controls ( Buzoainu et al., 2013a ). Fecal Firmicutes were more abundant in offspring fed GE maize. There were other inconsistent differences in mostly low-abundance microorganisms. On the basis of the overall results from their studies, the authors concluded that none of the changes seen in the animals was expected to have biologically relevant health effects on the animals.
Relatively few studies have examined the influence of plant secondary metabolites from any crop on the gut microbiota. The review of Valdés et al. (2015) highlighted investigations on polyphenol-rich foods—such as red wine, tea, cocoa, and blueberries—on the microbiota. Effects were considered minor. As discussed above (see the section “Endogenous Toxins in Plants”), current commercialized GE crops do not have distinctly different secondary metabolite profiles that would lead one to think that they would affect the gut microbiota.
No studies have shown that there are perturbations of the gut microbiota of animals fed foods derived from GE crops that are of concern. However, the committee concluded that this topic has not been adequately explored. It will be important to conduct research that leads to an understanding of whether GE foods or GE foods coupled with other chemicals have biologically relevant effects on the gut microbiota.
FINDING: On the basis of available evidence, the committee determined that the small perturbations found in the gut microbiota of animals fed foods derived from GE crops are not expected to cause health problems. A better understanding of this subject is likely as the methods for identifying and quantifying gut microorganisms mature.
Horizontal Gene Transfer to Gut Microorganisms or Animal Somatic Cells
Horizontal (or lateral) gene transfer is “the stable transfer of genetic material from one organism to another without reproduction or human intervention” ( Keese, 2008:123 ). Since GE crops were commercialized, concern has been voiced by some scientists and some members of the public that foreign DNA introduced into plants through genetic-engineering technologies might, after ingestion, be transferred to the human gut microbiota and directly or indirectly (that is, from bacteria) into human somatic cells. Although most of the concern regarding horizontal gene transfer has been focused on antibiotic-resistance genes used as markers of the transgenic event, other transgenes, such as those with Bt toxins, have also been of concern.
A prerequisite for horizontal gene transfer is that the recombinant DNA must survive the adverse conditions of both food processing and passage through the gastrointestinal tract. Netherwood et al. (2004) showed in patients with a surgically implanted exiting tube placed at the end of the small intestine (an ileostomy) that a small amount of the GE soybean transgene EPSPS passed through the upper gastrointestinal tract to the point of the distal ileum; in subjects without an ileostomy, no transgene was recovered from their feces. In their review on stability and degradation of DNA from foods in the gastrointestinal tract, Rizzi et al. (2012) noted that recombinant plant DNA fragments were detected in the gastrointestinal tracts of nonruminant animals but not detected in blood or other tissues, although some nonrecombinant plant DNA could be found. The authors concluded that some natural plant DNA fragments persist in the lumen of the gastrointestinal tract and in the bloodstream of animals and humans.
For an event to be considered horizontal gene transfer, DNA must be in the form of a functional (rather than fragmented) gene, enter into bacterial or somatic cells, and be incorporated into the genome with an appropriate promoter, and it must not adversely affect the competitiveness of the cells; otherwise, the effect would be short-lived.
Plant DNA has not been demonstrated to be incorporated into animal cells; however, it has been shown to be transferred in prokaryotes (bacteria). Indeed, molecular geneticists had to find genetic-engineering approaches for getting DNA to be taken into eukaryote cells and incorporated into a genome. The report A Decade of EU-Funded GMO Research (2001–2010) ( EC, 2010a ) described a study that shows that rumen ciliates (a type of microorganism) exposed to Bt 176 maize for 2 or 3 years did not incorporate the Bt 176 transgene. There are no reproducible examples of horizontal gene transfer of recombinant plant DNA into the human gastrointestinal microbiota or into human somatic cells. Three independent reviews of the literature on the topic ( van den Eede et al., 2004 ; Keese, 2008 ; Brigulla and Wackernagel, 2010 ) concluded that new gene acquisition by the gut bacteria through horizontal gene transfer would be rare and does not pose a health risk.
FINDING: On the basis of its understanding of the process required for horizontal gene transfer from plants to animals and data on GE organisms, the committee concludes that horizontal gene transfer from GE crops or conventionally bred crops to humans does not pose a substantial health risk.
Transfer of Transgenic Material Across the Gut Barrier into Animal Organs
Conflicting reports exist regarding the question of intact transgenes and transgenic proteins from foods crossing the gut barrier. Spisák et al. (2013) published results that indicate that complete genes in foods can pass into human blood. That is plausible, but Lusk (2014) examined the approach used by Spisák et al. and found it more likely that the findings were due to contaminants. Lusk emphasized the need for negative controls in such studies. Placental transfer of foreign DNA into mice was found by Schubbert et al. (1998) by detection in the mouse fetus, but a later report from the same laboratory ( Hohlweg and Doerfler, 2001 ) did not find the transfer in an eight-generation study.
Studies with dairy cows and goats did not find transgenes or GE proteins in milk, although chloroplast DNA fragments were detected in milk ( Phipps et al., 2003 ; Nemeth et al., 2004 ; Calsamiglia, et al., 2007; Rizzi et al., 2008 ; Guertler et al., 2009 , Einspanier, 2013 ; Furgał-Dierżuk et al., 2015 ). That makes it clear that there is no apparent potential for trangenes or transgenic proteins to be present in dairy products. However, these animals are ruminants, and their digestive systems are different from that of humans.
Walsh et al. (2012a) studied the fate of a Bt gene and protein in pigs that have digestive systems that are more similar to that of humans. They found no evidence of the gene or protein in any organs or blood after 110 days of feeding on Bt maize, but they did find them in the digestive contents of the stomach, cecum, and colon. Fragments of Cry1Ab transgene (as well as other common maize gene fragments) but not the intact Bt gene were found in blood, liver, spleen, and kidney of pigs raised on Bt maize ( Mazza et al., 2005 ).
FINDING: Experiments have found that Cry1Ab fragments but not intact Bt genes can pass into organs and that these fragments present concerns no different than other genes that are in commonly consumed non-GE foods and that pass into organs as fragments.
FINDING: There is no evidence that Bt transgenes or proteins have been found in the milk of ruminants. Therefore, the committee finds that there should be no exposure to Bt transgenes or proteins from consuming dairy products.
OVERALL FINDING ON PURPORTED ADVERSE EFFECTS ON HUMAN HEALTH OF FOODS DERIVED FROM GE CROPS: On the basis of detailed examination of comparisons of currently commercialized GE and non-GE foods in compositional analysis, acute and chronic animal-toxicity tests, long-term data on health of livestock fed GE foods, and human epidemiological data, the committee found no differences that implicate a higher risk to human health from GE foods than from their non-GE counterparts.
- ASSESSMENT OF HUMAN HEALTH BENEFITS FROM GENETICALLY ENGINEERED CROPS
There are now a number of examples of crops, either commercialized or in the pipeline toward commercialization, that have GE traits that could improve human health. Improvement of human health can be the sole motivation for development of a specific crop trait, or it can be the secondary effect of a crop trait that is developed primarily for another reason. For example, the genetic engineering of rice to have higher beta-carotene has the specific goal of reducing vitamin A deficiency. GE maize that produces Bt toxins is engineered to decrease insect-pest damage, but a secondary effect could be a decrease in contamination of maize kernels by fungi that produce mycotoxins, such as fumonisins, that at high concentrations could impair human health. Beyond the direct effects of the crops on improvement of human health, there is also a potential indirect benefit associated with a decline in the exposure of insecticide applicators and their families to some insecticides because some GE plants decrease the need for insecticidal control.
Foods with Additional Nutrients or Other Healthful Qualities
Improved micronutrient content.
According to WHO, some 250 million preschool children are vitamin A–deficient. Each year, 250,000–500,000 vitamin A–deficient children become blind, and half of them die within 12 months of losing their sight. 6 Unlike children in wealthier societies, those children have diets that are restricted mostly to poor sources of nutrients, such as rice ( Hefferon, 2015 ). Overall improvement of the diets of the children and their parents is a goal that has not been reached; measures that improve the nutritional quality of their food sources are desirable although not optimal, as a diverse, healthy diet would be.
Crop breeders have used conventional breeding to improve the concentrations of beta-carotene in maize ( Gannon et al., 2014 ; Lividini and Fiedler, 2015 ), cassava, banana and plantain ( Musa spp.) ( Saltzman et al., 2013 ), and sweet potato ( Ipomoea batatas ) ( Hotz et al., 2012a , b ). There is some loss of beta-carotene during storage and cooking, but bioavailability is still good ( Sanahuja et al., 2013 ; De Moura et al., 2015 ). The most rigorous assessments of the effects of those high–beta-carotene varieties were conducted with orange-fleshed sweet potato (high in beta-carotene) in farming areas of Mozambique and Uganda. In both countries, there was increased beta-carotene intake. In Uganda, there was a positive relationship between consumption of high–beta-carotene sweet potato and positive vitamin A status ( Hotz et al., 2012a ). A more recent study in Mozambique found a decrease in diarrhea prevalence associated with consumption of the high–beta-carotene sweet potato ( Jones and DeBrauw, 2015 ).
No reported experiments have tested any crop with high–beta-carotene for unintended effects. There has been concern about the potential for too high a concentration of beta-carotene in crops because of the hypervitaminosis A syndrome that can be caused by direct intake of too much vitamin A, but that is not a problem when the source is beta-carotene ( Gannon et al., 2014 ).
Golden Rice, which was produced through genetic engineering to increase beta-carotene content, is one of the most recognized examples of the use of genetic-engineering technology to improve a crop's nutritional value. It is based on the understanding that rice possesses the entire machinery to synthesize beta-carotene in leaves but not in the grain. The breakthrough in the development of Golden Rice was the finding that only two genes are required to synthesize beta-carotene in the endosperm of the rice grain ( Ye et al., 2000 ). The first version of Golden Rice had a beta-carotene content of 6 µg/g. To raise the content to a point where it could alleviate vitamin A deficiency without consumption of very large amounts of rice, a second version of Golden Rice was produced by transforming the plant with the psy gene from maize. The carotene content was thereby raised above 30 µg/g ( Paine et al., 2005 ). Varieties that yield well, have good taste and cooking qualities, and cause no adverse health effects from unintended changes in the rice could have highly important health effects ( Demont and Stein, 2013 ; Birol et al., 2015 ). There have been claims that Golden Rice was ready for public release for well over a decade ( Hefferon, 2015 ), but this is not the case.
There is a publication on a field test of the first version of Golden Rice ( Datta et al., 2007 ), but the committee could not find information on the newer, higher–beta-carotene Golden Rice in the peer-reviewed literature. Therefore, it contacted the International Rice Research Institute (IRRI) Golden Rice project coordinator, Violeta Villegas, for an update on the status of the project. In discussions with Dr. Villegas (IRRI, personal communication, 2015), it was clear that the project is progressing with a new lead transgenic event, GR2-E, because of difficulties with the previous lead event, GR2-R. The GR2-E event has been backcrossed into varieties that have been requested by several countries including the Philippines, Bangladesh, and Indonesia. As of March 2016, Golden Rice GR2-E in PSBRc82 and BRRI dhan20 genetic backgrounds was being grown in confined field tests in the Philippines and Bangladesh, respectively. Both Golden Rice varieties underwent preliminary assessment inside the greenhouse prior to planting in confined field tests. If performance is good, the varieties will be moved to open field-testing on multiple locations. Once a food regulatory approval is received in one of the participating countries, IRRI will supply the rice with the GR2-E event to an independent third party to assess its efficacy at alleviating vitamin A deficiency.
Past issues with persons and organizations opposed to Golden Rice for a myriad of reasons may have affected IRRI's work on the rice, but the overall project status 7 points out that development of Golden Rice varieties that meet the needs of farmers and consumers and that are in full compliance with the regulatory systems of the partnering countries remains the primary objective. IRRI's summary statement on its Golden Rice project was that “Golden Rice will only be made available broadly to farmers and consumers if it is successfully developed into rice varieties suitable for Asia, approved by national regulators, and shown to improve vitamin A status in community conditions. If Golden Rice is found to be safe and efficacious, a sustainable delivery program will ensure that Golden Rice is acceptable and accessible to those most in need.” 8
Increasing concentrations of beta-carotene is only one goal of conventional crop breeding and genetic engineering. Projects for increasing iron and zinc in crops as different as wheat, pearl millet ( Pennisetum glaucum ), and lentil ( Lens culinaris ) are at varied stages of development ( Saltzman et al., 2013 ).
FINDING: Experimental results with non-GE crop varieties that have increased concentrations of micronutrients demonstrate that both GE and non-GE crops with these traits could have favorable effects on the health of millions of people, and projects aimed at providing these crops are at various stages of completion and testing.
Altering Oil Composition
Substantial efforts have been made to increase the oxidative stability of soybean oil, a major cooking oil all over the world, as a means of avoiding trans-fats generated through the hydrogenation process and enhancing omega-3 fatty acid content of the oil for use in both food and feed applications. Soybean oil is composed principally of five fatty acids: palmitic acid (16:0, carbon number:double bond number), stearic acid (18:0), oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3) in approximate percentages of 10, 4, 18, 55, and 13. High content of unsaturated fats creates a disadvantage in industrial processing because they are susceptible to oxidation and trans-fat generation during hydrogenation, whereas oils with a high percentage of oleic acid (about 80 percent) require less processing and offer another route to decrease concentrations of trans-fats in food products. High-oleic acid-containing soybean was produced by downregulating expression of the fatty acid desaturating enzymes FAD2-1A and -1B to decrease the concentration of trans-fats in soybean ( EFSA, 2013 ). In 2015, high-oleic acid soybean was commercially available in North America and was produced on a small area in the United States for specialty-product contracts (C. Hazel, DuPont Pioneer, personal communication, December 14, 2015).
Canola ( Brassica napus ), known in Europe as rapeseed, is the major oilseed crop in Canada. Canola was developed through conventional breeding at the University of Manitoba, Canada, by Downey and Stefansson in the early 1970s and had a good nutritional profile—58-percent oleic acid and 36-percent polyunsaturated fatty acids—in addition to low erucic acid and a moderate concentration of saturated fatty acid (6 percent). Because of demand for saturated functional oils for the trans - fat–free market, high-lauric acid GE canola was created in 1995 through an “ Agrobacterium mediated transformation in which the transfer-DNA (T-DNA) contained the gene encoding the enzyme 12:0 ACP thioesterase ( bay TE ) from the California Bay tree ( Umbellularia californica ). In addition, the T-DNA contained sequences that encoded the enzyme neomycin phosphotransferase II (NPTII). The expression of NPTII activity was used as a selectable trait to screen transformed plants for the presence of the bay TE gene. No other translatable DNA sequences were incorporated into the plant genome” ( Health Canada, 1999:1 ). The presence of lauric acid (12:0) in the oil allows it to be used as a replacement for other types of oils with lauric acid (for example, coconut and palm kernel oil) in such products as “confectionery coatings and fillings, margarines, spreads, shortenings, and commercial frying oils. It has also been used as a substitute for cocoa butter, lard, beef fats, palm oil, and partially or fully hydrogenated soybean, maize, cottonseed, peanut, safflower, and sunflower oils” ( Health Canada, 1999:2 ). However, low yield and comparably poor agronomic traits have removed high-lauric acid canola from the commercial market. The long-term use of crops with altered oil content is uncertain.
FINDING: Crops with altered oil composition might improve human health, but this will depend on the specific alterations, how the crops yield, and how the products of the crops are used.
Genetically Engineered Foods with Lower Concentrations of Toxins
Acrylamide is produced in starchy foods when they are cooked at high temperatures. Processing of potatoes for French fries and potato chips generates acrylamide. Toasting bread also produces acrylamide. That is viewed as a problem because the U.S. National Toxicology Program (2014) concluded that acrylamide “is reasonably anticipated to be a human carcinogen based on sufficient evidence of carcinogenicity from studies in experimental animals” and causes neurological damage at high exposure. Acrylamide is produced from a chemical reaction between asparagine and a reducing sugar, so decreasing the concentration of either is expected to decrease acrylamide. A potato line was genetically engineered to have low amounts of free asparagine and in early tests had as little as 5 percent of the acrylamide compared with non-GE potatoes when cooked at high temperatures ( Rommens et al., 2008 ).
In 2014, USDA deregulated a low-acrylamide potato produced by Simplot Plant Sciences (USDA–APHIS, 2014c) on the basis of nonplant pest status. The company also provided information to FDA. No problems were found by FDA with respect to the company's assessment of composition or safety ( FDA, 2015 ). It should be noted that for many people reduced acrylamide in potatoes is expected to lower overall acrylamide intake substantially, but many foods contain acrylamide ( FDA, 2000b , revised 2006). An FDA survey of commonly consumed foods showed French fries at seven McDonald's locations had an average acrylamide concentration of 288 parts per billion (ppb), whereas Gerber Finger Foods Biter Biscuits had 130 ppb and Wheatena Toasted Wheat Cereal had 1,057 ppb, which is much more than from fast-food French fries ( FDA, 2002 , revised 2006). 9 Any toasted bread is expected to be high in acrylamide. Therefore, how much low-acrylamide potato decreases total exposure depends on individual diets. Furthermore, EPA has established limits for exposure to acrylamide, and current actual exposures are generally below the limits.
Although the low-acrylamide potato is the only GE crop with a lower food-toxin concentration that has been deregulated in the United States, other GE crops with lower natural toxin concentrations are in the pipeline. Potatoes and other crops in the “deadly nightshade” family (Solanaceae, which includes tomato and eggplant) produce glycoalkaloids, some of which have human toxicity, as described above (see the section “Endogenous Toxins in Plants” in this chapter). Langkilde et al. (2012) conducted a compositional and toxicological analysis of the potatoes with lower solanine and higher chaconine. The study used Syrian golden hamsters instead of rats because the hamsters are very sensitive to the glycoalkaloids. There were some statistically significant differences, but they were considered not of biological relevance. At this point, the evidence is not sufficient to conclude that a low-glycoalkaloid potato would be healthier for humans.
Highly toxic chemicals (aflatoxins and fumonisins) are produced by Fusarium and Aspergillis fungi on the kernels of maize ( Bowers et al., 2014 ). Aflatoxins are considered by the U.S. National Toxicology Program (2014) to be “human carcinogens based on sufficient evidence of carcinogenicity from studies in humans.” They are also associated with many other illnesses and considered a global health problem ( Wild and Gong, 2010 ). Fumonisins cause a number of physiological disorders and are considered possibly carcinogenic to humans ( IARC, 2002 ). Several investigators have reported a substantial decrease in fumonisins in Bt maize compared with conventionally bred varieties ( Munkvold and Desjardins, 1997 ; Bowers et al., 2014 ). However, there is no clear association between Bt maize and aflatoxin concentrations ( Wiatrak et al., 2005 ; Abbas et al., 2007 ; Bowen et al., 2014 ).
Research continues on how to use genetic engineering to develop varieties of maize and peanut ( Arachis hypogaea ) that inhibit aflatoxin production, but a GE solution has so far been elusive ( Bhatnagar-Mathur et al., 2015 ). A reduction in aflatoxin in both maize and peanut would have substantial health benefits in some developing countries ( Williams et al., 2004 ; Wild and Gong, 2010 ).
FINDING: It is possible that GE crops that would result in improved health by lowering exposure of humans to plant-produced toxins in foods could be developed, but there is insufficient information to assess the possibility. However, GE plants that indirectly or directly reduce fungal-toxin production and intake would offer substantial benefits to some of the world's poorest populations, which have the highest dietary intake of food-associated fungal toxins.
Health Effects of Farmer Exposure to Insecticides and Herbicides
Chapter 4 presents data that demonstrate substantially lower use of insecticides in some Bt crops than in conventionally bred crops. There is a logical expectation that a decrease in the number of insecticide applications would lead to lower farm-worker exposure and therefore lower health burden, especially in countries where acute poisonings due to applicator exposure are common. Racovita et al. (2015) reviewed five studies of Bt cotton in China, India, Pakistan, and South Africa that ranged from one to four growing seasons. All reported a decline in the number of insecticide applications to Bt versus non- Bt cotton. In a study in China by Huang et al. (2002) , Bt cotton was treated with insecticides 6.6 times and non- Bt cotton was treated 19.8 times during the growing season. The frequency of Bt and non- Bt cotton farmers reporting poisonings were 5 percent and 22 percent, respectively in 1999, 7 percent and 29 percent in 2000, 8 percent and 12 percent in 2001. Kouser and Qaim (2011) found fewer overall insecticide treatments in a study conducted in India: 1.5 treatments of Bt cotton and 2.2 treatments of non- Bt cotton. In this study, the farmers who used Bt cotton reported 0.19 poisonings per season while those with conventionally bred cotton reported 1.6 poisonings. Bennett et al. (2006) studied the same types of farmers in South Africa. Bt cotton was not yet widely available in the beginning of the experiment, but eventually some farmers adopted Bt cotton and decreased spraying. The study looked at overall poisonings according to hospital records over time; there were 20 poisonings in the year before common availability of Bt cotton and four in a later year, when there was 60 percent adoption of Bt cotton.
The findings of those and other studies (for example, Huang et al., 2005 ; Dev and Rao, 2007 ; Kouser and Qaim, 2013 ) are in line with an expectation of a decrease in poisonings when Bt cotton is grown instead of non- Bt cotton. However, Racovita et al. (2015:15) , who carefully assessed each of the studies, found many shortcomings that led them to conclude that “the link between [genetically modified] crop cultivation and a reduction in number of pesticide poisonings should be considered as still circumstantial.” The shortcomings include the fact that the number of poisonings is based on farmer recall of incidents sometimes more than a year after the field season or, in the Bennett et al. (2006) study, simply based on hospital cases. Another issue was that there may have been differences in risk–avoidance behavior between farmers who did and did not plant Bt cotton. Finally, the studies focused on farmers, not farm workers, who do not control farm operations. Racovita et al. (2015) called for more rigorous studies that would address the shortcomings of previous studies, given the politicized nature of the use of Bt crops.
Farm-worker exposure to insecticides and herbicides is lower in the United States and some other developed countries than is the case for farm workers on resource-poor farms. However, there is substantial exposure, and any effects seen in the United States would be of global concern. Prospective cohort studies of health are the high benchmark of epidemiology studies, and the Agricultural Health Study (AHS) funded by the U.S. National Institute of Environmental Health Sciences used this approach to evaluate private and commercial applicators in Iowa and North Carolina. The landmark study resulted in two peer-reviewed articles on glyphosate exposure and cancer incidence ( De Roos et al., 2005 ; Mink et al., 2012 ) and one on glyphosate exposure and non-cancer health outcomes ( Mink et al., 2011 ). De Roos et al. (2005:49) concluded that “glyphosate exposure was not associated with cancer incidence overall or with most cancer subtypes we studied.” The data suggested a weak association with multiple myeloma on the basis of a small number of cases, but that association was not found in a follow-up study ( DeRoos et al., 2005 ; Mink et al., 2012 ). Mink et al. (2012:440) reported on the continuation of the AHS cohort study and found “no consistent pattern of positive associations indicating a causal relationship between total cancer (in adults or children) or any site-specific cancer and exposure to glyphosate.” Mink et al. (2011) reviewed noncancer health outcomes that included respiratory conditions, diabetes, myocardial infarction, reproductive and developmental outcomes, rheumatoid arthritis, thyroid disease, and Parkinson's disease. They reviewed cohort, case–control, and cross-sectional studies within the AHS study and found “no evidence of a consistent pattern of positive associations indicating a causal relationship between any disease and exposure to glyphosate” ( Mink et al., 2011:172 ).
FINDING: There is evidence that use of Bt cotton in developing countries is associated with reduced insecticide poisonings. However, there is a need for more rigorous survey data addressing the shortcomings of existing studies.
FINDING: A major government-sponsored prospective study of farm-worker health in the United States does not show any significant increases in cancer or other health problems that are due to use of glyphosate.
- ASSESSMENT OF FOOD SAFETY OF CROPS TRANSFORMED THROUGH EMERGING GENETIC-ENGINEERING TECHNOLOGIES
Increased Precision and Complexity of Genetic-Engineering Alterations
At the time that the committee wrote its report, major commercialized GE crops had been engineered by using Agrobacterium tumefaciens mediated or gene gun-mediated transformation, both of which result in semirandom insertion of the transgene into the genome. Variation in expression of the transgene was routinely observed because of the specific genomic characteristics of the insertion sites. Because of that variation, there was a need to screen large numbers of transgenic plants to identify the optimal transgenic individual. Regulations in the United States require approval of each transformation event regardless of whether the transgene itself was previously approved for release in that crop. That is at least in part because of the potential for unintended effects of each insertion.
Precision genome-editing technologies now permit insertion of single or multiple genes into one targeted location in the genome and thereby eliminate variation that is due to position effects (see Chapter 7 ). Such precision is expected to decrease unintended effects of gene insertion, although it will not eliminate the effects of somaclonal variation (discussed in Chapter 7 ).
Consider, for example, the engineering of completely new metabolic pathways into a plant for nutritional enhancement. The simplest example would be a set of two genes, such as has been used to create Golden Rice to deliver precursors of vitamin A. A more complex example would be engineering of fish oils (very long-chain unsaturated fatty acids) to improve the health profile of plant oils; depending on the target species, this process has required introduction of at least of three and at most nine transgenes ( Abbadi et al., 2004 ; Wu et al., 2005 ; Ruiz-Lopez et al., 2014 ). If each of those transgenes is integrated into the genome on a different chromosome on the basis of separate insertion events, it will require a number of generations of crosses to put them all together in one plant. If, instead, all the transgenes could be targeted at the same site on a chromosome either simultaneously or one after another, they would not segregate from each other as they were moved into elite varieties. From a food-safety perspective, engineering transgenes into a single target locus also ensures that expression of the whole pathway is preserved so that the correct end product accumulates. Emerging genetic-engineering technologies currently enable insertion of a few genes in one construct, but in the future that number may increase dramatically.
In the future, the scale of genetic-engineering alterations may go much further than just manipulating oil profiles. The committee heard from speakers about projects aimed at changing the entire photosynthetic pathway of the rice plant ( Weber, 2014 ) to create an entirely novel crop ( Zhu et al., 2010 ; Ruan et al., 2012 ). The committee also heard from researchers interested in developing cereal crops with nitrogen fixation. Those projects are discussed further in Chapter 8 . Although the precision of future genetic-engineering alterations should decrease unintended effects of the process of engineering, the complexity of the changes in a plant may leave it not substantially equivalent to its non-GE counterpart.
It is also important to note that crops that use RNA interference (RNAi) were coming on the market when the committee was writing its report. EPA convened a science advisory panel to evaluate hazards that might arise from use of this genetic-engineering approach. The panel concluded that “dietary RNA is extensively degraded in the mammalian digestive system by a combination of ribonucleases (RNases) and acids that are likely to ensure that all structural forms of RNA are degraded throughout the digestive process. There is no convincing evidence that ingested [double-stranded] RNA is absorbed from the mammalian gut in a form that causes physiologically relevant adverse effects” ( EPA, 2014c:14 ). When the committee was writing its report, deployment of dietary RNAi was a new technology. EPA's panel made a number of recommendations, including investigating factors that may affect absorption and effects of dietary double-stranded RNAs and investigating the stability of double-stranded RNA in people who manifest diseases.
FINDING: The precision of emerging genetic-engineering technologies should decrease some sources of unintended changes in the plants, thus simplifying food-safety testing. However, engineering involving major changes in metabolic pathways or insertion of multiple resistance genes will complicate the determination of food safety because changes in metabolic pathways are known to have unexpected effects on plant metabolites.
Increased Diversity of Crops To Be Engineered
The most far-ranging effects of emerging genetic-engineering technologies may be the diversity of crops that will be engineered and commercialized. Commercial GE crops at the time the committee conducted its review were mainly high-production commodity crops (maize, soybean, and cotton) engineered with trans-kingdom genes, but the applications of emerging genetic-engineering technologies are much broader: these technologies can be easily applied to any plant species that can be regenerated from tissue culture. Furthermore, the emerging technologies described in Chapter 7 can focus on any gene in which an altered nucleotide sequence results in a desired trait.
As a consequence, the committee expects a sizable increase in the number of food-producing crop species that are genetically altered. Examples of new target crops include forages (grasses and legumes), beans, pulses, a wide array of vegetables, herbs, and spices, and plants grown for flavor compounds. New traits will probably include fiber content (either increased to add more fiber or decreased to improve digestibility), altered oil profiles, decreased concentrations of antinutrients, increased or more consistent concentrations of such phytochemicals as antioxidants (for example, flavonoids) and phytoestrogens (for example, isoflavones or lignans), and increased mineral concentrations. Some of these are considered further in Chapter 8 .
From a food-safety perspective, the increase in crops and traits presents a number of challenges. First is the need to develop better and more detailed baseline data on the general chemical composition and probably the transcriptomic profiles of currently marketed conventionally bred varieties of the crops (see Chapter 7 ). Perhaps more problematic will be designing whole-food animal-testing regimens if the food from the crop cannot be used as a major component of the test animals' diet. Maize, rice, soybean, and other grains can be added to diets at up to 30 percent without adverse effects on animal health. That is unlikely to be the case with new spices or some vegetables. It would be beneficial if new, publicly acceptable approaches for testing could be developed that do not require animal testing ( NRC, 2007 ; Liebsch et al., 2011 ; Marx-Stoelting et al., 2015 ). Chapter 9 addresses the potential need to move to an entirely product-based approach to regulation and testing based on the novelty of a new crop or food.
FINDING: Some future GE crops will be designed to be substantially different from current crops and may not be as amenable to animal testing as currently marketed GE crops.
RECOMMENDATION: There is an urgent need for publicly funded research on novel molecular approaches for testing future products of genetic engineering so that accurate testing methods will be available when the new products are ready for commercialization.
- CONCLUSIONS
The committee's objective in this chapter was to examine the evidence that supports or negates specific hypotheses and claims about the risks and benefits associated with foods derived from GE crops. As acknowledged at the beginning of the chapter, understanding the health effects of any food, whether non-GE or GE, can be difficult. The properties of most plant secondary metabolites are not understood, and isolating the effects of diet on animals, including humans, is challenging. Although there are well-developed methods for assessing potential allergenicity of novel foods, these methods could miss some allergens. However, the research that has been conducted in studies with animals and on chemical composition of GE foods reveals no differences that would implicate a higher risk to human health from eating GE foods than from eating their non-GE counterparts. Long-term epidemiological studies have not directly addressed GE food consumption, but available time-series epidemiological data do not show any disease or chronic conditions in populations that correlate with consumption of GE foods. The committee could not find persuasive evidence of adverse health effects directly attributable to consumption of GE foods.
New methods to measure food composition that involve transcriptomics, proteomics, and metabolomics provide a broad, nontargeted assessment of thousands of plant RNAs, proteins, and compounds. When the methods have been used, the differences found in comparisons of GE with non-GE plants have been small relative to the naturally occurring variation found in conventionally bred crop varieties. Differences that are detected by using -omics methods do not on their own indicate a safety problem.
There is some evidence that GE insect-resistant crops have had benefits to human health by reducing insecticide poisonings and decreasing exposure to fumonisins. Several crops had been developed or were in development with GE traits designed to benefit human health; however, when the committee was writing its report, commercialized crops with health benefits had been only recently introduced and were not widely grown, so the committee could not evaluate whether they had had their intended effects.
New crops developed with the use of emerging genetic-engineering technologies were in the process of being commercialized. The precision associated with the technologies should decrease some sources of unintended changes that occur when plants are genetically engineered and thus simplify food-safety testing. However, engineering involving major changes in metabolic pathways or insertion of multiple resistance genes will complicate the determination of food safety because changes in metabolic pathways are known to have unexpected effects on plant metabolites. Therefore, publicly funded research on novel approaches for testing future products of genetic engineering is needed so that accurate testing methods will be available when the new products are ready for commercialization.
The committee has compiled publicly available information on funding sources and first-author affiliation for the references cited in this chapter; the information is available at https://www .nationalacademies .org/ge-crops .
In November 2015, EPA took steps to withdraw the product's registration in light of new information that indicated there could be synergistic effects of the two herbicides, which could possibly result in greater toxicity to nontarget plants ( Taylor, 2015 ). A court ruling in January 2016 allowed the herbicide to remain on the market while EPA considered other administrative actions ( Callahan, 2016 ).
GE rice was not commercialized in 2015, but GE varieties in development have been tested.
OECD develops consensus documents that provide reference values for existing food crops ( OECD, 2015 ). These are publicly available online at http://www .oecd.org/science /biotrack/consensusdocumentsfortheworkonthesafetyofnovelfoodsandfeedsplants.htm (accessed May 9, 2016). The International Life Science Institute (ILSI) also maintains a crop composition database at www .cropcomposition.org (accessed May 9, 2016). ILSI reports that in 2013 the database contained more than 843,000 data points representing 3,150 compositional components.
Jung, R., W.-N. Hu, R.B. Meeley, V.J.H. Sewalt, and R. Nair. Grain quality through altered expression of seed proteins. U.S. Patent 8,546,646, filed September 14, 2012, and issued October 1, 2013.
Micronutrient deficiencies. Available at http://www .who.int/nutrition /topics/vad/en/ . Accessed October 30, 2015.
What is the status of the Golden Rice project coordinated by IRRI? Available at http://irri .org/golden-rice /faqs/what-is-the-status-of-the-golden-rice-project-coordinated-by-irri . Accessed October 30, 2015.
Acrylamide concentrations reported by FDA were for individual purchased food products and were not adjusted for unit-to-unit variation.
- Abbadi A, Domergue F, Bauer J, Napier JA, Welti R, Zähringer U, Cirpus P, Heinz E. Biosynthesis of very-long-chain polyunsaturated fatty acids in transgenic oilseeds: Constraints on their accumulation. Plant Cell. 2004; 16 :2734–2748. [ PMC free article : PMC520968 ] [ PubMed : 15377762 ]
- Abbas HK, Shier WT, Cartwright RD. Effect of temperature, rainfall and planting date on aflatoxin and fumonisin contamination in commercial Bt and non-Bt maize hybrids in Arkansas. Phytoprotection. 2007; 88 :41–50.
- Abraham TM, Pencina KM, Pecina MJ, Fox CS. Trends in diabetes incidence: The Framingham heart study. Diabetes Care. 2015; 38 :482–487. [ PMC free article : PMC4338506 ] [ PubMed : 25552418 ]
- ADAS. Strategy support for the post-market monitoring (PMM) of GM plants: Review of existing PPM strategies developed for the safety assessment of human and animal health. EFSA supporting publication. 2015; 2014 :EN–739.
- Ahuja I, Kissen R, Bones AM. Phytoalexins in defense against pathogens. Trends in Plant Science. 2012; 17 :73–90. [ PubMed : 22209038 ]
- American Association for the Advancement of Science. Statement by the AAAS Board of Directors on Labeling of Genetically Modified Foods. Oct 20, 2012. [October 13, 2015]. http://www .aaas.org/sites /default/files/AAAS_GM_statement .pdf .
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Fifth. Arlington, VA: American Psychiatric Publishing; 2013.
- Amos J. French GM-fed Rat Study Triggers Furore. Online. BBC News. Sep 19, 2012. [December 13, 2015]. http://www .bbc.com/news /science-environment-19654825 .
- An R. Educational disparity in obesity among U.S. adults 1984–2013. Annals of Epidemiology. 2015; 25 :637–642. [ PubMed : 26187624 ]
- Arjó G, Capell T, Matias-Guiu X, Zhu C, Christou P, Piñol C. Mice fed on a diet enriched with genetically engineered multivitamin corn show no sub-acute toxic effects and no sub-chronic toxicity. Plant Biotechnology Journal. 2012; 10 :1026–1034. [ PubMed : 22928600 ]
- Astwood JD, Leach JN, Fuchs RL. Stability of food allergens to digestion in vitro. Nature Biotechnology. 1996; 14 :1269–1273. [ PubMed : 9631091 ]
- Bartholomaeus A, Parrott W, Bondy G, Walker K. The use of whole food animal studies in the safety assessment of genetically modified crops: Limitations and recommendations. Critical Reviews in Toxicology. 2013; 43 :1–24. [ PMC free article : PMC3833814 ] [ PubMed : 24164514 ]
- Belknap JK, Mitchell SR, O'Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behavior Genetics. 1996; 26 :149–160. [ PubMed : 8639150 ]
- Bennett R, Morse S, Ismael Y. The economic impact of genetically modified cotton on South African smallholders: Yield, profit and health effects. Journal of Development Studies. 2006; 42 :662–677.
- Berry C. Letter to the Editor. Food and Chemical Toxicology. 2013; 53 :445–446. [ PubMed : 23142391 ]
- Bhatnagar-Mathur P, Sunkara S, Bhatnagar-Panwar M, Waliyar F, Sharma KK. Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Science. 2015; 234 :119–132. [ PubMed : 25804815 ]
- Birol E, Meenakshi JV, Oparinde A, Perez S, Tomlins K. Developing country consumers' acceptance of biofortified foods: A synthesis. Food Security. 2015; 7 :555–568.
- Bøhn T, Cuhra M, Traavik T, Sanden M, Fagan J, Primicerio R. Compositional differences in soybeans on the market: Glyphosate accumulates in Roundup Ready GM soybeans. Food Chemistry. 2014; 153 :207–215. [ PubMed : 24491722 ]
- Boobis AR, Ossendorp BC, Banasiak U, Hamey PY, Sebestyen I, Moretto A. Cumulative risk assessment of pesticide residues in food. Toxicology Letters. 2008; 180 :137–150. [ PubMed : 18585444 ]
- Bowen KL, Flanders KL, Hagan AK, Ortiz B. Insect damage, aflatoxin content, and yield of Bt corn in Alabama. Journal of Economic Entomology. 2014; 107 :1818–1827. [ PubMed : 26309272 ]
- Bowers E, Hellmich R, Munkvold G. Comparison of fumonisn contamination using HPLC and ELISA methods in Bt and near-isogenic maize hybrids infested with European corn borer or Western bean cutworm. Journal of Agricultural and Food Chemistry. 2014; 62 :6463–6472. [ PubMed : 24964132 ]
- Brigulla M, Wackernagel W. Molecular aspects of gene transfer and foreign DNA acquisition in prokaryotes with regard to safety issues. Applied Microbiology and Biotechnology. 2010; 86 :1027–1041. [ PubMed : 20191269 ]
- Brix AE, Nyska A, Haseman JK, Sells DM, Jokinen MP, Walker NJ. Incidences of selected lesions in control female Harlan Sprague–Dawley rates from two-year studies performed by the National Toxciology Program. Toxicologic Pathology. 2005; 33 :477–483. [ PubMed : 16036865 ]
- Brouk MJ, Cvetkovic B, Rice DW, Smith BL, Hinds MA, Owens FN, Iiams C, Sauber TE. Performance of lactating dairy cows fed corn as whole plant silage and grain produced from genetically modified corn containing event DAS-59122–7 compared to a nontransgenic, near-isogenic control. Journal of Dairy Science. 2011; 94 :1961–1966. [ PubMed : 21426987 ]
- Bush RK, Taylor SL, Nordlee JA, Busse WW. Soybean oil is not allergenic to soybean-sensitive individuals. Journal of Allergy and Clinical Immunology. 1985; 76 :242–245. [ PubMed : 3894482 ]
- Butler D. Hyped GM Maize Study Faces Growing Scrutiny. Online. Nature. Oct 11, 2012. [December 13, 2015]. http://www .nature.com /news/hyped-gm-maize-study-faces-growingscrutiny-1.11566 . [ PubMed : 23060167 ]
- Buzoianu SG, Walsh MC, Rea MC, O'Donovan O, Gelencsér E, Ujhelyi G, Szabó E, Nagy A, Ross RP, Gardiner GE, Lawlor PG. Effects of feeding Bt maize to sows during gestation and lactation on maternal and offspring immunity and fate of transgenic material. PLoS ONE. 2012a; 7 :e47851. [ PMC free article : PMC3473024 ] [ PubMed : 23091650 ]
- Buzoianu SG, Walsh MC, Rea MC, Cassidy JP, Ross RP, Gardiner GE, Lawlor PG. Effect of feeding genetically modified Bt MON810 maize to approximately 40-day-old pigs for 110 days on growth and health indicators. Animal. 2012b; 6 :1609–1619. [ PubMed : 23031560 ]
- Buzoianu SG, Walsh MC, Rea MC, O'Sullivan O, Crispie F, Cotter PD, Ross PR, Gardiner GE, Lawlor PG. The effect of feeding Bt MON810 maize to pigs for 110 days on intestinal microbiota. PLoS ONE. 2012c; 7 :e33668. [ PMC free article : PMC3344822 ] [ PubMed : 22574106 ]
- Buzoianu SG, Walsh MC, Rea MC, O'Sullivan O, Crispie F, Cotter PD, Ross PR, Gardiner GE, Lawlor PG. High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt maize) for 31 days. Applied and Environmental Microbiology. 2012d; 78 :4217–4224. [ PMC free article : PMC3370545 ] [ PubMed : 22467509 ]
- Buzoianu SG, Walsh MC, Rea MC, Quigley L, O'Sullivan O, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. Sequence-based analysis of the intestinal Microbiota of sows and their offspring fed genetically modified maize expressing a truncated form of Bacillus thuringiensis Cry1Ab protein (Bt Maize). Applied and Environmental Microbiology. 2013a; 79 :7735–7744. [ PMC free article : PMC3837803 ] [ PubMed : 24096421 ]
- Buzoianu SG, Walsh MC, Rea MC, Cassidy JP, Ryan TP, Ross PR, Gardiner GE, Lawlor PG. Transgenerational effects of feeding genetically modified maize to nulliparious sows and offspring on offspring growth and health. Journal of Animal Science. 2013b; 91 :318–330. [ PubMed : 23097397 ]
- CAC (Codex Alimentarius Commission). Guideline for the Conduct of Food Safety Assessment of Foods Using Recombinant DNA Plants. Doc CAC/GL 45-2003. Rome: World Health Organization and Food and Agriculture Organization; 2003.
- CAC (Codex Alimentarius Commission). Annex 2: Food Safety Assessment of Foods Derived from Recombinant-DNA Plants Modified for Nutritional or Health Benefits in Guideline for the Conduct of Food Safety Assessment of Foods Using Recombinant DNA Plants. Doc CAC/GL 45-2003. Rome: World Health Organization and Food and Agriculture Organization; 2008.
- CAC (Codex Alimentarius Commission). Foods Derived from Modern Biotechnology. Rome: World Health Organization and Food and Agriculture Organization; 2009.
- Callahan P. Court clears way for revival of worrisome weedkiller. EPA nixes approval of Enlist Duo weed killer. Online. Chicago Tribune. Jan 28, 2016. [March 21, 2016]. http://www .chicagotribune .com/news/watchdog /ct-dow-enlist-duo-court-ruling-20160127-
- Calsamiglia S, Hernandez B, Hartnell GF, Phipps R. Effects of corn silage derived from a genetically modified variety containing two transgenes on feed intake, milk production, and composition, and the absence of detectable transgenic deoxyribonucleic acid in milk in Holstein dairy cows. Journal of Dairy Science. 2007; 90 :4718–4723. [ PubMed : 17881694 ]
- Catassi C, Kryszak D, Bhatti B, Strugeon C, Helzlsouer K, Clipp SL, Gelfond D, Puppa E, Sferruzza A, Fasano A. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Annals of Medicine. 2010; 42 :530–538. [ PubMed : 20868314 ]
- CDC (Centers for Disease Control and Prevention). Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2010. Morbidity and Mortality Weekly Report. 2014; 63 :1–21. [ PubMed : 24670961 ]
- Coresh J, Astor BC, Greene T, Eknoyan G, Levey AS. Prevalence of chronic kidney disease and decreased kidney function in the adult US population: Third national health and nutrition examination survey. American Journal of Kidney Diseases. 2003; 41 :1–12. [ PubMed : 12500213 ]
- Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS. Prevalence of Chronic Kidney Disease in the United States. Journal of the American Medical Association. 2007; 298 :2038–2047. [ PubMed : 17986697 ]
- Costa J, Mafra I, Amaral JS, Oliveira MBPP. Detection of genetically modified soybean DNA in refined vegetable oils. European Food Research and Technology. 2010a; 230 :915–923.
- Costa J, Mafra I, Amaral JS, Oliveira MBPP. Monitoring genetically modified soybean along the industrial soybean oil extraction and refining processes by polymerase chain reaction techniques. Food Research International. 2010b; 43 :301–306.
- Council on Science and Public Health of the American Medical Association House of Delegates. Report 2 (A-12). Labeling of Bioengineered Foods (Resolutions 508 and 509-A-11). 2012. [March 12, 2016]. http: //factsaboutgmos .org/sites/default/files/AMA%20Report .pdf .
- Datta SK, Datta K, Parkhi V, Rai M, Baisakh N, Sahoo G, Rehana S, Bandyopadhyay A, Alamgir M, Ali MS, Abrigo E, Oliva N, Torrizo L. Golden Rice: Introgression, breeding, and field evaluation. Euphytica. 2007; 154 :271–278.
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014; 505 :559–563. [ PMC free article : PMC3957428 ] [ PubMed : 24336217 ]
- Davis RK, Stevenson GT, Busch KA. Tumor incidence in normal Sprague-Dawley female rats. Cancer Research. 1956; 16 :194–197. [ PubMed : 13304860 ]
- DeFrancesco L. How safe does transgenic food need to be? Nature Biotechnology. 2013; 31 :794–802. [ PubMed : 24022153 ]
- de Luis R, Lavilla M, Sanchez L, Calvo M, Perez MD. Immunochemical detection of Cry1A(b) protein in model processed foods made with transgenic maize. European Food Research and Technology. 2009; 229 :15–19.
- De Moura FF, Miloff A, Boy E. Retention of provitamin A carotenoids in staple crops targeted for biofortification in Africa: Cassava, maize and sweet potato. Critical Reviews in Food Science and Nutrition. 2015; 55 :1246–1269. [ PMC free article : PMC4353306 ] [ PubMed : 24915386 ]
- De Roos AJ, Blair A, Rusiecki JA, Hoppin JA, Svec M, Dosemeci M, Sandler DP, Alavanja MC. Cancer incidence among glyphosate-exposed pesticide applicators in the Agricultural Health Study. Environmental Health Perspective. 2005; 113 :49–54. [ PMC free article : PMC1253709 ] [ PubMed : 15626647 ]
- Demont M, Stein AJ. Global value of GM rice: A review of expected agronomic and consumer benefits. New Biotechnology. 2013; 30 :426–436. [ PubMed : 23628812 ]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108 :4554–4561. [ PMC free article : PMC3063582 ] [ PubMed : 20847294 ]
- Dev MS, Rao NC. Socio-economic Impact of Bt Cotton. Monograph No. 3. Hyderabad: Centre for Economic and Social Studies; 2007.
- Diaz C, Fernandez C, McDonald R, Yeung JM. Determination of Cry9C protein in processed foods made with StarLink™ corn. Journal of AOAC International. 2002; 85 :1070–1076. [ PubMed : 12374406 ]
- Dinse GE, Peddada SD, Harris SF, Elmore SA. Comparison of NTP historical control tumor incidence rates in female Harlan Sprague–Dawley and Fischer 344/N rats. Toxicologic Pathology. 2010; 38 :765–775. [ PMC free article : PMC4791045 ] [ PubMed : 20622195 ]
- Dixon RA. Natural products and disease resistance. Nature. 2001; 411 :843–847. [ PubMed : 11459067 ]
- Dixon RA. Phytoestrogens. Annual Review of Plant Biology. 2004; 55 :225–261. [ PubMed : 15377220 ]
- Domingo JL, Bordonaba JG. A literature review on the safety assessment of genetically modified plants. Environment International. 2011; 37 :734–742. [ PubMed : 21296423 ]
- Dona A, Arvanitoyannis IS. Health risks of genetically modified foods. Critical Reviews in Food Science and Nutrition. 2009; 49 :164–175. [ PubMed : 18989835 ]
- Duke SO, Rimando AM, Pace PF, Reddy KN, Smeda RJ. Isoflavone, glyphosate, and aminomethylphosphonic acid levels in seeds of glyphosate-treated, glyphosate resistant soybean. Journal of Agricultural and Food Chemistry. 2003; 51 :340–344. [ PubMed : 12502430 ]
- Dung LT, Ham LH. Comments on “Long term toxicity of a Roundup herbicide and a Roundup-tolerant genetically modified maize. Food and Chemical Toxicology. 2013; 53 :443–444. [ PubMed : 23142394 ]
- Dunn OJ. Multiple comparisons among means. Journal of the American Statistical Association. 1961; 56 :52–64.
- EC (European Commission). A Decade of EU-funded GMO Research (2001–2010). Brussels: European Commission; 2010a.
- EC (European Commission). Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Official Journal of the European Union. 2010b; 276 :33–79.
- EC (European Commission). Commission implementing regulation (EU) No 503/2013 of 3 April 2013 on applications for authorisation of genetically modified food and feed in accordance with Regulation (EC) No 1829/2003 of the European Parliament and of the Council and amending Commission Regulations (EC) No 641/2004 and (EC) No 1981/2006. Official Journal of the European Union. 2013; 157 :1–48.
- EFSA (European Food Safety Authority). Statement of the Scientific Panel on Genetically Modified Organisms on the Analysis of Data from a 90-day Rat Feeding Study with MON 863 Maize. 2007. [December 13, 2015]. http://www .efsa.europa .eu/sites/default/files /scientific_output /files/main_documents /GMO_statement_MON863%2C0.pdf .
- EFSA (European Food Safety Authority). Scientific opinion on the assessment of allergenicity of GM plants and microorganisms and derived food and feed. EFSA Journal. 2010; 8 :1700.
- EFSA (European Food Safety Authority). Guidance on risk assessment of food and feed from genetically modified plants. EFSA Journal. 2011a; 9 :2150.
- EFSA (European Food Safety Authority). Scientific opinion on guidance on conducting repeated-dose 90-day oral toxicity study in rodents on whole food/feed. EFSA Journal. 2011b; 9 :2438.
- EFSA (European Food Safety Authority). Statistical significance and biological relevance. EFSA Journal. 2011c; 9 :2372.
- EFSA (European Food Safety Authority). Review of the Séralini et al. (2012) publication on a 2-year rodent feeding study with glyphosate formulations and GM maize NK603 as published online on 19 September 2012 in Food and Chemical Toxicology. EFSA Journal. 2012; 10 :2910.
- EFSA (European Food Safety Authority). Scientific Opinion on application EFSA-GMO-NL-2007-45 for the placing on the market of herbicide-tolerant, high-oleic acid, genetically modified soybean 305423 for food and feed uses, import and processing under Regulation (EC) No 1829/2003 from Pioneer. EFSA Journal. 2013; 11 :3499.
- EFSA (European Food Safety Authority). Conclusion on the peer review of the pesticide risk assessment for the active substance glyphosate. EFSA Journal. 2015; 13 :4302.
- Einspanier R. The fate of transgenic DNA and newly expressed proteins. In: Flachowsky G, editor. Animal Nutrition with Transgenic Plants. Oxfordshire: UK: CABI Biotechnology source; 2013. pp. 130–139.
- El Ouakfaoui S, Miki B. The stability of the Arabidopsis transcriptome in transgenic plants expressing the marker genes nptII and uidA. Plant Journal. 2005; 41 :791–800. [ PubMed : 15743445 ]
- Entine J. A Science-based Look at Genetically Engineered Crops. Presentation to the National Academy of Sciences' Committee on Genetically Engineered Crops: Past Experience and Future Prospects, September 16. Washington, DC: 2014.
- EPA (U.S. Environmental Protection Agency). Good laboratory practice standards. Federal Register. 1989; 54 :34067.
- EPA (U.S. Environmental Protection Agency). Bacillus thuringiensis subspecies Cry1F Protein and the Genetic Material Necessary for Its Production (Plasmid Insert PHI 8999) in Corn. 2001a. [October 10, 2015]. http://ofmpub .epa.gov /apex/pesticides/f?p =chemicalsearch:3:0 ::no:1,3,31,7,12,25:p3_xchemical_id:1322 .
- EPA (U.S. Environmental Protection Agency). EPA Releases Draft Report on Starlink corn. Online. EPA Press Release. 2001b. [March 12, 2016]. https://yosemite .epa .gov/opa/admpress.nsf /blab9f485b098972852562e7004dc686 /cd9013801973259885256a0800710574?OpenDocument .
- EPA (U.S. Environmental Protection Agency). Glyphosate; Pesticide tolerances. Federal Register. 2013; 78 :25396–25401.
- EPA (U.S. Environmental Protection Agency). Final Registration of Enlist Duo™ Herbicide. Oct 15, 2014a. [October 13, 2015]. http://www2 .epa.gov/sites /production/files /2014-10/documents /final_registration_-_enlist_duo.pdf .
- EPA (U.S. Environmental Protection Agency). Memorandum: Response to Public Comments Received Regarding New Uses of Enlist Duo™ on Corn and Soybeans. Oct 14, 2014b. [October 10, 2015]. http://www2 .epa.gov/sites /production/files /2014-10/documents /response_to_comments.pdf .
- EPA (U.S. Environmental Protection Agency). SAP Minutes No. 2014-02, A Set of Scientific Issues Being Considered by the Environmental Protection Agency Regarding: RNAi Technology: Problem Formulation for Human Health and Ecological Risk Assessment. 2014c. [March 13, 2016]. https://www .epa.gov/sites /production/files /2015-06/documents/012814minutes.pdf .
- EPA (U.S. Environmental Protection Agency). EDSP Weight of Evidence Analysis of Potential Interaction with the Estrogen, Androgen, or Thyroid Pathway; Chemical: Glyphosate. 2015. [March 13, 2016]. https://www .epa.gov/sites /production/files /2015-06/documents /glyphosate-417300_2015-06-29 _txr0057175.pdf .
- Fagan J, Antoniou M, Robinson C. GMO Myths and Truths. London: Earth Open Source; 2014.
- Fasano A, Berti I, Gerarduzzi T, Not T, Colletti RB, Drago S, Elitsur Y, Green PH, Guandalini S, Hill ID, Pietzak M. Prevalence of celiac disease in at-risk and not-at-risk groups in the United States: A large multicenter study. Archives of Internal Medicine. 2003; 163 :286–292. [ PubMed : 12578508 ]
- FDA (U.S. Food and Drug Administration). Good Laboratory Practice Regulations Management Briefings: Post Conference Report. Rockville, MD: FDA; 1979.
- FDA (U.S. Food and Drug Administration). revised 2007. Guidance for Industry and Other Stakeholders: Toxicological Principles for the Safety Assessment of Food Ingredients (Redbook 2000). 2000a. [October 29, 2015]. http://www .fda.gov/Food /GuidanceRegulation /GuidanceDocumentsRegulatoryInformation /IngredientsAdditivesGRASPackaging /ucm2006826.htm .
- FDA (U.S. Food and Drug Administration). revised 2006. Survey Data on Acrylamide in Food: Total Diet Study Results. 2000b. [October 30, 2015]. http://www .fda.gov/Food /FoodborneIllnessContaminants /ChemicalContaminants /ucm053566.htm .
- FDA (U.S. Food and Drug Administration). revised 2006. Survey Data on Acrylamide in Food: Individual Food Products. 2002. [December 22, 2015]. http://www .fda.gov/food /foodborneillnesscontaminants /chemicalcontaminants /ucm053549.htm .
- FDA (U.S. Food and Drug Administration). Biotechnology Consultation Note to the File BNF No 000133. Dec 16, 2013. [October 29, 2015]. http://www .fda.gov/Food /FoodScienceResearch /GEPlants/Submissions/ucm382207.htm .
- FDA (U.S. Food and Drug Administration). Biotechnology Consultation Agency Response Letter BNF No 000141. Mar 20, 2015. [October 30, 2015]. http://www .fda.gov/Food /FoodScienceResearch /GEPlants/Submissions/ucm436169.htm .
- Fernandez A, Mills ENC, Lovik M, Spök A, Germini A, Mikalsen A, Wal JM. Endogenous allergens and compositional analysis in the allergenicity assessment of genetically modified plants. Food and Chemical Toxicology. 2013; 62 :1–6. [ PubMed : 23959104 ]
- Feron VJ, Groten JP. Toxicological evaluation of chemical mixtures. Food and Chemical Toxicology. 2002; 40 :825–839. [ PubMed : 11983277 ]
- Ferruzzi MG. The influence of beverage composition on delivery of phenolic compounds from coffee and tea. Physiology & Behavior. 2010; 100 :33–41. [ PubMed : 20138903 ]
- Finamore A, Roselli M, Britti S, Monastra G, Ambra R, Turrini A, Mengheri E. Intestinal and peripheral immune response to MON810 maize ingestion in weaning and old mice. Journal of Agricultural and Food Chemistry. 2008; 56 :11533–11539. [ PubMed : 19007233 ]
- Folta K. Letter to the Editor. Food and Chemical Toxicology. 2014; 65 :392. [ PubMed : 24394484 ]
- Fonseca C, Planchon S, Renaut J, Oliveira MM, Batista R. Characterization of maize allergens—MON810 vs. its non-transgenic counterpart. Journal of Proteomics. 2012; 75 :2027–2037. [ PubMed : 22270010 ]
- Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJL, Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: A systematic analysis. Lancet. 2011; 378 :1461–1484. [ PubMed : 21924486 ]
- Franz JE, Mao MK, Sikorski JA. Glyphosate: A Unique Global Herbicide. ACS Monograph 189. Washington, DC: American Chemical Society; 1997.
- Friedman M. Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. Journal of Agricultural and Food Chemistry. 2006; 54 :8655–8681. [ PubMed : 17090106 ]
- Fryar CD, Carroll MD, Ogden CL. Prevalence of overweight, obesity, and extreme obesity among adults: United States, 1960–1962 through 2011–2012. 2014. [October 13, 2015]. http://www .cdc.gov/nchs /data/hestat/obesity_adult_11_12 /obesity_adult_11_12.pdf .
- FSANZ (Food Standards Australia New Zealand). GM Food Labelling. 2013. [December 22, 2015]. http://www .foodstandards .gov.au/consumer /gmfood/labelling/Pages/default.aspx .
- Fukushima A, Kusano M, Mejia RF, Iwasa M, Kobayashi M, Hayashi M, Watanabe-Takahashi A, Narisawa T, Tohge T, Hur M, Sykin Wurtele E, Nikolau BJ, Saito K. Metabolomic characterization of knockout mutants in Arabidopsis: Development of a metabolite profiling database for knockout mutants in Arabidopsis. Plant Physiology. 2014; 165 :948–961. [ PMC free article : PMC4081348 ] [ PubMed : 24828308 ]
- Furgał-Dierżuk I, Strzetelski J, Twardowska M, Kwiatek K, Mazur M. The effect of genetically modified feeds on productivity, milk composition, serum metabolite profiles and transfer of tDNA into milk of cows. Journal of Animal and Feed Sciences. 2015; 24 :19–30.
- Gannon B, Kaliwile C, Arscott SA, Schmaelzle S, Chileshe J, Kalungwana N, Mosonda M, Pixley K, Masi C, Tanumihardjo SA. Biofortified orange maize is as efficacious as a vitamin A supplement in Zambian children even in the presence of high liver reserves of vitamin A: A community-based, randomized placebo-controlled trial. American Journal of Clinical Nutrition. 2014; 100 :1541–1550. [ PMC free article : PMC4232019 ] [ PubMed : 25411289 ]
- García-Villalba R, León C, Dinelli G, Segura-Carretero A, Fernández-Gutiérrez A, Garcia-Cañas V, Cifuentes A. Comparative metabolomic study of transgenic versus conventional soybean using capillary electrophoresis-time-of-flight mass spectrometry. Journal of Chromatography A. 2008; 1195 :164–173. [ PubMed : 18508066 ]
- Gasnier C, Dumont C, Benachour N, Clair E, Chagnon MC, Séralini G-É. Glyphosate-based herbicides are toxic and endocrine disruptors in human cell lines. Toxicology. 2009; 262 :184–191. [ PubMed : 19539684 ]
- Goodman R. Evaluating GE Food Sources for Risks of Allergy: Methods, Gaps and Perspective. Presentation to the National Academy of Sciences' Committee on Genetically Engineered Crops: Past Experience and Future Prospects, March 5. Washington, DC: 2015.
- Goodman RE, Panda R, Ariyarathna H. Evaluation of endogenous allergens for the safety evaluation of genetically engineered food crops: Review of potential risks, test methods, examples and relevance. Journal of Agricultural and Food Chemistry. 2013; 61 :8317–8332. [ PubMed : 23848840 ]
- Graf L, Hayder H, Mueller U. Endogenous allergens in the regulatory assessment of genetically engineered crops. Food and Chemical Toxicology. 2014; 73 :17–20. [ PubMed : 25128445 ]
- Green PH, Cellier C. Celiac disease. New England Journal of Medicine. 2007; 357 :1731–1743. [ PubMed : 17960014 ]
- Guertler P, Paul V, Albrecht C, Meyer HHD. Sensitive and highly specific quantitative real-time PCR and ELISA for recording a potential transfer of novel DNA and Cry1Ab protein from feed into bovine milk. Analytical and Bioanalytical Chemistry. 2009; 393 :1629–1638. [ PubMed : 19225766 ]
- Gupta R, Sheikh A, Strachan DP, Anderson HR. Time trends in allergic disorders in the UK. Thorax. 2007; 62 :91–96. [ PMC free article : PMC2111268 ] [ PubMed : 16950836 ]
- Guyton KZ, Loomis D, Grosse Y, Ghissassi FE, Benbrahim-Tallaa L, Guha N, Scoccianti C, Mattock H, Straif K. Carcinogenicity of tetrachlorvinphos, parathion, malathion, diazinon, and glyphosate. Lancet Oncology. 2015; 16 :490–491. [ PubMed : 25801782 ]
- Halle I, Flachowsky G. A four-generation feeding study with genetically modified (Bt) maize in laying hens. Journal of Animal and Feed Sciences. 2014; 23 :58–63.
- Hammond B, Dudek R, Lemen J, Nemeth M. Results of a 13 week safety assurance study with rats fed grain from glyphosate tolerant corn. Food and Chemical Toxicology. 2004; 42 :1003–1014. [ PubMed : 15110110 ]
- Hammond BG, Dudek R, Lemen JK, Nemeth MA. Results of a 90-day safety assurance study with rats fed grain from corn borer-protected corn. Food and Chemical Toxicology. 2006; 44 :1092–1099. [ PubMed : 16487643 ]
- Hammond B, Goldstein DA, Saltmiras D. Response to original research article, ‘Long term toxicity of a Roundup herbicide and a Roundup-tolerant genetically modified maize.’ Food and Chemical Toxicology. 2013; 53 :459–464. [ PubMed : 23142397 ]
- Hayes AW. Retraction notice to “Long term toxicity of a Roundup herbicide and a Roundup-tolerant genetically modified maize.” Food and Chemical Toxicology. 2014; 63 :244. [ PubMed : 24490213 ]
- He XY, Huang KL, Li X, Qin W, Delaney B, Luo YB. Comparison of grain from corn rootworm resistant transgenic DAS-59122-7 maize with non-transgenic maize grain in a 90-day feeding study in Sprague-Dawley rats. Food and Chemical Toxicology. 2008; 46 :1994–2002. [ PubMed : 18381227 ]
- He XY, Mao Z, Tang YB, Luo X, Li SS, Cao JY, Delaney B, Kun LH. A 90-day toxicology study of transgenic lysine-rich maize grain (Y642) in Sprague-Dawley rats. Food and Chemical Toxicology. 2009; 47 :425–432. [ PubMed : 19073230 ]
- Health Canada. Novel Food Information—Food Biotechnology, High Lauric Acid Canola Lines 23-198, 23-18-17. 1999. [May 11, 2016]. http://www .hc-sc.gc.ca /fn-an/alt_formats /hpfb-dgpsa/pdf/gmf-agm /ofb-096-100-a-eng.pdf .
- Health Canada. Proposed Re-evaluation Decision PRVD2015-01, Glyphosate. 2015. [March 13, 2016]. http://www .hc-sc.gc.ca /cps-spc/pest/part /consultations/_prvd2015-01 /prvd201501-eng.php .
- Hefferon KL. Nutritionally enhanced food crops; Progress and perspectives. International Journal of Molecular Sciences. 2015; 16 :3895–3914. [ PMC free article : PMC4346933 ] [ PubMed : 25679450 ]
- Hellenas KE, Branzell C, Johnsson H, Slanina P. High levels of glycoalkaloids in the established Swedish potato variety Magnum Bonum. Journal of the Science of Food and Agriculture. 1995; 23 :520–523.
- Herman RA, Price WD. Unintended compositional changes in genetically modified (GM) crops: 20 years of research. Journal of Agricultural and Food Chemistry. 2013; 61 :11695–11701. [ PubMed : 23414177 ]
- Herman RA, Storer NP, Gao Y. Digestion assays in allergenicity assessment of transgenic proteins. Environmental Health Perspectives. 2006; 114 :1154–1157. [ PMC free article : PMC1552003 ] [ PubMed : 16882518 ]
- Hernández AF, Parrón T, Tsatsakis AM, Requena M, Alarcón R, López-Guarnido O. Toxic effects of pesticide mixtures at a molecular level: Their relevance to human health. Toxicology. 2013; 307 :136–145. [ PubMed : 22728724 ]
- Hidalgo FJ, Zamora R. Peptides and proteins in edible oils: Stability, allergenicity, and new processing trends. Trends in Food Science & Technology. 2006; 17 :56–63.
- Hilbeck A, Binimelis R, Defarge N, Steinbrecher R, Székács A, Wickson F, Antoniou M, Bereano PL, Clark EA, Hansen M, Novotny E, Heinemann J, Meyer H, Shiva V, Wynne B. No scientific consensus on GMO safety. Environmental Sciences Europe. 2015; 27 :4.
- Hohlweg U, Doerfler W. On the fate of plant or other foreign genes upon the uptake in food or after intramuscular injection in mice. Molecular Genetics and Genomics. 2001; 265 :225–233. [ PubMed : 11361332 ]
- Hotz C, Loechl C, Lubowa A, Tumwine JK, Ndeezi G, Nandutu Masawi A, Baingana R, Carriquiry A, de Brauw A, Meenakshi JV, Gilligan DO. Introduction of beta-carotene-rich orange sweet potato in rural Uganda resulted in increased vitamin A intakes among children and women and improved vitamin A status among children. Journal of Nutrition. 2012a; 142 :1871–1880. [ PubMed : 22875553 ]
- Hotz C, Loechl C, de Brauw A, Eozenou P, Gilligan D, Moursi M, Munhaua B, van Jaarsveld P, Carriquiry A, Meenakshi JV. A large-scale intervention to introduce orange sweet potato in rural Mozambique increases vitamin A intakes among children and women. British Journal of Nutrition. 2012b; 108 :163–176. [ PubMed : 22018075 ]
- Huang J, Hu R, Fan C, Pray CE, Rozelle S. Bt cotton benefits, costs, and impacts in China. AgBioForum. 2002; 5 :153–166.
- Huang J, Hu R, Rozelle S, Pray C. Insect-resistant GM rice in farmers' fields: Assessing productivity and health effects in China. Science. 2005; 308 :688–690. [ PubMed : 15860626 ]
- IARC (International Agency for Research on Cancer). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 82: Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene, and Styrene. Lyon, France: IARC; 2002. [ PMC free article : PMC4781602 ] [ PubMed : 12687954 ]
- IARC (International Agency for Research on Cancer). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 88: Formaldehyde, 2-Butoxyethanol, and 1-tert-Butoxypropan-2-ol. Lyon, France: IARC; 2006. [ PMC free article : PMC4781641 ] [ PubMed : 17366697 ]
- IARC (International Agency for Research on Cancer). Glyphosate. Part of Volume 112 in International Agency for Research on Cancer Monographs on the Evaulation of Carcinogenic Risks to Humans: Some Organophosphate Insecticides and Herbicides: Diazinon, Glyphosate, Malathion, Parathion, and Tetrachlorvinphos. 2015. [December 20, 2015]. http://monographs .iarc .fr/ENG/Monographs/vol112/mono112-09 .pdf .
- Ibáñez C, Simó C, García-Cañas V, Acunha T, Cifuentes A. The role of direct high-resolution mass spectrometry in foodomics. Analytical and Bioanalytical Chemistry. 2015; 407 :6275–6287. [ PubMed : 26143059 ]
- Jackson KD, Howie LD, Akinbami LJ. Trends in allergenic conditions among children: United States 1997–2011. NCHS Data Brief. 2013; 121 :1–8. [ PubMed : 23742874 ]
- Jaffe G. Issues for the Committee on Genetically Engineered Crops to Consider. Presentation to the National Academy of Sciences' Committee on Genetically Engineered Crops: Past Experience and Future Prospects, September 16. Washington, DC: 2014.
- John B. Letter to the Editor. Food and Chemical Toxicology. 2014; 65 :391. [ PubMed : 24394483 ]
- Johnson N. Retracted Roundup-fed Rat Research Republished. Online. The Grist. Jul 1, 2014. [December 13, 2015]. http://grist .org/food /retracted-roundup-fed-rat-research-republished/
- Jones YM, De Brauw A. Using Agriculture to Improve Child Health: Promoting Orange Sweet Potatoes Reduces Diarrhea. World Development. 2015; 74 :15–24.
- Joshi L, van Eck JM, Mayo K, Silvestro RG, Blake ME, Ganapathi T, Haridas V, Gutterman JU, Arntzen CJ. Metabolomics of plant saponins: Bioprospecting triterpene glycoside diversity with respect to mammalian cell targets. OMICS A Journal of Integrative Biology. 2002; 6 :235–246. [ PubMed : 12427275 ]
- Keese P. Risks from GMOs due to horizontal gene transfer. Environmental Biosafety Research. 2008; 7 :123–149. [ PubMed : 18801324 ]
- Kiliç A, Akay MT. A three-generation study with genetically modified Bt corn in rats: Biochemical and histopathological investigation. Food and Chemical Toxicology. 2008; 46 :1164–1170. [ PubMed : 18191319 ]
- Knudsen I, Poulsen M. Comparative safety testing of genetically modified foods in a 90-day rat feeding study design allowing the distinction between primary and secondary effects of the new genetic event. Regulatory Toxicology and Pharmacology. 2007; 49 :53–62. [ PubMed : 17719159 ]
- Kouser S, Qaim M. Impact of Bt cotton on pesticide poisoning in smallholder agriculture: A panel data analysis. Ecological Economics. 2011; 70 :2105–2113.
- Kouser S, Qaim M. Valuing financial, health, and environmental benefits of Bt cotton in Pakistan. Agricultural Economics. 2013; 44 :323–335.
- Krimsky S. An illusory consensus behind GMO health assessment. Science, Technology, & Human Values. 2015; 40 :883–914.
- Krishnan HB, Kerley MS, Allee GL, Jang S, Kim WS, Fu CJ. Maize 27 kDa gamma-zein is a potential allergen for early weaned pigs. Journal of Agricultural and Food Chemistry. 2010; 58 :7323–7328. [ PubMed : 20491474 ]
- Kuc J. Phytoalexins from the Solanaceae. In: Bailey JA, Mansfield JW, editors. Phytoalexins. New York: Wiley; 1982. pp. 81–105.
- Kuiper HA, Kok EJ, Davies HV. New EU legislation for risk assessment of GM food: No scientific justification for mandatory animal feeding trials. Plant Biotechnology Journal. 2013; 11 :781–784. [ PubMed : 23786622 ]
- Ladics GS, Selgrade MK. Identifying food proteins with allergenic potential: Evolution of approaches to safety assessment and research to provide additional tools. Regulatory Toxicology and Pharmacology. 2009; 54 :S2–S6. [ PubMed : 19028539 ]
- Ladics GS, Budziszewski GJ, Herman RA, Herouet-Guicheney C, Joshi S, Lipscomb EA, McClain S, Ward JM. Measurement of endogenous allergens in genetically modified soybeans—Short communication. Regulatory Toxicology and Pharmacology. 2014; 70 :75–79. [ PubMed : 24945742 ]
- Langkilde S, Schrøder M, Frank T, Shepherd LVT, Conner S, Davies HV, Meyer O, Danier J, Rychlik M, Belknap WR, McCue KF, Engel K-H, Stewart D, Knudsen I, Poulsen M. Compositional and toxicological analysis of a GM potato line with reduced α-solanine content—A 90-day feeding study in the Syrian Golden hamster. Regulatory Toxicology and Pharmacology. 2012; 64 :177–185. [ PubMed : 22796474 ]
- Lee SH, Hamaker BR. Cys 155 of 27 kDa maize γ-zein is a key amino acid to improve its in vitro digestibility. FEBS Letters. 2006; 580 :5803–5806. [ PubMed : 17045266 ]
- Ley RE. Obesity and the human microbiome. Current Opinion in Gastroenterology. 2010; 26 :5–11. [ PubMed : 19901833 ]
- Liebsch M, Grune B, Seiler A, Butzke D, Oelgeschläger M, Pirow R, Adler S, Riebeling C, Luch A. Alternatives to animal testing: Current status and future perspectives. Archives of Toxicology. 2011; 85 :841–858. [ PMC free article : PMC3149673 ] [ PubMed : 21607681 ]
- Litten-Brown JC, Corson AM, Clarke L. Porcine models for the metabolic syndrome digestive and bone disorders: A general overview. Animal. 2010; 4 :899–920. [ PubMed : 22444262 ]
- Liu P, He X, Chen D, Luo Y, Cao S, Song H, Liu T, Huang K, Xu W. A 90-day subchronic feeding study of genetically modified maize expressing Cry1Ac-M protein in Sprague–Dawley rats. Food and Chemical Toxicology. 2012; 50 :3215–3221. [ PubMed : 22709787 ]
- Lividini K, Fielder JL. Assessing the promise of biofortification: A case study of high provitamin A maize in Zambia. Food Policy. 2015; 54 :65–77.
- Livingston M, Fernandez-Cornejo J, Unger J, Osteen C, Schimmelpfennig D, Park T, Lambert D. The Economics of Glyphosate Resistance Management in Corn and Soybean Production. Washington, DC: U.S. Department of Agriculture–Economic Research Service; 2015.
- Ludvigsson JF, Rubio-Tapia A, CT van Dyke, Lelton LJ, Zinsmeister AR, Lahr BD, Murray JA. Increasing incidence of celiac disease in a North American population. American Journal of Gastroenterology. 2013; 108 :818–824. [ PMC free article : PMC3686116 ] [ PubMed : 23511460 ]
- Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS ONE. 2014; 9 :e110808. [ PMC free article : PMC4213012 ] [ PubMed : 25354084 ]
- MacKenzie SA, Lamb I, Schmidt J, Deege L, Morrisey MJ, Harper M, Layton RJ, Prochaska LM, Sanders C, Locke M, Mattsson JL, Fuentes A, Delaney B. Thirteen week feeding study with transgenic maize grain containing event DAS-01507-1 in Sprague-Dawley rats. Food and Chemical Toxicology. 2007; 45 :551–562. [ PubMed : 17097206 ]
- Macpherson AJ, McCoy KD, Johansen F-E, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunology. 2008; 1 :11–22. [ PubMed : 19079156 ]
- Magana-Gomez JA, de la Barca AMC. Risk assessment of genetically modified crops for nutrition and health. Nutrition Reviews. 2009; 67 :1–16. [ PubMed : 19146501 ]
- Martin C, Zhang Y, Tonelli C, Petroni K. Plants, diet, and health. Annual Review of Plant Biology. 2013; 64 :19–46. [ PubMed : 23451785 ]
- Martín-Hernández C, Bénet S, Obert L. Determination of proteins in refined and nonrefined oils. Journal of Agricultural and Food Chemistry. 2008; 56 :4348–4351. [ PubMed : 18512931 ]
- Marx-Stoelting P, Braeuning A, Buhrke T, Lampen A, Niemann L, Oelgeschlaeger M, Rieke S, Schmidt F, Heise T, Pfeil R, Solecki R. Application of omics data in regulatory toxicology: Report of an international BfR expert workshop. Archives in Toxicology. 2015; 89 :2177–2184. [ PubMed : 26486796 ]
- Mazza R, Soave M, Morlacchini M, Piva G, Marocco A. Assessing the transfer of genetically modified DNA from feed to animal tissues. Transgenic Research. 2005; 14 :775–784. [ PubMed : 16245168 ]
- Miller ER, Ullrey DE. The pig as a model for human nutrition. Annual Review of Nutrition. 1987; 7 :361–382. [ PubMed : 3300739 ]
- Miller HI. Substantial equivalence: Its uses and abuses. Nature Biotechnology. 1999; 17 :1042–1043. [ PubMed : 10545866 ]
- Millstone E, Brunner E, Mayer S. Beyond “substantial equivalence.” Nature. 1999; 401 :525–526. [ PubMed : 10524614 ]
- Mink PJ, Mandel JS, Lundin JI, Sceurman BK. Epidemiologic studies of glyphosate and non-cancer health outcomes: A review. Regulatory Toxicology and Pharmacology. 2011; 61 :172–184. [ PubMed : 21798302 ]
- Mink PJ, Mandel JS, Sceurman BK, Lundin JI. Epidemiologic studies of glyphosate and cancer: A review. Regulatory Toxicology and Pharmacology. 2012; 63 :440–452. [ PubMed : 22683395 ]
- Munkvold GP, Desjardins AE. Fumonisins in maize: Can we reduce their occurrence? Plant Disease. 1997; 81 :556–565. [ PubMed : 30861834 ]
- Murray JA, Van Dyke C, Plevak MF, Dierkhising RA, Zinsmeister AR, Melton LJ. Trends in the identification and clinical features of celiac disease in a North American community, 1950–2001. Clinical Gastroenterology and Hepatology. 2003; 1 :19–27. [ PubMed : 15017513 ]
- Murthy HN, Georgiev MI, Park S-Y, Dandin VS, Paek K-Y. The safety assessment of food ingredients derived from plant cell, tissue and organ cultures: A review. Food Chemistry. 2015; 176 :426–432. [ PubMed : 25624252 ]
- Muzzalupo I, Pisani F, Greco F, Chiappetta A. Direct DNA amplification from virgin olive oil for traceability and authenticity. European Food Research and Technology. 2015; 241 :151–155.
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant Journal. 2014; 77 :367–379. [ PMC free article : PMC4282528 ] [ PubMed : 24274116 ]
- National Toxicology Program. Report on Carcinogens, Thirteenth Edition. Research Triangle. Park, NC: U.S. Department of Health and Human Services, Public Health Service; 2014.
- NCI (National Cancer Institute). Surveillance, Epidemology and End Results (SEER) Program. 2014. [October 29, 2015]. http://www .cancer.org /research/cancerfactsstatistics /cancerfactsfigures2015 /index .
- Nemeth A, Wurz A, Artim L, Charlton S, Dana G, Glenn K, Hunst P, Jennings J, Shilito R, Song P. Sensitive PCR analysis of animal tissue samples for fragments of endogenous and transgenic plant DNA. Journal of Agricultural and Food Chemistry. 2004; 52 :6129–6135. [ PubMed : 15453677 ]
- Netherwood T, Martin-Orue SM, O'Donnell AG, Gockling S, Graham J, Mathers JC, Gilbert HJ. Assessing the survival of transgenic plant DNA in the human gastrointestinal tract. Nature Biotechnology. 2004; 22 :204–209. [ PubMed : 14730317 ]
- Nicolia A, Manzo A, Veronesi F, Rosellini D. An overview of the last 20 years of genetically engineered crop safety research. Critical Reviews in Biotechnology. 2014; 34 :77–88. [ PubMed : 24041244 ]
- Nordlee JA, Taylor SL, Townsend JA, Thomas LA, Bush RK. Identification of a Brazil-nut allergen in transgenic soybean. New England Journal of Medicine. 1996; 334 :688–692. [ PubMed : 8594427 ]
- Novak WK, Haslberger AG. Substantial equivalence of antinutritional and inherent plant toxins in genetically modified novel foods. Food and Chemical Toxicology. 2000; 38 :473–483. [ PubMed : 10828499 ]
- NRC (National Research Council). Genetically Modified Pest-Protected Plants: Science and Regulation. Washington, DC: National Academy Press; 2000. [ PubMed : 25032472 ]
- NRC (National Research Council). Environmental Effects of Transgenic Plants: The Scope and Adequacy of Regulation. Washington, DC: National Academy Press; 2002. [ PubMed : 25032287 ]
- NRC (National Research Council). Safety of Genetically Engineered Foods: Approaches to Assessing Unintended Health Effects. Washington, DC: National Academies Press; 2004. [ PubMed : 25009871 ]
- NRC (National Research Council). Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press; 2007. [ PubMed : 24901193 ]
- Nwaru BI, Hickstein L, Panesar SS, Roberts G, Muraro A, Sheikh A. Prevalence of common food allergies in Europe: A systematic review and meta-analysis. Allergy. 2014; 69 :992–1007. [ PubMed : 24816523 ]
- OECD (Organisation for Economic Co-operation and Development). Safety Evaluation of Foods Derived by Modern Biotechnology: Concepts and Principles. Paris: OECD; 1993.
- OECD (Organisation for Economic Co-operation and Development). Test No. 408: Repeated Dose 90-Day Oral Toxicity Study in Rodents in OECD Guidelines for the Testing of Chemicals. Paris: OECD; 1998a.
- OECD (Organisation for Economic Co-operation and Development). Principles of Good Laboratory Practice and Compliance Monitoring. 1998b. [October 13, 2015]. http://www .oecd.org/officialdocuments /publicdisplaydocumentpdf /?cote=env/mc/chem(98)17&doclanguage=en .
- OECD (Organisation for Economic Co-operation and Development). Report of the Task Force for the Safety of Novel Foods and Feeds. 2000. [October 12, 2015]. http://www .oecd.org/officialdocuments /publicdisplaydocumentpdf /?cote=env/mc/chem(98)17&doclanguage=enhttp://www .biosafety .be/ARGMO/Docments /report_taskforce.pdf .
- OECD (Organisation for Economic Co-operation and Development). An Introduction to the Food/Feed Safety Consensus Documents of the Task Force. Series on the Safety of Novel Foods and Feeds, No 14. Paris: OECD; 2006.
- OECD (Organisation for Economic Co-operation and Development). Safety Assessment of Foods and Feeds Derived from Transgenic Crops, Volume 2, Novel Food and Feed Safety. Paris: OECD; 2015.
- Oguchi T, Onishi M, Chikagawa Y, Kodama T, Suzuki E, Kasahara M, Akiyama H, Teshima R, Futo S, Hino A, Furui S, Kitta K. Investigation of residual DNAs in sugar from sugar beet (Beta vulgaris L.). Journal of the Food Hygenic Society of Japan. 2009; 50 :41–46. [ PubMed : 19325225 ]
- Onose J, Imai T, Hasumura M, Ueda M, Ozeki Y, Hirose M. Evaluation of subchronic toxicity of dietary administered Cry1Ab protein from Bacillus thuringiensis var. Kurustaki HD-1 in F344 male rats with chemically induced gastrointestinal impairment. Food and Chemical Toxicology. 2008; 46 :2184–2189. [ PubMed : 18381229 ]
- Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL, Drake R. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nature Biotechnology. 2005; 23 :482–487. [ PubMed : 15793573 ]
- Panchin AY, Tuzhikov AI. Published GMO studies find no evidence of harm when corrected for multiple comparisons. Critical Reviews in Biotechnology, Early Online. 2016:1–5. [ PubMed : 26767435 ]
- Patisaul HB, Jefferson W. The pros and cons of phytoestrogens. Frontiers in Neuroendocrinology. 2010; 31 :400–419. [ PMC free article : PMC3074428 ] [ PubMed : 20347861 ]
- Patterson JK, Lei XG, Miller DD. The pig as an experimental model for elucidating the mechanisms governing dietary influence on mineral absorption. Experimental Biology and Medicine. 2008; 233 :651–664. [ PubMed : 18408137 ]
- Pecetti L, Tava A, Romani A, De Benedetto MG, Corsi P. Variety and environment effects on the dynamics of saponins in lucerne (Medicago sativa L.). European Journal of Agronomy. 2006; 25 :187–192.
- Phipps RH, Deaville ER, Maddison BC. Detection of transgenic DNA and endogenous plant DNA in rumen fluid, duodenal digesta, milk, blood and feces of lactating dairy cows. Journal of Dairy Science. 2003; 86 :4070–4078. [ PubMed : 14740846 ]
- Poulsen M, Schrøder M, Wilcks A, Kroghsbo S, Lindecrona RH, Miller A, Frenzel T, Danier J, Rychlik M, Shu Q, Emami K, Taylor M, Gatehouse A, Engel KH, Knudsen I. Safety testing of GM-rice expressing PHA-E lectin using a new animal test design. Food Chemistry and Toxicology. 2007; 45 :364–377. [ PubMed : 17052831 ]
- Racovita M, Oboryo DN, Craig W, Ripandelli R. What are the non-food impacts of GM crop cultivation on farmers' health. Environmental Evidence. 2015; 4 :17.
- Ren Y, Wang T, Peng Y, Xia B, Qu LJ. Distinguishing transgenic from nontransgenic Arabidopsis plants by (1)H NMR-based metabolic fingerprinting. Journal of Genetics and Genomics. 2009; 36 :621–628. [ PubMed : 19840760 ]
- Rhee GS, Cho DH, Won YH, Seok JH, Kim SS, Kwack SJ, Lee RD, Chae SY, Kim JW, Lee BM, Park KL, Choi KS. Multigenerational reproductive and developmental toxicity study of bar gene inserted into genetically modified potato on rats. Journal of Toxicology and Environmental Health, Part A: Current Issues. 2005; 68 :2263–2276. [ PubMed : 16326439 ]
- Ricroch AE. Assessment of GE food safety using “-omics” techniques and long-term animal feeding studies. New Biotechnology. 2013; 30 :349–354. [ PubMed : 23253614 ]
- Ricroch A, Bergé JB, Kuntz M. Evaluation of genetically engineered crops using transcriptomic, proteomic and metabolomic profiling techniques. Plant Physiology. 2011; 155 :1752–1761. [ PMC free article : PMC3091128 ] [ PubMed : 21350035 ]
- Ricroch AE, Berheim A, Snell C, Pascal G, Paris A, Kuntz M. Long-term and multi-generational animal feeding studies. In: Flachowsky G, editor. Animal Nutrition with Transgenic Plants. Oxfordshire, UK: CABI Biotechnology Series; 2013a. pp. 112–127.
- Ricroch A, Berheim A, Pascal G, Paris A, Kuntz M. Assessment of the health impact of GE plant diets in long term and multigenerational animal feeding trials. In: Flachowsky G, editor. Animal Nutrition with Transgenic Plants. Oxfordshire, UK: CABI Biotechnology Series; 2013b. p. 234.
- Ricroch AE, Boisron A, Kuntz M. Looking back at safety assessment of GM food/feed: An exhaustive review of 90-day animal feeding studies. International Journal of Biotechnology. 2014; 13 :230–256.
- Riddle MS, Murray JA, Porter CK. The incidence and risk of celiac disease in a healthy US population. American Journal of Gastroenterology. 2012; 107 :1248–1255. [ PMC free article : PMC3493152 ] [ PubMed : 22584218 ]
- Rizzi A, Brusetti L, Arioli S, Nielsen KM, Tamagnini I, Tamburini A, Sorlini C, Daffonchio D. Detection of feed-derived maize DNA in goat milk and evaluation of the potential of horizontal transfer to bacteria. European Food Research and Technology. 2008; 227 :1699–1709.
- Rizzi A, Raddadi N, Sorlini C, Nordgård L, Nielsen KM, Daffonchio D. The stability and degradation of dietary DNA in the gastrointestinal tract of mammals: Implications for horizontal gene transfer and the biosafety of GMOs. Critical Reviews in Food Science and Nutrition. 2012; 52 :142–161. [ PubMed : 22059960 ]
- Roberfroid M. Letter to the Editor. Food and Chemical Toxicology. 2014; 65 :390. [ PubMed : 24394492 ]
- Rommens CM, Yan H, Swords K, Richael C, Ye J. Low-acrylamide French fries and potato chips. Plant Biotechnology Journal. 2008; 6 :843–853. [ PMC free article : PMC2607532 ] [ PubMed : 18662372 ]
- Ruan CJ, Shao HB, Teixeira da Silva JA. A critical review on the improvement of photosynthetic carbon assimilation in C3 plants using genetic engineering. Critical Reviews in Biotechnology. 2012; 32 :1–21. [ PubMed : 21699437 ]
- Rubio-Tapia A, Ludvigsson JF, Brantner TL, Murray JA, Everhart JE. The prevalence of celiac disease in the United States. American Journal of Gastroenterology. 2012; 107 :1538–1544. [ PubMed : 22850429 ]
- Ruiz-Lopez N, Haslam RP, Napier JA, Sayanova O. Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant Journal. 2014; 77 :198–208. [ PMC free article : PMC4253037 ] [ PubMed : 24308505 ]
- Saltzman A, Birol E, Bouis HE, Boy E, De Moura FF, Islam Y, Pfeiffer WH. Biofortification: Progress toward a more nourishing future. Global Food Security. 2013; 2 :9–17.
- Sanahuja G, Farré G, Berman J, Zorrilla-López U, Twyman RM, Capell T, Christou P, Zhu C. A question of balance: Achieving appropriate nutrient levels in biofortified staple crops. Nutritional Research Reviews. 2013; 26 :235–245. [ PubMed : 24134863 ]
- Sanders D. Letter to the Editor. Food and Chemical Toxicology. 2013; 53 :450–453. [ PubMed : 23142680 ]
- Schubbert R, Hohlweg U, Renz D, Doerfler W. On the fate of orally ingested foreign DNA in mice: Chromosomal association and placental transmission to the fetus. Molecular Genetics and Genomics. 1998; 259 :569–576. [ PubMed : 9819049 ]
- Séralini GE. Presentation to the National Academy of Sciences' Committee on Genetically Engineered Crops: Past Experience and Future Prospects, September 16. Washington, DC: 2014.
- Séralini GE, Cellier D, de Vendomois JS. New analysis of a rat feeding study with a genetically modified maize reveals signs of hepatorenal toxicity. Archives of Environmental Contamination and Toxicology. 2007; 52 :596–602. [ PubMed : 17356802 ]
- Séralini GE, Clair E, Mesnage R, Gress S, Defarge N, Malatesta M, Hennequin D, de Vendômois JS. Long term toxicity of a Roundup herbicide and a Roundup-tolerant genetically modified maize. Food and Chemical Toxicology. 2012; 50 :4221–4231. [ PubMed : 22999595 ]
- Séralini GE, Clair E, Mesnage R, Gress S, Defarge N, Malatesta M, Hennequin D, de Vendômois JS. Republished study: Long-term toxicity of a Roundup herbicide and a Roundup-tolerant genetically modified maize. Environmental Sciences Europe. 2014; 26 :14. [ PMC free article : PMC5044955 ] [ PubMed : 27752412 ]
- Shepherd LVT, Hackett CA, Alexander CJ, McNicol JW, Sungurtas JA, Stewart D, McCue KF, Belknap WR, Davies HV. Modifying glycoalkaloid content in transgenic potato—Metabolome impacts. Food Chemistry. 2015; 187 :437–443. [ PubMed : 25977048 ]
- Simó C, Ibáñez C, Valdés A, Cifuentes A, García-Cañas V. Metabolomics of genetically modified crops. International Journal of Molecular Sciences. 2014; 15 :18941–18966. [ PMC free article : PMC4227254 ] [ PubMed : 25334064 ]
- Sinden SL, Webb RE. Effect of variety and location on the glycoalkaloid content of potatoes. American Potato Journal. 1972; 49 :334–338.
- Singhal KK, Tyagi AK, Rajput YS, Singh M, Kaur H, Perez T, Hartnell GF. Feed intake, milk production and composition of crossbred cows fed with insect-protected Bollgard II ® cottonseed containing Cry1Ac and Cry2Ab proteins. Animal. 2011; 5 :1769–1773. [ PubMed : 22440417 ]
- Small E. Adaptations to herbivory in alfalfa (Medicago sativa). Canadian Journal of Botany. 1996; 74 :807–822.
- Smith JM. Seeds of Deception: Exposing Industry and Government Lies about the Safety of the Genetically Engineered Foods You're Eating. Fairfield, IA: Yes! Books; 2003.
- Smith JM. Are genetically modified foods a gut-wrenching combination? Institute for Responsible Technology. 2013. [October 12, 2015]. http: //responsibletechnology .org/glutenintroduction/
- Smith JM. Recommendations for the Committee on Genetically Engineered Crops. Presentation to the National Academy of Sciences' Committee on Genetically Engineered Crops: Past Experience and Future Prospects, September 16. Washington, DC: 2014.
- Snell C, Bernheim A, Berge JB, Kuntz M, Pascal G, Paris A, Ricroch AE. Assessment of the health impact of GM plant diets in long-term and multigenerational animal feeding trials: A literature review. Food and Chemical Toxicology. 2012; 50 :1134–1148. [ PubMed : 22155268 ]
- Spisák S, Solymosi N, Ittzés P, Bodor A, Kondor D, Vattay G, Barták B, Sipos F, Galamb O, Tulassay Z, Szállási Z, Rasmussen S, Sicheritz-Ponten T, Brunak S, Molnár B, Csabai I. Complete genes may pass from food to human blood. PLoS ONE. 2013; 8 :e69805. [ PMC free article : PMC3728338 ] [ PubMed : 23936105 ]
- Springob K, Kutchan TM. Introduction to the different classes of natural products. In: Osbourn AE, Lanzotti V, editors. Plant-Derived Natural Products. New York: Springer-Verlag; 2009. pp. 3–50.
- Steinke K, Guertler P, Paul V, Wiedemann S, Ettle T, Albrecht C, Meyer HHD, Spiekers H, Schwarz FJ. Effects of long-term feeding of genetically modified corn (event MON810) on the performance of lactating dairy cows. Journal of Animal Physiology and Animal Nutrition. 2010; 94 :e185–e193. [ PubMed : 20579187 ]
- Swiatkiewicz S, Swiatkiewicz M, Arczewska-Wlosek A, Jozefiak D. Genetically modified feeds and their effect on the metabolic parameters of food-producing animals: A review of recent studies. Animal Feed Science and Technology. 2014; 198 :1–19.
- Taylor A. EPA nixes approval of Enlist Duo weed killer. Online. The Des Moines Register. Nov 25, 2015. [December 13, 2015]. http://www .desmoinesregister .com/story/money /agriculture/2015 /11/25/epa-nixes-approval-enlist-duo-weed-killer/76386952/
- Taylor B, Jick H, MacLaughlin D. Prevalence and incidence rates of autism in the UK: Time trend from 2004–2010 in children aged 8 years. BMJ. 2013; 3 :e003219. [ PMC free article : PMC3808754 ] [ PubMed : 24131525 ]
- Thayer KA, Heindel JJ, Bucher JR, Gallo MA. Role of environmental chemicals in diabetes and obesity: A National Toxicology Program workshop review. Environmental Health Perspectives. 2012; 120 :779–789. [ PMC free article : PMC3385443 ] [ PubMed : 22296744 ]
- Trabalza-Marinucci M, Brandi G, Rondini C, Avellini L, Giammarini C, Costarelli S, Acuti G, Orlandi C, Filippini G, Chiaradia E, Malatesta M, Crotti S, Antonini C, Amagliani G, Manuali E, Mastrogiacomo AR, Moscati L, Haouet MN, Gaiti A, Magnani M. A three-year longitudinal study on the effects of a diet containing genetically modified Bt176 maize on the health status and performance of sheep. Livestock Science. 2008; 113 :178–190.
- Treutter D. Significance of flavonoids in plant resistance: A review. Environmental Chemistry Letters. 2006; 4 :147–157.
- Trikha A, Baillargeon JA, Kuo YF, Tan A, Pierson K, Sharma G, Wilkinson G, Bonds RS. Development of food allergies in patients with Gastroesophageal Reflux Disease treated with gastric acid suppressive medications. Pediatric Allergy and Immunology. 2013; 24 :582–588. [ PMC free article : PMC4528619 ] [ PubMed : 23905907 ]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009; 457 :480–484. [ PMC free article : PMC2677729 ] [ PubMed : 19043404 ]
- Untersmayr E, Jensen-Jarolim E. The role of protein digestibility and antacids on food allergy outcomes. Journal of Allergy and Clinical Immunology. 2008; 121 :1301–1308. [ PMC free article : PMC2999748 ] [ PubMed : 18539189 ]
- U.S. Census Bureau. 65+ in the United States: 2010. Washington, DC: U.S. Government Printing Office; 2014.
- U.S. Pharmacopeia. Pharmacopeia, simulated gastric fluid, TS, simulated intestinal fluid, TS. United States Pharmacopeial Convention, v 24. The National Formulary 9 (US Pharmacopiea Board of Trustees). Rockville, MD: 2000. 2235.
- USDA–APHIS (U.S. Department of Agriculture–Animal and Plant Health Inspection Service). Dow AgroSciences Petitions (09-233-01p, 09-349-01p, and 11-234-01p) for Determinations of Nonregulated Status for 2,4-D-Resistant Corn and Soybean Varieties. Final Environmental Impact Statement—August 2014. 2014a. [May 9, 2016]. https://www .regulations .gov/document?D=APHIS-2013-0042-10218 .
- USDA–APHIS (U.S. Department of Agriculture–Animal and Plant Health Inspection Service). Record of Decision: Dow AgroSciences Petitions (09-233-01p, 09-349-01p, and 11-234-01p) for Determination of Nonregulated Status for 2,4-D-Resistant Corn and Soybean Varieties. 2014b. [December 13, 2015]. https://www .aphis.usda .gov/brs/aphisdocs/24d_rod.pdf .
- USDA–APHIS (U.S. Department of Agriculture–Animal and Plant Health Inspection Service). Determinations of Nonregulated Status: J.R. Simplot Co.; Potato Genetically Engineered for Low Acrylamide Potential and Reduced Black Spot Bruise. 2014c. [December 22, 2015]. http://www .regulations .gov/#!documentDetail;D =APHIS-2012-0067-0384 .
- USRDS (United States Renal Data System). CKD in the general population. 2014 USRDS Annual Data Report Volume 1. 2014. [October 13, 2015]. pp. 12–22. http://www .usrds.org/2014/view/Default .aspx .
- Valdés L, Cuervo A, Salazr N, Ruas-Madiedo P, Gueimondea M, González S. The relationship between phenolic compounds from diet and microbiota: Impact on human health. Food & Function. 2015; 6 :2424–2439. [ PubMed : 26068710 ]
- van den Eede G, Aarts H, Buhk HJ, Corthier G, Flint HJ, Hammes W, Jacobsen B, Midtvedt T, van der Vossen J, von Wright A, Wackernagel W, Wilcks A. The relevance of gene transfer to the safety of food and feed derived from genetically modified (GM) plants. Food and Chemical Toxicology. 2004; 42 :1127–1156. [ PubMed : 15123384 ]
- Van Eenennaam AL, Young AE. Prevalence and impacts of genetically engineered feedstuffs on livestock populations. Journal of Animal Science. 2014; 92 :4255–4278. [ PubMed : 25184846 ]
- VanEtten H, Mansfield JW, Bailey JA, Farmer EE. Two classes of plant antibiotics: Phytoalexins versus “phytoanticipins.” The Plant Cell. 1994; 6 :1191–1192. [ PMC free article : PMC160512 ] [ PubMed : 12244269 ]
- Verhoeckx KCM, Vissers YM, Baumert JL, Faludi R, Feys M, Flanagan S, Herouet-Guicheney C, Holzhauser T, Shimojo R, van der Bolt N, Wichers H, Kimber I. Food processing and allergenicity. Food and Chemical Toxicology. 2015; 80 :223–240. [ PubMed : 25778347 ]
- Wal JM. Assessing and managing allergenicity of genetically modified (GM) foods. In: Flanagan S, editor. Handbook of Food Allergen Detection and Control. Cambridge, UK: Woodhead Publishing; 2015. pp. 161–178.
- Walsh MC, Buzoianu SG, Gardiner GE, Rea MC, Gelencsér E, Jánosi A, Epstein MM, Ross RP, Lawlor PG. Fate of transgenic DNA from orally administered Bt MON810 maize and effects on immune response and growth in pigs. PLoS ONE. 2011; 6 :e27177. [ PMC free article : PMC3223173 ] [ PubMed : 22132091 ]
- Walsh MC, Buzoianu SG, Rea MC, O'Donovan O, Gelencsér E, Ujhelyi G, Ross RG, Gardiner GE, Lawlor PG. Effects of feeding Bt MON810 maize to pigs for 110 days on peripheral immune response and digestive fate of the Cry1Ab gene and truncated Bt toxin. PLoS ONE. 2012a; 7 :e36141. [ PMC free article : PMC3345032 ] [ PubMed : 22574138 ]
- Walsh MC, Buzoianu SG, Gardiner GE, Rea MC, Ross RP, Cassidy JP, Lawlor PG. Effects of short-term feeding of Bt MON810 maize on growth performance, organ morphology and function in pigs. British Journal of Nutrition. 2012b; 107 :364–371. [ PubMed : 21733303 ]
- Walsh MC, Buzoianu SG, Gardiner GE, Rea MC, O'Donovan O, Ross RP, Lawlor PG. Effects of feeding Bt MON810 maize to sows during first gestation and lactation on maternal and offspring health indicators. British Journal of Nutrition. 2013; 109 :873–881. [ PubMed : 23168255 ]
- Wang X, Chen X, Xu J, Dai C, Shen W. Degradation and detection of transgenic Bacillus thuringiensis DNA and proteins in flour of three genetically modified rice events submitted to a set of thermal processes. Food and Chemical Toxicology. 2015; 84 :89–98. [ PubMed : 26277627 ]
- Weber A. C 4 Photosynthesis—A Target for Genome Engineering. Presentation to the National Academy of Sciences' Committee on Genetically Engineered Crops: Past Experience and Future Prospects, December 10. Washington, DC: 2014.
- West J, Fleming KM, Tata LJ, Card TR, Crooks CJ. Incidence and prevalence of celiac disease and dermatitis herpetiformis in the UK over two decades: Population-based study. American Journal of Gastroenterology. 2014; 109 :757–768. [ PMC free article : PMC4012300 ] [ PubMed : 24667576 ]
- Wiatrak PJ, Wright DL, Marois JJ, Wilson D. Influence of planting date on aflatoxin accumulation in Bt, non-Bt, and tropical non-Bt hybrids. Agronomy Journal. 2005; 97 :440–445.
- Wiener JB, Rogers MD, Hammitt JK, Sand PH, editors. The Reality of Precaution: Comparing Risk Regulation in the United States and Europe. New York: RFF Press; 2011.
- Wild CP, Gong YY. Mycotoxins and human disease: A largely ignored global health issue. Carcinogensis. 2010; 31 :71–82. [ PMC free article : PMC2802673 ] [ PubMed : 19875698 ]
- Williams JH, Phillips TD, Jolly PE, Stiles JK, Jolly CM, Aggarwal D. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. American Journal of Clinical Nutrition. 2004; 80 :1106–1122. [ PubMed : 15531656 ]
- World Health Organization. Frequently Asked Questions on Genetically Modified Foods. 2014. [March 12, 2016]. http://www .who.int/foodsafety /areas_work /food-technology/Frequently _asked_questions_on_gm_foods.pdf .
- Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X. Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nature Biotechnology. 2005; 23 :1013–1017. [ PubMed : 15951804 ]
- Wu Y, Holding DR, Messing J. γ-Zeins are essential for endosperm medication in quality protein maize. Proceedings of the National Academy of Sciences of the United States of America. 2010; 107 :12810–12815. [ PMC free article : PMC2919962 ] [ PubMed : 20615951 ]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I. Engineering the provitamin A (β-Carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000; 287 :303–305. [ PubMed : 10634784 ]
- Yudina TG, Brioukhanov AL, Zalunin IA, Revina LP, Shestakov AI, Voyushina NE, Chestukhina GG, Netrusov AI. Antimicrobial activity of different proteins and their fragments from Bacillus thuringiensis parasporal crystals against clostridia and archaea. Anaerobe. 2007; 13 :6–13. [ PubMed : 17126041 ]
- Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, Zhang M, Wang L, Houb Y, Ouyang H, Zhang Y, Zheng Y, Wang J, Lv X, Wang Y, Zhang F, Zeng B, Li W, Yan F, Zhao Y, Pang X, Zhang X, Fu H, Chen F, Zhao N, Hamaker BR, Bridgewater LC, Weinkove D, Clement K, Dore J, Holmes E, Xiao H, Zhao G, Yang S, Bork P, Nicholson JK, Wei H, Tang H, Zhang X, Zhao L. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine. 2015; 2 :968–984. [ PMC free article : PMC4563136 ] [ PubMed : 26425705 ]
- Zhu X-G, Shan L, Wang Y, Quick WP. C4 rice—an ideal arena for systems biology research. Journal of Integrative Plant Biology. 2010; 52 :762–770. [ PubMed : 20666931 ]
- Cite this Page National Academies of Sciences, Engineering, and Medicine; Division on Earth and Life Studies; Board on Agriculture and Natural Resources; Committee on Genetically Engineered Crops: Past Experience and Future Prospects. Genetically Engineered Crops: Experiences and Prospects. Washington (DC): National Academies Press (US); 2016 May 17. 5, Human Health Effects of Genetically Engineered Crops.
- PDF version of this title (15M)
In this Page
Related information.
- PMC PubMed Central citations
- PubMed Links to PubMed
Recent Activity
- Human Health Effects of Genetically Engineered Crops - Genetically Engineered Cr... Human Health Effects of Genetically Engineered Crops - Genetically Engineered Crops
Your browsing activity is empty.
Activity recording is turned off.
Turn recording back on
Connect with NLM
National Library of Medicine 8600 Rockville Pike Bethesda, MD 20894
Web Policies FOIA HHS Vulnerability Disclosure
Help Accessibility Careers

IMAGES
VIDEO
COMMENTS
Genetic engineering is the act of modifying the genetic makeup of an organism. Modifications can be generated by methods such as gene targeting, nuclear transplantation, transfection of synthetic ...
Genetic engineering is the use of molecular biology technology to modify DNA sequence(s) in genomes, using a variety of approaches. For example, homologous recombination can be used to target specific sequences in mouse embryonic stem (ES) cell genomes or other cultured cells, but it is cumbersome, poorly efficient, and relies on drug positive/negative selection in cell culture for success.
Introduction. Gene therapy is a therapeutic strategy using genetic engineering techniques to treat various diseases. 1, 2) In the early 1960s, gene therapy first progressed with the development of recombinant DNA (rDNA) technology, 1) and was further developed using various genetic engineering tools, such as viral vectors. 3 - 5) More than ...
was published by Springer between 2019-2023. is devoted to rapid publication of full-length research papers that lead to significant contribution in advancing knowledge in genetic engineering and biotechnology and provide novel perspectives in this research area. JGEB includes all major themes related to genetic engineering and recombinant DNA.
Manar A. Basheer, Khaled Abutaleb, Nermine N. Abed and Amal A. I. Mekawey. Journal of Genetic Engineering and Biotechnology 2023 21 :127. Research Published on: 21 November 2023. The Correction to this article has been published in Journal of Genetic Engineering and Biotechnology 2023 21 :164. Full Text.
Genomic research has evolved from seeking to understand the fundamentals of the human genetic code to examining the ways in which this code varies among people, and then applying this knowledge to ...
Dipender Gill. Objective To investigate the association of genetically proxied (using a surrogate biomarker) inhibition of phosphodiesterase 5 (PDE5), an established drug target for erectile ...
Reverse genetics is a method in molecular genetics that is used to help understanding the function of a gene by analyzing the phenotypic effects of specific engineered gene sequences. Robb et al. [ 68] defined and compared the three terms: "genome engineering", "genome editing", and "gene editing".
Genetic engineering can be used in a diverse range of contexts, including research (e.g., to build model organisms), pharmacology (e.g., for insulin production) and agriculture (e.g., to improve ...
Genetic engineering opens new possibilities for biomedical enhancement requiring ethical, societal and practical considerations to evaluate its implications for human biology, human evolution and our natural environment. In this Commentary, we consider human enhancement, and in particular, we explore genetic enhancement in an evolutionary context.
Genetic engineering (GE) is often termed as gene manipulatio n or recombinant DNA technology with all. three often used interchangeably -- implying to t he m anipulation and alteration of the ...
Background. Genome editing is a type of genetic engineering in which DNA is deliberately inserted, removed, or modified in living cells. 1 The name CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) refers to the unique organization of short, partially repeated DNA sequences found in the genomes of prokaryotes. CRISPR and its associated protein (Cas-9) is a method of adaptive ...
The engineered phage-mediated industrial food processing and biocontrol, advanced wastewater treatment, phage-based nano-medicines, and their use as a bio-recognition element for the detection of bacterial pathogens are also part of this review. The genetic engineering approaches hold great potential to accelerate translational phages and research.
This paper discusses the use of genetic engineering applications in animal breeding, including a description of the methods, their potential and current uses and ethical issues.
7.21 Conclusion. Genetic engineering is a paradigmatic case for application of the PP to environmental and public health policy, due to the scientific uncertainty related to the consequences of genetic engineering and the moral uncertainty concerning those consequences. In this chapter I have applied the PP to genetic engineering of microbes ...
The introduction of genome engineering technology has transformed biomedical research, making it possible to make precise changes to genetic information. However, creating an efficient gene-editing system requires a deep understanding of CRISPR technology, and the complex experimental systems under investigation. While Large Language Models (LLMs) have shown promise in various tasks, they ...
He stated that his research could help tamp down the HIV/AIDS epidemic; the most hard‐hit areas (such as Africa), however, would likely not gain much benefit from gene‐editing technologies. Per a December 2018 poll, 78 Americans draw the line at so‐called enhancement, but favor the use of genetic engineering to address disease and ...
Genetic engineering techniques have been used in research to increase the understanding of genetic function, diseases, and productivity. Early attempts of genetic engineering were mainly in laboratory animals such as mice, where genetic engineering was carried out by using exogenous recombinant vector form or packing vector that would be ...
Abstract. Genetic engineering is the best technology that is promoting the world and this technology is applied to many plants, animals and microorganisms. It has wider applications in the field of Biology, Medicine, Industry, Research, Agriculture and many other fields of science. In this research paper I update the Roles of Genetic ...
Introduction to Genetic Engineering. April 2020. Authors: Osama Rahil Shaltami. University of Benghazi. Citations (1)
Discussions and debates over some of these topics have been held numerous times in the last three decades, especially within the context of in vitro fertilization, transgenic animals, cloning, pre-implantation genetic diagnosis (PGD), research with stem cells and induced pluripotent stem cells, as well as related to the large scope of ...
COMPARING GENETICALLY ENGINEERED CROPS WITH THEIR COUNTERPARTS. An oft-cited risk of GE crops is that the genetic-engineering process could cause "unnatural" changes in a plant's own naturally occurring proteins or metabolic pathways and result in the unexpected production of toxins or allergens in food (Fagan et al., 2014).Because analysis of risks of the product of the introduced ...