Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
Enter the email address you signed up with and we'll email you a reset link.
- We're Hiring!
- Help Center


QUALITY CONTROL IN MICROBIOLOGY

Quality control (QC) is a process intended to review the quality of all factors involved in production. It ensures manufactured products and means of service/production strictly follows a defined set of quality criteria and meets the requirements of the consumers. Quality control in microbiology is the process of recording what happens throughout a process in a microbiology lab and pinpoint issues that affect those processes. Each laboratory must have qualified supervisory and technical personnel, standard operating procedures, be well lit, dust free and have an air conditioned environment. All materials, equipment and procedures must be adequately controlled. Quality of the media directly affects the observation and inferences drawn from the cultural characteristics of microbiology, checking of different parameters of media such as growth supporting characteristics, gel strength and batch contamination can help to assess their quality. The meticulous performance of quality control of culture can assure precision in reporting. Quality control is applied in almost all aspects of life and activities such as Pharmaceutical, Agriculture, Medical, Food microbiology and other areas involved in production and related to man. Hospital, industry and research are fields in which microbiologists are active and are responsible for assuring quality of products through all day to day operations. Microbiologists evaluate biomanufacturing plants by assessing raw materials, other supplies and the finished/ packaged products. Compliance to common Good Manufacturing Practices (GMP) is followed. Quality control in a microbiology lab may be observed by outside regulatory bodies. The recall of drugs, products or equipments are time consuming are some of the limitations affecting quality control. Procedures to maintain quality control in microbiology are setting quality standards, making improvements which reduce the risk to the consumer and inadvertently to the manufacturer.
Related Papers
African Journal of Clinical and Experimental Microbiology
S. Fayemiwo
Developments in Plant Pathology
Carlo Leifert
American Journal of Clinical Pathology
Alexander Duncan
Brazilian Journal of Microbiology
Fabiana Pereira
Stringent quality control protocols must be used in order to guaranty that a particular medium is able to recover all sort of organism that may be present in clinical samples. Our aim was to evaluate an alternative protocol that would allow us to detect medium failure to yield quantitative growth of selected pathogens, and compare with the document M22-A2 from NCCLS. The detection limit of Haemophilus influenzae was significantly different depending on media source. We conclude that for some fastidious microorganisms, quantitative verification of the growth capacity of the culture medium is advised.
International Journal for Research in Applied Science & Engineering Technology (IJRASET)
IJRASET Publication
Quality assurance is part of quality management that ensured the pharmaceutical products are of required quality. It includes GMP, GLP, GCP, product design and development. The finished product is tested & checked according to their procedure. The confidence delivered by quality assurance is twice intrinsically to management and extrinsically to clients, government agencies, regulators, certifiers, and third parties Quality control is a procedure which contemplates on performing the quality demand. Quality control intend to distinguish (and dead-on) imperfection in the finished product. Quality control, in consequence, is a reactive procedure. Quality control can be delineated as "section of quality management emphasized on furnishing quality must-have".
John Olusola OLAYIWOLA
The Quality Control (QC) laboratory, Chittagong is basically designed to analysis the biological and chemical characteristics of fish and fishery products in view of sustaining Bangladesh export performance in world seafood markets through integration of qualified personnel, standard methods, appropriate equipment and quality assurance system are in place for necessary testing and interpretation of tested results. We describe herein, facilities on microbiological test at biosafety level 2 (BSL-2) containment laboratories, with regard to biosecurity regulations, safety considerations, necessary space, and physical aids in executing ISO standards. These competences can positive impact the number of testing samples investigating microbial pathogens of biodefense concern. Acquisition, use, storage and transfer of microbiological pathogenic bacteria such as reference strains are highly regulated due to their potential to pose a severe threat to export of fish and fishery products. All fe...
North American Journal of Medical Sciences
Ezekiel Nwose
Kakurinov Vladimir
Transplantation Proceedings
Monica Saldias
Loading Preview
Sorry, preview is currently unavailable. You can download the paper by clicking the button above.
RELATED PAPERS
Nune Hambardzumyan
Current Requirements and Future Perspectives
kyriacos tsimillis
Angelos Evangelopoulos
International Journal of Pharmaceutical Compounding
Kate Douglass
Gudisa Bereda
Kirsti Derome , A. Fuerst , Nicholas John Clarke
Journal of Nepal Medical Association
ROSHAN KHATRI
Lorena Ivona Stefan
Trends in Biotechnology
rudolf bliem
Alberto Lumibao
Ahmed Badr Eldin
Mohammad Miyan
Microorganisms
Nicolina Dias
icp-forests.org
Nicholas John Clarke
Applications and Experiences of Quality Control
Journal of Oral …
Vandana Sharma
International Journal of Research Studies in Biosciences
Zerihun Tsegaye
Journal of Analytical & Pharmaceutical Research
vandana saklani
Scott Sutton
Open Access Macedonian Journal of Medical Sciences
Sonja Stefanov
pkarkalousos.gr
RELATED TOPICS
- We're Hiring!
- Help Center
- Find new research papers in:
- Health Sciences
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
- Academia ©2024
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

Internal quality assurance in diagnostic microbiology: A simple approach for insightful data
Valentin scherz.
Institute of Microbiology, University of Lausanne and University Hospital Center, Lausanne, Switzerland
Christian Durussel
Gilbert greub, associated data.
In accordance with our ethics statement and the particular context of quality assurance, explicit patient agreements were not obtained and this quality assurance project was exempted of ethical committee approval. Since particular patterns of microbiology results could lead to patients’ recognition, our institute’s quality committee, which is the initiator of this project and is responsible for all ethical aspects of our quality control projects, enforced that data cannot be made publicly available. A representative of this committee, Madame Sarah Chappuis, Rue du Bugnon 48, 1011 Lausanne, will answer to questions regarding this decision ( [email protected] ). This same representative will also make data available upon motivated request and commitment to keep the obtained data confidential.
Given the importance of microbiology results on patient care, high quality standards are expected. Internal quality assurance (IQA) could mitigate the limitations of internal quality control, competency assessment and external quality assurance, adding a longitudinal insight, including pre- and post-analytical steps. Here, we implemented an IQA program in our clinical microbiology facilities with blind resubmission of routine samples during 22 months. One-hundred-and-twenty-one out of 123 (98.4%) serological analyses and 112 out of 122 (91.8%) molecular analyses were concordant. Among the discordances in molecular biology analyses, 6 results were low positive samples that turned out negative, likely due to stochastic repartition of nucleic acids. Moreover, one identified retranscription error led us to implement automated results transmission from the Applied Biosystems instruments to the laboratory information system (LIS). Regarding Gram stain microscopy, 560 out of 745 (75.2%) of compared parameters were concordant. As many as 67 out of 84 (79.8%) pairs of culture results were similar, including 16 sterile pairs, 27 having identical identification or description and semi-quantification and 24 only showing variations in semi-quantification with identical description or identification of colonies. Seventeen pairs had diverging identification or description of colonies. Culture was twice only done for one member of the pairs. Regarding antibiotic susceptibility testing, a major discrepancy was observed in 5 out of 48 results (10.4%). In conclusion, serological tests were highly reproducible. Molecular diagnosis also revealed to be robust except when the amounts of nucleic acids present in the sample were close to the limits of detection. Conventional microbiology was less robust with major discrepancies reaching 39.5% of the samples for microscopy. Similarly, culture and antibiotic susceptibility testing were prone to discrepancies. This work was ground for reconsidering multiples aspects of our practices and demonstrates the importance of IQA to complete the other quality management procedures.
Introduction
Microbiology laboratories are nowadays facing numerous challenges. The spread of antimicrobial drugs resistances demands rapid and yet reliable characterization of causative agents to prevent treatment failures and evitable occurrences of resistances [ 1 – 3 ]. Regular identification of emerging pathogens [ 3 ] and implementation of new molecular techniques [ 4 ] bring greater technical complexity, while prompt results and better cost-efficiency are expected [ 5 – 7 ]. Besides these evolutions, the awareness about medical errors and their consequences emphasizes the great importance of quality in health [ 8 , 9 ]. Indeed, clinical laboratories and related quality management programs have a role in preventing deaths due to evitable medical errors [ 10 ].
Quality management went along a constant evolution from the sixties to reach the current state of practice [ 11 ]. It is now generally accepted [ 12 ] and legally prescribed in Switzerland [ 13 ] and in the USA [ 14 ] that a minimal quality assurance (QA) program is composed of quality control (QC), external quality assessment (EQA), standard operating procedure (SOP) and competency assessment (CA) of coworkers. These procedures, despite being needed and regulated by certification processes such as ISO 15189 [ 15 ], have major limitations discussed below.
First, QC focuses on the intrinsic performances of laboratory tests, reviewing their performance (specificity & sensitivity), the regular maintenance of all used materials, the constant quality of reactants and media, as well as the use of negative and positives controls [ 12 ]. Despite their established role, QC are insufficient [ 11 ], especially because they do not cover pre- and post-analytical steps [ 10 , 16 , 17 ].
EQA includes external audits as well as proficiency testing (PT) with samples sent by reference nodal laboratories to be analyzed and described [ 12 ]. Audits can reveal structural dysfunctions and possible points to improve. PT with unknown samples probes the ability of the laboratory to treat and characterize samples and to identify pathogens. Both audits and PT provide valuable information on the functioning of the laboratories but are limited by their rare occurrence. PT has shown several additional limitations:
- It only partially evaluates pre- and post-analytical steps [ 18 ]; for example, PT samples are often coming in modified forms (e.g. dried, synthetically produced, pre-treated as with serum samples instead of whole blood), which can prevent their inclusion into regular lines of sample handling.
- PT are not available for all the different tests offered by our laboratories (for instance no PT is available for unusual pathogens such as T . whipplei and W . chondrophila ) [ 19 ].
- It has been shown that their recognition as EQA can lead to falsely reassuring results because of the extra caution given in their processing [ 20 , 21 ]; indeed, samples labeled as EQA were shown to provide more reliable results than EQA submitted blindly [ 22 ].
Competency assessment (CA) probes the ability of the laboratory personnel to complete properly their tasks, in good adequacy with SOP. CA programs can be simple, such as direct observation of technicians and quizzes to complete, or more sophisticated, with for instance, submission of blind challenge samples [ 23 – 28 ]. Despite the different approaches evaluated in the literature, most suffer of similar limitations as QC, with a primary focus on individuals performing defined analytical phases and limited insight on the overall quality of laboratory processes. CA programs can also certainly be biased when the workers identify the time of evaluation. Interestingly, blind resubmissions of previously analyzed samples, which is one of the methods suggested in literature for CA purposes, seem to suffer of less limitations but are only seldom used [ 24 ].
Internal quality assurance (IQA) programs based on resubmissions of replicated samples could address the mentioned limitations but lack of supporting definitions and data. The potential of this type of procedures is recognized by reference organizations such as the Eurachem and the Standard Unit, Microbiology Services of Public Health England, which recommend them to assess internally the overall reproducibility of samples testing and handling [ 29 , 30 ]. However, there is very few original publications in clinical microbiology setting supporting these recommendations, the only ones being the studies conducted in Cambridge in the nineties by Constantine et al [ 31 ] and Gray et al [ 32 ]. The authors concluded then that their works showed the capacity of this system to detect error, highlighted point of improvements and produced relevant data on the reproducibility of their results and the overall quality of their laboratories [ 31 – 33 ].
Thus, in the present work, we implemented an internal quality assessment program using blind split samples. This provided data on the reproducibility of the results in our microbiology laboratory and helped us to define specific corrective measures.
Materials and methods
Study design.
A new IQA procedure was initiated in October 2014 in the three clinical laboratories (serology, molecular microbiology and conventional microbiology) of the Institute of Microbiology of the University Hospital of Lausanne (Switzerland). This procedure was designed and implemented by the quality committee of our laboratories and exempted from formal ethic committee approval based on its regulation regarding quality control studies [ 34 ]. Indeed, the scheme which was applied here is in line with IQA procedure recommended by reference organizations in routine practice [ 29 , 30 ]. The present work reviews the data gathered over a 22-month period and evaluated the discrepancies, including their possible origin and their putative clinical impact (see below).
Data collection
The head technicians of our three laboratories were instructed to secretly withdraw samples out of the already processed ones. In serology and molecular biology laboratories, samples were chosen to be proportionally representative of the analyses done by our facilities, but also ideally, to test at least once every test offered by our service. In the classical microbiology laboratory, the head technician generally selected samples that were expected to be challenging, because they were positive at Gram stain examination of by culture after overnight incubation. This approach was retained to increase the number of samples for which the adequacy of bacterial identification and antibiotics susceptibility could be investigated. The analysis request forms were replicated with anonymous identification data, which are commonly used in our institution for clinical research purposes (and thus not assimilated by our technician to a quality control). Part of the original sample material was taken into a new container and handed back in secret at the reception of the laboratories. The correspondence between the patient identification code and the replicate identification code was reported by the head technicians into a database for later comparison of the results. Results of the two matching samples were later reported into an Access database and data analyses were conducted with Microsoft Excel and PRISM (GraphPad Software). All data were processed anonymously.
Serology and molecular biology
Pairs of serology results were compared for agreement between qualitative results (positive, indeterminate or negative) and variations in titers when relevant. Similarly, results of qPCR pairs were compared for differences in (i) qualitative results (positive versus negative), (ii) number of copies/ml with a margin of acceptance of one log 10 between the two results and (iii) cycle thresholds (Ct) with a margin of acceptance of 3.3 Ct between the two results.
Gram stain examination
Gram stains results were compared regarding semi-quantification of the following parameters: leukocytes, erythrocytes, epithelial cells, yeasts, Gram-positive cocci, bacilli and diplococci and Gram-negative cocci, bacilli and coccobacilli. The given interpretations of the Gram results were also compared when relevant (e.g. suspicion of urines contamination, salivary flora in sputa, dominant bacterial flora in vaginal swabs). For each parameter, the amplitude of discrepancies was classified according to S1A Table . The most important discrepancy between all parameters of a pair of samples was recorded. We examined the discrepancies for each parameter by linearly weighted Kappa score using GraphPad QuickCalcs website [ 35 ]. Kappa score quantifies the level of correspondence between the ratings of two observers, considering the natural probability to obtain the same level of concordance only by chance [ 36 ]. The weighted version of Kappa score is used to consider the partial agreements within ordered scales [ 37 ]. We adopted the weighting suggested by Cicchetti et Allison [ 38 ], which sanctions discrepancies in a linear and not in a quadratic method [ 39 ].
Culture and antibiotic susceptibility testing (AST)
Culture results were compared for description and/or identification and semi-quantification and discrepant cases were described. Quantitative variations were scored according to S1B Table .
Sets of AST were compared when conducted for the same identified bacteria within a pair of replicates. Discrepancies were classified following S1C Table . The most significant discrepancy within all tested antibiotics was retained to grade the concordance of the whole AST sets (e.g. if the AST pairs included a single major discrepancy, the pair of sets were considered as being majorly discrepant). If AST were not performed for the same species on both replicates, the cause of the discrepancy was analyzed and described.
Evaluation of clinical impact and putative cause
Interpretation of the clinical impact and putative cause of discordant results was done by a data review committee of three persons (VS, CD and GG). Discrepancies were classified as clinically negligible or significant.
Among 123 serological tests performed twice on 35 sera, a total of 121 (98.4%) were concordant ( Table 1 ). The first discordant test was a measure of IgG against cytomegalovirus (CMV) by enzyme immuno-assay (EIA), which was initially positive (index of 1.20) and intermediate when redone (0.953 for a cut-off for positivity of 1 and for negativity of 0.5). The 123 tests were often completed within panels of multiple tests detecting antibody reactivity for the same pathogen (in total 76 different pathogen-specific panels), sometimes reducing the negative impact of discordant results. Thus, this first discordant result was accompanied by electrochemiluminescence (ECLIA) results, which were both times positive for anti-CMV IgG antibodies (4.84 and 4.78 U/ml with a cutoff of 1 for positivity). The second discordant pair of results was in the titers of a syphilis test (TPPA) obtained manually, being twice positive but with titers of 1/2'560 initially and 1/10’240 when redone. The clinical impact was considered significant in this 2 nd case since a high titer of 1/10’240 generally reflects an acute infection and since this difference of two dilutions might be considered as clinically relevant [ 40 , 41 ]. The limited number of discrepant results did not allow to investigate the effect of the time delays between the management of the original samples and their replicates.
| Percentage of agreement | Discordant analyses | Clinically significant discordances | Time between replicates | |
|---|---|---|---|---|
| Mean (SD) | ||||
| p25/p50/p75 (in hours) | ||||
| 98.4% | 2/123 | 1/2 | 28 (19.1) | |
| 19.1/24.6/41.6 | ||||
| 91.8% | 10/122 | 6/10 | ||
| Qualitative results | 95.1% | 6/122 | 6/6 | 182.8 (45.8) |
| Quantitative results | 82.6% | 4/23 | 0/4 | 169.1/191.5/215.8 |
| 75.2% | 185/745 | 13/71 | 10.3 (9.5) | |
| 50.0% | 43/86 | 13/43 | ||
| Aggregated by panels | 75.0% | 12/48 | 1/12 | 5.0/6.5/8.2 |
| Single antibiotics | 96.4% | 25/759 | 1/25 |
This table shows an overview of the agreement, discordance and clinical significance of discordances for the three laboratories. The column “ Time between replicates ” indicates the distribution of time, in hours, between the management of the original sample and its replicate. For AST, results were analyzed once to see whether the panels done for the same bacteria had identical results ( Aggregated by panels ) and once to see whether every single antibiotic tested had the same results twice ( Single antibiotics ).
* 1 overall agreement for all parameters;
* 2 results aggregated per samples;
SD = standard deviation; p25 = percentile 25; p50 = percentile 50, p75 = percentile 75.
Molecular diagnosis
Among 122 tests achieved on 52 replicated samples, 112 (91.8%) molecular tests were concordant ( Table 1 ). Six tests were qualitatively different, being once positive and once negative. Of the 23 results pairs that included quantitative results, three were diverging in the number of copies/ml (difference in copies/ml over one log 10 ) and one was quantitatively different only considering the Ct values (difference in Ct over 3.3).
Regarding the qualitative differences, four discordant molecular results corresponded to low positive samples (132–175 copies/ml) that turned out to be negative when retested ( Fig 1a ) whereas one initially negative result turned out to be positive at 100 copies/ml when retested ( Fig 1b ). These 5 discordant results corresponding to EBV in blood (n = 3) and BKV in blood and urine (n = 2) DNA amplifications could be explained by stochastic repartition of nucleic acids in the samples. The remaining qualitatively discordant result corresponded to the detection of a coronavirus in a nasopharyngeal secretion, initially positive at 2280 copies/ml with a Ct of 36.6, and negative when redone ( Fig 1c ). Even though all these six qualitative discrepancies were expected since being below the level of sensitivity of our PCRs, we considered that such results may have a significantly different clinical impact ( Table 1 ). The analysis conducted on the effect of time on the discrepancy level was inconclusive because of the small number of discordant results. The average time between original testing and replication was longer for concordant results than for discrepant.

Straight line represents the line of identity, expressing perfect match. Pointed lines delaminate a ± 3.3 Ct tolerance margin. Represented are low positive versus negative results (n = 5, diamonds); moderate positive versus negative results (n = 1, square), quantitative discrepancies with over one log 10 difference in copies/ml (straight triangles, n = 3), quantitative discrepancy with difference over 3.3Ct but less than one log10 (reversed triangle, n = 1); agreeing positive results (rounds, n = 20). 92 negative agreeing results are not represented on this figure. Note that these results here include two negative pairs and one positive pair of tests that were not quantitatively reported in copies/ml but only reported in Ct number by the laboratory.
Concerning the four quantitative discordances out of the 23 samples pairs that included quantitative results, a Bordetella pertussis detection in a nasopharyngeal secretion was discordant both considering the bacterial load (2700 copies/ml versus 260 copies/ml) and the Ct values (34.6 versus 39.5) ( Fig 1d ). However, this difference was not considered clinically significant ( Table 1 ). A Parainfluenza 2 also tested in a nasopharyngeal secretion was initially reported at 505’000 copies/ml and then reported at only 50’000 copies/ml when redone, a difference not considered to be associated with a significant clinical impact; this one log 10 difference in viral load quantification was likely due to a technical error when redone since the CT difference was of 2.4 (31.0 versus 33.4) ( Fig 1e and Table 1 ). The third quantitatively discordant result corresponded to the detection of HSV-1 in a vaginal swab with a viral load of 9.6×10 10 copies/ml versus 1.6×10 8 copies/ml, a difference identified to be due to a human transcription error since the Ct values were similar (19.8 and 19.0) ( Fig 1f ). This error was sufficient to lead to a change of process with implementation of automated data transfer from the PCR automated system to the Laboratory Information System (LIS), but was not considered to have a significant clinical impact ( Table 1 ). The fourth quantitative discordance had little clinical impact ( Table 1 ) despite the Ct difference (39.1 versus 35.4) since the calculated EBV viral load exhibited a difference of less than one log 10 (182 versus 771 copies/ml) ( Fig 1g ).
Conventional microbiology
Ninety-seven samples were duplicated and resubmitted to the conventional microbiology laboratory, requiring variable combinations of Gram staining, culture and AST. Four samples pairs results were excluded of our analysis. Two because they were including only parasites in stool analyses, too few to be considered; one because it was possibly being subject to an inversion in IQA sample preparation since staining and cultures results were incompatible; one because the request for analysis forms were not demanding for the same analyses.
Regarding microscopy of the Gram stains, 32 out of 81 (39.5%) pairs exhibited at least one major discrepancy (Tables (Tables1 1 and and2), 2 ), mainly due to discordances in the reported quantities of red blood cells (19.8%), leukocytes (8.6%) and Gram-positive cocci (8.6%) ( Table 2 ). Major discordances were observed in 5.3%, 4.9% and 3.7% for Gram-negative coccobacilli, Gram-negative bacilli and Gram-positive bacilli, respectively ( Table 2 ). Overall agreement of the microscopy was of 75.2% (560/745) of all evaluated parameters, whereas the occurrence of a major error occurred in 6.4% of cases (48/745) (Tables (Tables1 1 and and2). 2 ). However, this relatively high overall agreement rate should be weighted by the common negativity of some parameters such as Gram-positive diplococci (100% of negativity), yeasts (96.3%) and Gram-negative coccobacilli (93.0%). Thus, after excluding those parameters, the obtained overall agreement would drop significantly from 75.2% to 67.6%. True rate of concordance is better assessed by weighted Kappa scores, shown in Table 2 for each parameter. As many as 13 out of 71 (18.3%) samples exhibiting at least one discrepancy were considered as having a clinically relevant discordance, corresponding to a rate of 16.0% (13/81) for all Gram stain results investigated ( Table 1 ). The discrepancies were considered by our experts to be possibly related to reading or reporting errors in 8 out of 13 significant and in 11 out of 58 negligible discrepant pairs of samples, while the rest seemed compatible with natural biological variations (e.g. sample and coloration heterogeneity). It should also be noted that for two samples, the Gram stain reading was done only for one of the replicates because of pre-analytical disagreement on the aspect of urine (urines are examined under microscopy only if considered cloudy). Regarding the effect of the time delay between the management of the original samples and their replicates, no significant differences were observed, when comparing the identical and negligible discrepant results versus the minor and major discrepant results. This absence of difference was recovered both when comparing all parameters together and bacteria and yeasts only.
| Identical result | Negligible discrepancy | Minor discrepancy | Major discrepancy | Total | Rate of double negativity | Weighted Kappa | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 37 | . | 29 | . | 8 | . | 7 | . | 81 | . | 0.49 | |
| 51 | . | 9 | . | 5 | . | 16 | . | 81 | . | 0.55 | |
| 58 | . | 11 | . | 6 | . | 6 | . | 81 | . | 0.62 | |
| 49 | . | 21 | . | 4 | . | 7 | . | 81 | . | 0.66 | |
| 57 | . | 0 | . | 0 | . | 0 | . | 57 | . | N.R. | |
| 60 | . | 3 | . | 0 | . | 1 | . | 64 | . | 0.69 | |
| 48 | . | 18 | . | 11 | . | 4 | . | 81 | . | 0.63 | |
| 53 | . | 1 | . | 0 | . | 3 | . | 57 | . | N.R. | |
| 69 | . | 8 | . | 1 | . | 3 | . | 81 | . | 0.79 | |
| 78 | . | 2 | . | 0 | . | 1 | . | 81 | . | N.R. | |
| 560 | . | 102 | . | 35 | . | 48 | . | 745 | |||
Gram staining of routine samples and IQA samples were compared and discrepancy amplitudes were categorized according to S1B Table for each parameter. Rate of double negativity represents the proportion of results that were negative for both the routine and IQA sample. Weighted Kappa score express the level of concordance between two rates, not only due to natural probability of agreement. Score of -1 expresses perfect disagreement; 0 expresses agreement corresponding to random; probability; 1 expresses perfect agreement. [ 36 , 37 ]
GP = Gram-positive; GN = Gram-negative; NR = non-relevant
Culture results were perfectly identical for 43 pairs of samples, 16 of which were twice sterile (Tables (Tables1 1 and and3). 3 ). For 24 other pairs of samples, the results showed only negligible or minor discrepancies within semi-quantitative scoring of similarly identified or described colonies, whereas 17 pairs showed discrepancies within the identification or description of colonies, as detailed in Table 3 . To be noted, two pairs of samples for which cultures were accomplished only for one member of the pair: once due to diverging microscopy results with a potentially significant impact and once to an error in procedure with negligible impact ( Table 3 ). Also noteworthy, three cases in which the differences in microscopy results motivated the additional inoculation of Gram-positive selective agar, explaining why supplementary clinically-significant strains ( Enterococcus faecalis or Staphylococcus aureus ) were recovered from cultures ( Table 3 ). Interestingly, when comparing the time delay from original samples processing to management of the replicates, the delay was significantly longer for samples that included minor or major discrepancies in culture growth versus the ones that were identical or negligibly discrepant (14.68 hours ± 2.45 versus 9.19 hours ± 1.13, p = 0.039).
| Unnecessary cultivation of vaginal swab for the IQA sample, with otherwise agreeing microscopy results showing no potential pathogen. Difference of procedure considered as of negligible impact. | |
| Respiratory sample was not cultivated for patient’s sample because of microscopy results suggestive of a contamination, whereas the IQA microcopy results led to cultivation and growth of moderate quantities of along a physiological mixed oropharyngeal flora. Difference of procedure considered of significant impact. | |
| Low quantities of recovered only in the patient’s respiratory sample but not in the IQA replicate, with otherwise agreeing results ( and oropharyngeal flora) | |
| Low quantities of recovered only in the patient’s respiratory sample, with otherwise agreeing results ( , yeast other than ) | |
| Low quantities of recovered only in the patient’s pharyngeal swab sample, with otherwise agreeing results (oropharyngeal flora) | |
| spp (10 bacteria/ml) recovered only in the patient’s urinary sample, with otherwise agreeing results ( ) | |
| Two strains of detected in the respiratory IQA sample, whereas only one strain was reported in the patient’s sample, with diverging antibiotic susceptibility between the two strains only for penicillin | |
| Similar growth on plates but different interpretation with only a description of the grown colonies for the patient’s respiratory sample (low quantities of , 3 different morphologies and oropharyngeal flora); this mixed bacterial flora turned out to include low quantities of and of group bacteria, with a physiological oropharyngeal flora, when the Gram-negative colonies were properly identified for the IQA sample | |
| Similar growth but different interpretation with only a partial identification for the patient’s respiratory sample (low quantifies of group and 3 other morphologies) which turned out to include low quantities of when properly identified for the IQA sample | |
| Similar growth but different interpretation with only description for IQA respiratory sample (low quantities of Gram-negative with oropharyngeal flora) which turned out to include low quantities of when properly identified for the patient’s sample. This difference of procedure could be the consequence of differences in the Gram stain results (slightly more abundant leucocytes and Gram-negative cocci may have motivated formal identification for the patient’s sample) | |
| Low quantities of and moderate quantities of recovered only in IQA ascites liquid sample, while initial patient sample was sterile. The Enterococcus grew on a Gram-positive selective plate only used for the IQA sample because of abundant Gram-positive cocci seen under microscopy. Possible error in samples handling, during the preparation of the IQA sample, since Gram results were also discordant | |
| Low quantities of recovered only in IQA ascites liquid sample, while patient’s sample stayed sterile | |
| (10 germs/ml) and (10 germs/ml) recovered only in patient’s urinary sample, while IQA sample stayed sterile. Suspicion of error in sample handling | |
| (10 germs/ml) recovered only in IQA urinary sample, in otherwise agreeing results ( ). This difference was explained by the usage of a Gram-positive selective agar plate since abundant Gram-positive cocci were observed under microscopy for IQA sample | |
| Moderate quantities of recovered only in the patient’s sample (ear swab), with otherwise agreeing results ( ). This difference was explained by the usage of Gram-positive selective agar plate since abundant Gram-positive cocci were observed under microscopy for the patient’s sample | |
| Low quantities of recovered only in the patient’s respiratory sample, in otherwise agreeing results ( group and ) | |
| Low quantities of group, and recovered only in the patient’s sample (a biliary liquid), with otherwise agreeing results ( spp). | |
| Low quantities of group recovered only in the IQA sample (a superficial wound swab), with otherwise agreeing results ( ) | |
| High quantities of s fragilis group recovered only in the patient’s urine sample, with otherwise agreeing results ( and ) | |
* indicates the colonies that led to an AST when present.
Regarding AST panels, at least one major discrepancy was observed in five (10.4%) out of 48 comparable AST panels, whereas 7 (14.6%) included at least one minor discrepancy, the remaining 36 AST panels (75.0%) being perfectly identical ( Table 4 ). These discordances had little clinical impact since only one minor discrepancy concerned an antibiotic which would potentially have been used in this situation (a Pseudomonas aeruginosa in a respiratory sample which was once susceptible and once intermediate to levofloxacin) ( Table 1 ).
| Identical AST results | 36 | |
| AST including at least one minor discrepancy | 7 | |
| AST including at least one major discrepancy | 5 | |
Major and minor categories as defined in S1C Table .
As many as 23 AST panels, done on the patient sample or the IQA replicate, had no matching AST panel for its counterpart, either because of different interpretation of similar culture results (n = 8) or because of different culture results (n = 15) (Tables (Tables3 3 and and5). 5 ). The clinical significance of the 8 AST panels not performed in presence of identical culture results is negligible, especially since 5 of them were accompanied in transmitted results by a comment suggesting to the clinician to ask for AST via telephone if needed. Nevertheless, this difference in attitude could possibly influence the clinical interpretation by unexperimented practitioners. On the contrary, 14 out of 15 of the identified bacteria that led to diverging presence of AST were considered as significant (species marked by * in Tables Tables3 3 and and5 5 )
| ( ) | |
| ( ) | |
When AST were compared individually, each tested antibiotic one by one, the overall agreement was as high as 96.7% (734/759). There were 15 minor (sensitive versus intermediate or intermediate versus resistant) and 10 major (sensitive versus resistant) discrepancies. Piperacillin-tazobactam was the antibiotic the most often discrepant, with 3 major and 2 minor discrepancies. Detailed analysis of AST results, ordered by antibiotics, is shown in Fig 2 , which represents concordances and discordances for antibiotics tested at least 10 times. All other results are available in S2 Table .

Counts of concordant pairs of AST results are in green, minor discrepancies (sensitive versus intermediate or intermediate versus resistant) are in yellow and major discrepancies (sensitive versus resistant) are in red. Only antibiotics tested more than 10 times are represented. Complete results are available in S2 Table . AMC = amoxicillin-clavulanic acid; AMK = amikacin; AMP = ampicillin; AMX = amoxicillin; CAZ = ceftazidime; CIP = ciprofloxacin; CLI = clindamycin; CLR = clarithromycin; CRO = ceftriaxone; CXM = cefuroxime; ERY = erythromycin; ETP = ertapenem; FEP = cefepime; FOF = fosfomycin; GEN = gentamicin; IPM = imipenem; LVX = levofloxacin; MEM = meropenem; NIT = nitrofurantoin; PEN = penicillin; SXT = trimethoprim-sulfamethoxazole; TEC = teicoplanin; TET = tetracycline; TOB = tobramycin; TZP = piperacillin-tazobactam; VAN = vancomycin.
Nine pairs of sample included rapid testing such as tests for C . difficile antigens and inducible beta-lactamase. All these tests were concordant.
Mycobacteria
Five pairs of samples submitted to conventional microbiology or molecular biology laboratories also included auramine stains, culture and AST for mycobacteria. All the results were concordant, showing agreement on two positive samples and three negative samples.
This work demonstrates the feasibility and usefulness of an internal quality assurance program as part of the global quality management in serology, molecular biology and conventional microbiology laboratories. Looking at the level of agreement of the different laboratories, we first observe the high robustness of serological tests, which is partially due to automated systems and commercial kits which are mostly used, enhancing the reproducibility of processes. Moreover, serological tests are achieved on sera, which are by nature homogenous, limiting the variability between different fractions of a given sample.
In molecular biology, the usage of IQA samples shows the common occurrence of discrepancies (10/122 analyses, Fig 1 ). Most discrepant results were due to the stochastic variability of qPCR occurring around the limit of detection, where by definition qPCRs lose part of their sensitivity [ 42 ]. Accordingly, these discrepancies should not be considered as technical errors or insufficiencies. Yet, this variability for weakly positive samples may be overlooked by clinicians who often give too much importance to negative results, even when the negative predictive value is known to be low (i.e. for M . tuberculosis detection and for Listeria PCR in cerebrospinal fluid). While such discrepancies are of limited implication in terms of quality assurance, the single quantitative discrepancy of a 610-folds factor between copies/ml, with almost identical Ct number, was worrisome. Investigations revealed that this difference was the consequence of an error in the manual transcription of the result. In this specific situation, this error did not have clinical impact, since both results would be interpreted as highly positive by clinicians. However, this error highlighted a weak point in our procedures and led us to integrate our automated PCR system with a middleware capable of two directions data transfers from the Applied Biosystems PCR machine to the LIS, in order to definitively abolish any risk of human errors during results retranscription. Made worthy, no false positive seem to have resulted from PCR contaminations. This is particularly remarkable considering the high sensitivity of PCR and likely results from the advanced automatization of our qPCR platform [ 19 ]. Moreover, our IQA process proved itself useful to detect potential errors in this laboratory.
Gram stain reading is well established as a complex procedure that has number of pitfalls and its poor reliability has been described at multiple occasions [ 43 – 47 ]. It is still worrisome to observe that almost 40% of the compared results included at least one discrepancy of more than two relative units, which we categorized as major ( Table 2 ). This rate cannot be directly compared to any other data available in literature, the only similar experiment being the study of Constantine et al. (14), which had only 2% of discrepancies for microscopy, but using a very different definition of discrepancy which allowed much larger differences in the semi-quantitative results. The other studies on the reproducibility of Gram stain use very different methodologies, such as comparison of the concordance between Gram stain and culture results or between different observers reading identical smears, thus providing non-comparable results [ 44 – 47 ].
Observing the results by parameters in Table 2 , the repartition of reported results should be considered. For example, the very high Gram-positive diplococci or yeasts concordance is associated with a very high level of negativity. Kappa score partially helps to compare parameters between them, taking into account the natural proportion of concordance explained by random probability [ 36 ]. According to this score, we conclude that in our laboratory the counts of host cells (leukocytes, erythrocytes) is less reliable than the counts of bacteria. This questions the sense of reporting parameters such as erythrocytes, with such a high level of variability and low clinical impact. Those data also support the need for continuous staff training in this domain and the integration of automatization in Gram reading to improve reproducibility.
In the evaluation of error rates in Gram staining examination, it is important to emphasize that our study included no gold-standard. Thus, our experts (GGR and CDU) could only subjectively assess the most likely origin of the discrepancies. Errors were typically suspected when discrepancies were not compatible with biological variations because they were of very large amplitude or when confusion was suspected (e.g. ++++ Gram-positive cocci versus ++++ erythrocytes). Nevertheless, the factors leading to result heterogeneity and errors are numerous for Gram stains. Indeed, the important heterogeneity of samples such as sputa or fragments and the heterogeneity induced by the staining process can already bring a lot of variability before the reading phase itself, which is a task known as subjective and complex. Moreover, despite the high proportion of discrepant cases, our experts concluded that most of them would have no clinical impact, mainly because they involved host cell quantifications and not detection of bacteria ( Table 1 ). Furthermore, the clinical impact of Gram stains itself is subject to discussion, as for the diagnosis of pneumonia [ 48 , 49 ]. Nevertheless, if the importance of Gram results can be relativized, our work also showed that discrepancies in microscopy results can influence subsequent analytical steps, with significant impact on final culture and AST results transmitted to the clinicians (Tables (Tables3 3 and and5 5 ).
Reviewing the comments transmitted with the Gram stain results, an interesting observation was made concerning 4 out of 22 urinary samples that had contradictory comments which concluded once to negativity and once to positivity for the corresponding replicate. These discrepancies were caused by relative quantifications of bacteria that were scored “few” for one sample and “+” or “++” for the other, with an automatic comment announcing positivity in the second case only. Comparing those results with the culture results performed on the same samples, all 4 samples were confirmed positive. This observation questions the usefulness of the “few” category.
The results of culture shown in Table 3 , with 12 significant differences out of 84 cultures pairs (85.7% accuracy), are representative of its complexity. Culture is a dynamic process influenced by many factors, including heterogeneity of samples, Gram stains results, selected culture media, stochastic growth variability and interpretation of the clinical significance of the presence of colonies with choices to identify or not the grown colonies. All these factors contribute to explain the observed discrepancies. Better homogenization of the samples, reconsideration of how Gram results are used to select culture media and training of the personnel concerning analytical choices are all possible ways to improve reliability of the culture steps. Furthermore, regarding the design of an IQA program, longer time between the treatment of the original samples and their replicates was in our data associated with higher level of discrepancy in cultures results. Even this is to be expected, it underlines the importance of shortening the time delay between the inoculation of the original samples and the replicates.
Interestingly, we observed a low rate of significant discrepancies within the AST performed on identical bacteria ( Table 4 , Fig 2 and S2 Table ). On the other hand, steps preceding accomplishment of AST (Gram staining, cultivation, identification and interpretation of culture) often influence significantly the AST transmitted to the clinicians (Tables (Tables3 3 and and5). 5 ). These observations make us consider that the accuracy of the analytical phase of AST is satisfactory and that we should focus on the pre-analytical factors that are influencing their realization. It could be argued that the clinical information available concerning the IQA samples were different (e.g. the age) or lacking (e.g. the service of hospitalization), influencing the interpretation of the culture. If this may explain some discrepancies in the interpretation of similar culture results, discrepancies originating from diverging interpretation of similar cultures concerned only a minority of samples ( Table 5 ).
Our study, which included only a limited number of samples, provided interesting data on reproducibility. However, samples were not totally chosen in blind but sometimes selected because they addressed a particular analytical question. For instance, cloudy urinary samples were preferably chosen over limpid samples and positive samples are certainly over-represented, increasing the probability of diverging results.
Another limitation of our IQA program concerns specifically the serology laboratory. First, in this facility, blood samples are coming in gel-based serum separating tubes. They are centrifuged and the supernatant is directly transferred into new tubes. This implies that the replicates could only come from already treated samples and that their visual aspect was different from almost all routine samples. The technicians of this facility are routinely checking anterior results from the tested patients to integrate the new results within their serological status. Because we were using a limited pool of anonymous patient profiles, our technicians eventually faced multiple anterior results corresponding to IQA samples processed earlier. Both the visual difference of samples and the anterior results from anonymous patient’s profile led to strong suspicions of the staff working in the serology laboratory about our ongoing experimental IQA process, which finally obliged us to discontinue the IQA in this laboratory. This specific issue did not concern the two other facilities where our process could be completed in secret and on samples that were not pretreated. Despite this limitation, we consider that our process could still have an interest for IQA in serology laboratories, but that it would need to be partly redesigned. A proposition would be to duplicate samples before their arrival in the laboratory. It would solve the problem of visual impact and improve the coverage of pre-analytical steps. It would also be necessary to use more sophisticated fake patient identities, without previous IQA results to prevent their recognition by our coworkers.
Finally, even if our results cover more steps than traditional quality processes, it is still not covering all steps from prescription to interpretation of results by clinicians. Thus, we intend to extend this quality assurance project by collaborating with three other Swiss hospital laboratories. Each laboratory would challenge the others with split samples sent into containers accompanied by etiquettes and identifications used by the receiving laboratories for their external demands. This way, each laboratory would play the role of reference laboratory for one fourth of all samples. This approach would resolve the limitations of our scheme concerning the pre-treatment of the serology samples, the recognition of the samples and the lack of reference results. Such interlaboratory shared quality assurance would also help to provide “external assurance” for some serological and molecular parameters for which there is currently no EQA available (i.e. Tropheryma whipplei ).
In conclusion, this study revealed insightful points to improve and questioned our current practices, such as the use of Gram results to select culture media. Also, this study confirms the great interest of an IQA scheme based on split samples reprocessed in blind. It represents only few hundreds supplementary analyses, on the over 25’000 analyses run every year in our laboratory and this allowed us to generate interesting data on our reproducibility. This design showed itself to be perfectly complementary to other quality management procedures already ongoing in our laboratory and it should be further developed to better delineate the identified limitations and to settle its place in the overall quality assurance program.
Part of this publication was the subject of a master thesis done by Valentin Scherz under the direct supervision of Professor Greub at the School of Medicine of the University of Lausanne. Christian Durussel is the senior laboratory technician, acting at the Institute of Microbiology as the quality officer.
Supporting information
Categories defining the amplitude of the discrepancies observed between A. semi-quantification of parameters pairs of Gram reading; B. semi-quantification of grown colonies; C. Antibiotic susceptibility testing (AST) all performed twice, first as part of the routine diagnostic procedure and then as part of the IQA program.
Complete results of per antibiotics analysis of antibiotics sensitivity testing concordances and discrepancies. Minor discrepancies are either sensitive versus intermediate or intermediate versus resistant. Major discrepancies are sensitive versus resistant.
Acknowledgments
We are grateful to the following laboratory technicians: Sarah Chappuis, Maria Senra-Ortiz and René Brouillet for the replication of the samples. We are also grateful to all technicians for reprocessing the samples and to Tytti Heinonen for her help in correcting this manuscript.
Funding Statement
Costs of this study were covered by the quality assurance budget of our microbiology institute and accordingly no specific funding were obtained for this work.
Data Availability
Quality Control in a Microbiology Laboratory
- First Online: 27 February 2024
Cite this chapter

- Sanjay Jain 4 ,
- Aarzoo Jahan 5 &
- Sompal Singh 5
337 Accesses
The quality control (QC) practice in a microbiology laboratory is a bit different than what we have covered till now. This chapter provides an overview of quality control specific to microbiology. Pre-analytical and analytical phases emphasis on QC in culture media, stains, processing of samples for culture, and various bacteriological techniques. Special attention is given to the QC for antimicrobial susceptibility testing (AST). The chapter also covers QC in serology, sterilization and disinfection, and the maintenance of reference QC stocks. Sections on specific microbiological domains such as bacteriology, mycology, mycobacteriology, virology, and parasitology provide insights into quality control in each area. QC in post-analytical phase, i.e., reporting and interpretation and external quality assurance contributing to the precision and reliability of microbial analyses.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Durable hardcover edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Suggested Reading
World Health Organization. Regional Office for South-East Asia. Quality assurance in bacteriology and immunology. 3rd ed. WHO Regional Office for South-East Asia; 2012. Available from: https://apps.who.int/iris/handle/10665/205730
Google Scholar
Download references
Author information
Authors and affiliations.
Department of Microbiology, North Delhi Municipal Corporation Medical College and Hindu Rao Hospital, New Delhi, India
Sanjay Jain
Department of Pathology, North Delhi Municipal Corporation Medical College and Hindu Rao Hospital, New Delhi, India
Aarzoo Jahan & Sompal Singh
You can also search for this author in PubMed Google Scholar
Editor information
Editors and affiliations.
Department of Pathology and Lab Medicine, All India Institute of Medical Sciences, Bhopal, Madhya Pradesh, India
Shakti Kumar Yadav
Division of Cytopathology, ICMR-National Institute of Cancer Prevention and Research, Noida, India
Ruchika Gupta
Sompal Singh
Rights and permissions
Reprints and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Jain, S., Jahan, A., Singh, S. (2023). Quality Control in a Microbiology Laboratory. In: Yadav, S.K., Gupta, R., Singh, S. (eds) Clinical Laboratory Management . Springer, Cham. https://doi.org/10.1007/978-3-031-46420-1_27
Download citation
DOI : https://doi.org/10.1007/978-3-031-46420-1_27
Published : 27 February 2024
Publisher Name : Springer, Cham
Print ISBN : 978-3-031-46419-5
Online ISBN : 978-3-031-46420-1
eBook Packages : Medicine Medicine (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
- Search Menu
- Sign in through your institution
- Advance articles
- Editor's Choice
- Supplements
- Virtual Roundtables
- Journal of Antimicrobial Chemotherapy
- JAC-Antimicrobial Resistance
- Author Guidelines
- Submission Site
- Open Access
- About the Journal of Antimicrobial Chemotherapy
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Self-Archiving Policy
- Dispatch Dates
- Contact BSAC
- Journals on Oxford Academic
- Books on Oxford Academic

Article Contents
- < Previous
Quality Assurance: Principles and Practice in the Microbiology Laboratory: J. J. S. Snell, D. F. J. Brown & C. Roberts, Eds. Public Health Laboratory Service, UK, 2000. ISBN 0-091144-452, £25.00.
- Article contents
- Figures & tables
- Supplementary Data
R. A. Howe, Quality Assurance: Principles and Practice in the Microbiology Laboratory: J. J. S. Snell, D. F. J. Brown & C. Roberts, Eds. Public Health Laboratory Service, UK, 2000. ISBN 0-091144-452, £25.00., Journal of Antimicrobial Chemotherapy , Volume 46, Issue 5, November 2000, Page 865, https://doi.org/10.1093/jac/46.5.865
- Permissions Icon Permissions
Quality issues are becoming increasingly important in diagnostic laboratories. The fact of quality is no longer sufficient and we must now develop mechanisms to assure consumers, the public and, most importantly, ourselves of the continuing quality of our service. Moving towards a quality-assured system is not easy, requiring a meticulous attention to detail in all areas of a laboratory's working and organization. Thus this book is welcome as an excellent practical guide to the institution of quality assurance (QA) in microbiology.
Since it was first published in 1991 under the title Quality Control: Principles and Practice in the Microbiology Laboratory there has been considerable revision, with the addition of several sections and many new contributors. The backbone of the book is a series of chapters addressing quality assurance issues in each area of the laboratory, including computing and infection control. In addition there are chapters on internal and external quality assessment, and preparing a laboratory for CPA or UKAS accreditation.
A great strength of this book is the practical style in which it has been written by contributors who clearly have experience of instituting the principles of QA in their laboratories. Thus, most chapters have clear descriptions of all of the factors that must be covered for quality assurance with examples. The new chapter on internal quality assessment gives a particularly good account of how to set up such a system. Obviously with many authors contributing on the same subject from slightly different angles there is a degree of repetition in their discussion of the principles of QA. A detailed explanation of the Westgard rules is probably not required both in the chapter on QA of antimicrobial assays and that on internal quality control in serodiagnosis. However, this is only a minor point against a book that can be recommended not only to its purported target audience of those with responsibility for the planning, implementation and maintenance of quality systems, but equally to both medical and scientific trainees.
| Month: | Total Views: |
|---|---|
| December 2016 | 3 |
| January 2017 | 1 |
| February 2017 | 10 |
| March 2017 | 11 |
| April 2017 | 8 |
| May 2017 | 3 |
| June 2017 | 5 |
| July 2017 | 10 |
| August 2017 | 14 |
| September 2017 | 10 |
| October 2017 | 12 |
| November 2017 | 18 |
| December 2017 | 76 |
| January 2018 | 49 |
| February 2018 | 59 |
| March 2018 | 81 |
| April 2018 | 113 |
| May 2018 | 67 |
| June 2018 | 65 |
| July 2018 | 58 |
| August 2018 | 47 |
| September 2018 | 56 |
| October 2018 | 21 |
| November 2018 | 54 |
| December 2018 | 34 |
| January 2019 | 27 |
| February 2019 | 38 |
| March 2019 | 53 |
| April 2019 | 87 |
| May 2019 | 36 |
| June 2019 | 32 |
| July 2019 | 81 |
| August 2019 | 45 |
| September 2019 | 56 |
| October 2019 | 67 |
| November 2019 | 47 |
| December 2019 | 39 |
| January 2020 | 84 |
| February 2020 | 89 |
| March 2020 | 137 |
| April 2020 | 98 |
| May 2020 | 46 |
| June 2020 | 59 |
| July 2020 | 45 |
| August 2020 | 58 |
| September 2020 | 50 |
| October 2020 | 36 |
| November 2020 | 61 |
| December 2020 | 50 |
| January 2021 | 50 |
| February 2021 | 55 |
| March 2021 | 22 |
| April 2021 | 21 |
| May 2021 | 18 |
| June 2021 | 30 |
| July 2021 | 32 |
| August 2021 | 22 |
| September 2021 | 24 |
| October 2021 | 34 |
| November 2021 | 22 |
| December 2021 | 27 |
| January 2022 | 17 |
| February 2022 | 24 |
| March 2022 | 19 |
| April 2022 | 27 |
| May 2022 | 28 |
| June 2022 | 12 |
| July 2022 | 19 |
| August 2022 | 37 |
| September 2022 | 21 |
| October 2022 | 18 |
| November 2022 | 29 |
| December 2022 | 26 |
| January 2023 | 23 |
| February 2023 | 17 |
| March 2023 | 21 |
| April 2023 | 30 |
| May 2023 | 37 |
| June 2023 | 29 |
| July 2023 | 31 |
| August 2023 | 28 |
| September 2023 | 22 |
| October 2023 | 18 |
| November 2023 | 21 |
| December 2023 | 62 |
| January 2024 | 33 |
| February 2024 | 22 |
| March 2024 | 86 |
| April 2024 | 67 |
| May 2024 | 64 |
| June 2024 | 24 |
| July 2024 | 16 |
| August 2024 | 5 |
Email alerts
Citing articles via.
- About Journal of Antimicrobial Chemotherapy
- Recommend to your Library
Affiliations
- Online ISSN 1460-2091
- Print ISSN 0305-7453
- Copyright © 2024 British Society for Antimicrobial Chemotherapy
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Click through the PLOS taxonomy to find articles in your field.
For more information about PLOS Subject Areas, click here .
Loading metrics
Open Access
Peer-reviewed
Research Article
Internal quality assurance in diagnostic microbiology: A simple approach for insightful data
Roles Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing
Affiliation Institute of Microbiology, University of Lausanne and University Hospital Center, Lausanne, Switzerland
Roles Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation
Roles Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing
* E-mail: [email protected]
- Valentin Scherz,
- Christian Durussel,
- Gilbert Greub

- Published: November 14, 2017
- https://doi.org/10.1371/journal.pone.0187263
- Reader Comments
Given the importance of microbiology results on patient care, high quality standards are expected. Internal quality assurance (IQA) could mitigate the limitations of internal quality control, competency assessment and external quality assurance, adding a longitudinal insight, including pre- and post-analytical steps. Here, we implemented an IQA program in our clinical microbiology facilities with blind resubmission of routine samples during 22 months. One-hundred-and-twenty-one out of 123 (98.4%) serological analyses and 112 out of 122 (91.8%) molecular analyses were concordant. Among the discordances in molecular biology analyses, 6 results were low positive samples that turned out negative, likely due to stochastic repartition of nucleic acids. Moreover, one identified retranscription error led us to implement automated results transmission from the Applied Biosystems instruments to the laboratory information system (LIS). Regarding Gram stain microscopy, 560 out of 745 (75.2%) of compared parameters were concordant. As many as 67 out of 84 (79.8%) pairs of culture results were similar, including 16 sterile pairs, 27 having identical identification or description and semi-quantification and 24 only showing variations in semi-quantification with identical description or identification of colonies. Seventeen pairs had diverging identification or description of colonies. Culture was twice only done for one member of the pairs. Regarding antibiotic susceptibility testing, a major discrepancy was observed in 5 out of 48 results (10.4%). In conclusion, serological tests were highly reproducible. Molecular diagnosis also revealed to be robust except when the amounts of nucleic acids present in the sample were close to the limits of detection. Conventional microbiology was less robust with major discrepancies reaching 39.5% of the samples for microscopy. Similarly, culture and antibiotic susceptibility testing were prone to discrepancies. This work was ground for reconsidering multiples aspects of our practices and demonstrates the importance of IQA to complete the other quality management procedures.
Citation: Scherz V, Durussel C, Greub G (2017) Internal quality assurance in diagnostic microbiology: A simple approach for insightful data. PLoS ONE 12(11): e0187263. https://doi.org/10.1371/journal.pone.0187263
Editor: Holger Rohde, Universitatsklinikum Hamburg-Eppendorf, GERMANY
Received: May 30, 2017; Accepted: October 17, 2017; Published: November 14, 2017
Copyright: © 2017 Scherz et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: In accordance with our ethics statement and the particular context of quality assurance, explicit patient agreements were not obtained and this quality assurance project was exempted of ethical committee approval. Since particular patterns of microbiology results could lead to patients’ recognition, our institute’s quality committee, which is the initiator of this project and is responsible for all ethical aspects of our quality control projects, enforced that data cannot be made publicly available. A representative of this committee, Madame Sarah Chappuis, Rue du Bugnon 48, 1011 Lausanne, will answer to questions regarding this decision ( [email protected] ). This same representative will also make data available upon motivated request and commitment to keep the obtained data confidential.
Funding: Costs of this study were covered by the quality assurance budget of our microbiology institute and accordingly no specific funding were obtained for this work.
Competing interests: The authors have declared that no competing interests exists.
Introduction
Microbiology laboratories are nowadays facing numerous challenges. The spread of antimicrobial drugs resistances demands rapid and yet reliable characterization of causative agents to prevent treatment failures and evitable occurrences of resistances [ 1 – 3 ]. Regular identification of emerging pathogens [ 3 ] and implementation of new molecular techniques [ 4 ] bring greater technical complexity, while prompt results and better cost-efficiency are expected [ 5 – 7 ]. Besides these evolutions, the awareness about medical errors and their consequences emphasizes the great importance of quality in health [ 8 , 9 ]. Indeed, clinical laboratories and related quality management programs have a role in preventing deaths due to evitable medical errors [ 10 ].
Quality management went along a constant evolution from the sixties to reach the current state of practice [ 11 ]. It is now generally accepted [ 12 ] and legally prescribed in Switzerland [ 13 ] and in the USA [ 14 ] that a minimal quality assurance (QA) program is composed of quality control (QC), external quality assessment (EQA), standard operating procedure (SOP) and competency assessment (CA) of coworkers. These procedures, despite being needed and regulated by certification processes such as ISO 15189 [ 15 ], have major limitations discussed below.
First, QC focuses on the intrinsic performances of laboratory tests, reviewing their performance (specificity & sensitivity), the regular maintenance of all used materials, the constant quality of reactants and media, as well as the use of negative and positives controls [ 12 ]. Despite their established role, QC are insufficient [ 11 ], especially because they do not cover pre- and post-analytical steps [ 10 , 16 , 17 ].
EQA includes external audits as well as proficiency testing (PT) with samples sent by reference nodal laboratories to be analyzed and described [ 12 ]. Audits can reveal structural dysfunctions and possible points to improve. PT with unknown samples probes the ability of the laboratory to treat and characterize samples and to identify pathogens. Both audits and PT provide valuable information on the functioning of the laboratories but are limited by their rare occurrence. PT has shown several additional limitations:
- It only partially evaluates pre- and post-analytical steps [ 18 ]; for example, PT samples are often coming in modified forms (e.g. dried, synthetically produced, pre-treated as with serum samples instead of whole blood), which can prevent their inclusion into regular lines of sample handling.
- PT are not available for all the different tests offered by our laboratories (for instance no PT is available for unusual pathogens such as T . whipplei and W . chondrophila ) [ 19 ].
- It has been shown that their recognition as EQA can lead to falsely reassuring results because of the extra caution given in their processing [ 20 , 21 ]; indeed, samples labeled as EQA were shown to provide more reliable results than EQA submitted blindly [ 22 ].
Competency assessment (CA) probes the ability of the laboratory personnel to complete properly their tasks, in good adequacy with SOP. CA programs can be simple, such as direct observation of technicians and quizzes to complete, or more sophisticated, with for instance, submission of blind challenge samples [ 23 – 28 ]. Despite the different approaches evaluated in the literature, most suffer of similar limitations as QC, with a primary focus on individuals performing defined analytical phases and limited insight on the overall quality of laboratory processes. CA programs can also certainly be biased when the workers identify the time of evaluation. Interestingly, blind resubmissions of previously analyzed samples, which is one of the methods suggested in literature for CA purposes, seem to suffer of less limitations but are only seldom used [ 24 ].
Internal quality assurance (IQA) programs based on resubmissions of replicated samples could address the mentioned limitations but lack of supporting definitions and data. The potential of this type of procedures is recognized by reference organizations such as the Eurachem and the Standard Unit, Microbiology Services of Public Health England, which recommend them to assess internally the overall reproducibility of samples testing and handling [ 29 , 30 ]. However, there is very few original publications in clinical microbiology setting supporting these recommendations, the only ones being the studies conducted in Cambridge in the nineties by Constantine et al [ 31 ] and Gray et al [ 32 ]. The authors concluded then that their works showed the capacity of this system to detect error, highlighted point of improvements and produced relevant data on the reproducibility of their results and the overall quality of their laboratories [ 31 – 33 ].
Thus, in the present work, we implemented an internal quality assessment program using blind split samples. This provided data on the reproducibility of the results in our microbiology laboratory and helped us to define specific corrective measures.
Materials and methods
Study design.
A new IQA procedure was initiated in October 2014 in the three clinical laboratories (serology, molecular microbiology and conventional microbiology) of the Institute of Microbiology of the University Hospital of Lausanne (Switzerland). This procedure was designed and implemented by the quality committee of our laboratories and exempted from formal ethic committee approval based on its regulation regarding quality control studies [ 34 ]. Indeed, the scheme which was applied here is in line with IQA procedure recommended by reference organizations in routine practice [ 29 , 30 ]. The present work reviews the data gathered over a 22-month period and evaluated the discrepancies, including their possible origin and their putative clinical impact (see below).
Data collection
The head technicians of our three laboratories were instructed to secretly withdraw samples out of the already processed ones. In serology and molecular biology laboratories, samples were chosen to be proportionally representative of the analyses done by our facilities, but also ideally, to test at least once every test offered by our service. In the classical microbiology laboratory, the head technician generally selected samples that were expected to be challenging, because they were positive at Gram stain examination of by culture after overnight incubation. This approach was retained to increase the number of samples for which the adequacy of bacterial identification and antibiotics susceptibility could be investigated. The analysis request forms were replicated with anonymous identification data, which are commonly used in our institution for clinical research purposes (and thus not assimilated by our technician to a quality control). Part of the original sample material was taken into a new container and handed back in secret at the reception of the laboratories. The correspondence between the patient identification code and the replicate identification code was reported by the head technicians into a database for later comparison of the results. Results of the two matching samples were later reported into an Access database and data analyses were conducted with Microsoft Excel and PRISM (GraphPad Software). All data were processed anonymously.
Serology and molecular biology
Pairs of serology results were compared for agreement between qualitative results (positive, indeterminate or negative) and variations in titers when relevant. Similarly, results of qPCR pairs were compared for differences in (i) qualitative results (positive versus negative), (ii) number of copies/ml with a margin of acceptance of one log 10 between the two results and (iii) cycle thresholds (Ct) with a margin of acceptance of 3.3 Ct between the two results.
Gram stain examination
Gram stains results were compared regarding semi-quantification of the following parameters: leukocytes, erythrocytes, epithelial cells, yeasts, Gram-positive cocci, bacilli and diplococci and Gram-negative cocci, bacilli and coccobacilli. The given interpretations of the Gram results were also compared when relevant (e.g. suspicion of urines contamination, salivary flora in sputa, dominant bacterial flora in vaginal swabs). For each parameter, the amplitude of discrepancies was classified according to S1A Table . The most important discrepancy between all parameters of a pair of samples was recorded. We examined the discrepancies for each parameter by linearly weighted Kappa score using GraphPad QuickCalcs website [ 35 ]. Kappa score quantifies the level of correspondence between the ratings of two observers, considering the natural probability to obtain the same level of concordance only by chance [ 36 ]. The weighted version of Kappa score is used to consider the partial agreements within ordered scales [ 37 ]. We adopted the weighting suggested by Cicchetti et Allison [ 38 ], which sanctions discrepancies in a linear and not in a quadratic method [ 39 ].
Culture and antibiotic susceptibility testing (AST)
Culture results were compared for description and/or identification and semi-quantification and discrepant cases were described. Quantitative variations were scored according to S1B Table .
Sets of AST were compared when conducted for the same identified bacteria within a pair of replicates. Discrepancies were classified following S1C Table . The most significant discrepancy within all tested antibiotics was retained to grade the concordance of the whole AST sets (e.g. if the AST pairs included a single major discrepancy, the pair of sets were considered as being majorly discrepant). If AST were not performed for the same species on both replicates, the cause of the discrepancy was analyzed and described.
Evaluation of clinical impact and putative cause
Interpretation of the clinical impact and putative cause of discordant results was done by a data review committee of three persons (VS, CD and GG). Discrepancies were classified as clinically negligible or significant.
Among 123 serological tests performed twice on 35 sera, a total of 121 (98.4%) were concordant ( Table 1 ). The first discordant test was a measure of IgG against cytomegalovirus (CMV) by enzyme immuno-assay (EIA), which was initially positive (index of 1.20) and intermediate when redone (0.953 for a cut-off for positivity of 1 and for negativity of 0.5). The 123 tests were often completed within panels of multiple tests detecting antibody reactivity for the same pathogen (in total 76 different pathogen-specific panels), sometimes reducing the negative impact of discordant results. Thus, this first discordant result was accompanied by electrochemiluminescence (ECLIA) results, which were both times positive for anti-CMV IgG antibodies (4.84 and 4.78 U/ml with a cutoff of 1 for positivity). The second discordant pair of results was in the titers of a syphilis test (TPPA) obtained manually, being twice positive but with titers of 1/2'560 initially and 1/10’240 when redone. The clinical impact was considered significant in this 2 nd case since a high titer of 1/10’240 generally reflects an acute infection and since this difference of two dilutions might be considered as clinically relevant [ 40 , 41 ]. The limited number of discrepant results did not allow to investigate the effect of the time delays between the management of the original samples and their replicates.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.pone.0187263.t001
Molecular diagnosis
Among 122 tests achieved on 52 replicated samples, 112 (91.8%) molecular tests were concordant ( Table 1 ). Six tests were qualitatively different, being once positive and once negative. Of the 23 results pairs that included quantitative results, three were diverging in the number of copies/ml (difference in copies/ml over one log 10 ) and one was quantitatively different only considering the Ct values (difference in Ct over 3.3).
Regarding the qualitative differences, four discordant molecular results corresponded to low positive samples (132–175 copies/ml) that turned out to be negative when retested ( Fig 1a ) whereas one initially negative result turned out to be positive at 100 copies/ml when retested ( Fig 1b ). These 5 discordant results corresponding to EBV in blood (n = 3) and BKV in blood and urine (n = 2) DNA amplifications could be explained by stochastic repartition of nucleic acids in the samples. The remaining qualitatively discordant result corresponded to the detection of a coronavirus in a nasopharyngeal secretion, initially positive at 2280 copies/ml with a Ct of 36.6, and negative when redone ( Fig 1c ). Even though all these six qualitative discrepancies were expected since being below the level of sensitivity of our PCRs, we considered that such results may have a significantly different clinical impact ( Table 1 ). The analysis conducted on the effect of time on the discrepancy level was inconclusive because of the small number of discordant results. The average time between original testing and replication was longer for concordant results than for discrepant.
Straight line represents the line of identity, expressing perfect match. Pointed lines delaminate a ± 3.3 Ct tolerance margin. Represented are low positive versus negative results (n = 5, diamonds); moderate positive versus negative results (n = 1, square), quantitative discrepancies with over one log 10 difference in copies/ml (straight triangles, n = 3), quantitative discrepancy with difference over 3.3Ct but less than one log10 (reversed triangle, n = 1); agreeing positive results (rounds, n = 20). 92 negative agreeing results are not represented on this figure. Note that these results here include two negative pairs and one positive pair of tests that were not quantitatively reported in copies/ml but only reported in Ct number by the laboratory.
https://doi.org/10.1371/journal.pone.0187263.g001
Concerning the four quantitative discordances out of the 23 samples pairs that included quantitative results, a Bordetella pertussis detection in a nasopharyngeal secretion was discordant both considering the bacterial load (2700 copies/ml versus 260 copies/ml) and the Ct values (34.6 versus 39.5) ( Fig 1d ). However, this difference was not considered clinically significant ( Table 1 ). A Parainfluenza 2 also tested in a nasopharyngeal secretion was initially reported at 505’000 copies/ml and then reported at only 50’000 copies/ml when redone, a difference not considered to be associated with a significant clinical impact; this one log 10 difference in viral load quantification was likely due to a technical error when redone since the CT difference was of 2.4 (31.0 versus 33.4) ( Fig 1e and Table 1 ). The third quantitatively discordant result corresponded to the detection of HSV-1 in a vaginal swab with a viral load of 9.6×10 10 copies/ml versus 1.6×10 8 copies/ml, a difference identified to be due to a human transcription error since the Ct values were similar (19.8 and 19.0) ( Fig 1f ). This error was sufficient to lead to a change of process with implementation of automated data transfer from the PCR automated system to the Laboratory Information System (LIS), but was not considered to have a significant clinical impact ( Table 1 ). The fourth quantitative discordance had little clinical impact ( Table 1 ) despite the Ct difference (39.1 versus 35.4) since the calculated EBV viral load exhibited a difference of less than one log 10 (182 versus 771 copies/ml) ( Fig 1g ).
Conventional microbiology
Ninety-seven samples were duplicated and resubmitted to the conventional microbiology laboratory, requiring variable combinations of Gram staining, culture and AST. Four samples pairs results were excluded of our analysis. Two because they were including only parasites in stool analyses, too few to be considered; one because it was possibly being subject to an inversion in IQA sample preparation since staining and cultures results were incompatible; one because the request for analysis forms were not demanding for the same analyses.
Regarding microscopy of the Gram stains, 32 out of 81 (39.5%) pairs exhibited at least one major discrepancy (Tables 1 and 2 ), mainly due to discordances in the reported quantities of red blood cells (19.8%), leukocytes (8.6%) and Gram-positive cocci (8.6%) ( Table 2 ). Major discordances were observed in 5.3%, 4.9% and 3.7% for Gram-negative coccobacilli, Gram-negative bacilli and Gram-positive bacilli, respectively ( Table 2 ). Overall agreement of the microscopy was of 75.2% (560/745) of all evaluated parameters, whereas the occurrence of a major error occurred in 6.4% of cases (48/745) (Tables 1 and 2 ). However, this relatively high overall agreement rate should be weighted by the common negativity of some parameters such as Gram-positive diplococci (100% of negativity), yeasts (96.3%) and Gram-negative coccobacilli (93.0%). Thus, after excluding those parameters, the obtained overall agreement would drop significantly from 75.2% to 67.6%. True rate of concordance is better assessed by weighted Kappa scores, shown in Table 2 for each parameter. As many as 13 out of 71 (18.3%) samples exhibiting at least one discrepancy were considered as having a clinically relevant discordance, corresponding to a rate of 16.0% (13/81) for all Gram stain results investigated ( Table 1 ). The discrepancies were considered by our experts to be possibly related to reading or reporting errors in 8 out of 13 significant and in 11 out of 58 negligible discrepant pairs of samples, while the rest seemed compatible with natural biological variations (e.g. sample and coloration heterogeneity). It should also be noted that for two samples, the Gram stain reading was done only for one of the replicates because of pre-analytical disagreement on the aspect of urine (urines are examined under microscopy only if considered cloudy). Regarding the effect of the time delay between the management of the original samples and their replicates, no significant differences were observed, when comparing the identical and negligible discrepant results versus the minor and major discrepant results. This absence of difference was recovered both when comparing all parameters together and bacteria and yeasts only.
https://doi.org/10.1371/journal.pone.0187263.t002
Culture results were perfectly identical for 43 pairs of samples, 16 of which were twice sterile (Tables 1 and 3 ). For 24 other pairs of samples, the results showed only negligible or minor discrepancies within semi-quantitative scoring of similarly identified or described colonies, whereas 17 pairs showed discrepancies within the identification or description of colonies, as detailed in Table 3 . To be noted, two pairs of samples for which cultures were accomplished only for one member of the pair: once due to diverging microscopy results with a potentially significant impact and once to an error in procedure with negligible impact ( Table 3 ). Also noteworthy, three cases in which the differences in microscopy results motivated the additional inoculation of Gram-positive selective agar, explaining why supplementary clinically-significant strains ( Enterococcus faecalis or Staphylococcus aureus ) were recovered from cultures ( Table 3 ). Interestingly, when comparing the time delay from original samples processing to management of the replicates, the delay was significantly longer for samples that included minor or major discrepancies in culture growth versus the ones that were identical or negligibly discrepant (14.68 hours ± 2.45 versus 9.19 hours ± 1.13, p = 0.039).
https://doi.org/10.1371/journal.pone.0187263.t003
Regarding AST panels, at least one major discrepancy was observed in five (10.4%) out of 48 comparable AST panels, whereas 7 (14.6%) included at least one minor discrepancy, the remaining 36 AST panels (75.0%) being perfectly identical ( Table 4 ). These discordances had little clinical impact since only one minor discrepancy concerned an antibiotic which would potentially have been used in this situation (a Pseudomonas aeruginosa in a respiratory sample which was once susceptible and once intermediate to levofloxacin) ( Table 1 ).
https://doi.org/10.1371/journal.pone.0187263.t004
As many as 23 AST panels, done on the patient sample or the IQA replicate, had no matching AST panel for its counterpart, either because of different interpretation of similar culture results (n = 8) or because of different culture results (n = 15) (Tables 3 and 5 ). The clinical significance of the 8 AST panels not performed in presence of identical culture results is negligible, especially since 5 of them were accompanied in transmitted results by a comment suggesting to the clinician to ask for AST via telephone if needed. Nevertheless, this difference in attitude could possibly influence the clinical interpretation by unexperimented practitioners. On the contrary, 14 out of 15 of the identified bacteria that led to diverging presence of AST were considered as significant (species marked by * in Tables 3 and 5 )
https://doi.org/10.1371/journal.pone.0187263.t005
When AST were compared individually, each tested antibiotic one by one, the overall agreement was as high as 96.7% (734/759). There were 15 minor (sensitive versus intermediate or intermediate versus resistant) and 10 major (sensitive versus resistant) discrepancies. Piperacillin-tazobactam was the antibiotic the most often discrepant, with 3 major and 2 minor discrepancies. Detailed analysis of AST results, ordered by antibiotics, is shown in Fig 2 , which represents concordances and discordances for antibiotics tested at least 10 times. All other results are available in S2 Table .
Counts of concordant pairs of AST results are in green, minor discrepancies (sensitive versus intermediate or intermediate versus resistant) are in yellow and major discrepancies (sensitive versus resistant) are in red. Only antibiotics tested more than 10 times are represented. Complete results are available in S2 Table . AMC = amoxicillin-clavulanic acid; AMK = amikacin; AMP = ampicillin; AMX = amoxicillin; CAZ = ceftazidime; CIP = ciprofloxacin; CLI = clindamycin; CLR = clarithromycin; CRO = ceftriaxone; CXM = cefuroxime; ERY = erythromycin; ETP = ertapenem; FEP = cefepime; FOF = fosfomycin; GEN = gentamicin; IPM = imipenem; LVX = levofloxacin; MEM = meropenem; NIT = nitrofurantoin; PEN = penicillin; SXT = trimethoprim-sulfamethoxazole; TEC = teicoplanin; TET = tetracycline; TOB = tobramycin; TZP = piperacillin-tazobactam; VAN = vancomycin.
https://doi.org/10.1371/journal.pone.0187263.g002
Nine pairs of sample included rapid testing such as tests for C . difficile antigens and inducible beta-lactamase. All these tests were concordant.
Mycobacteria
Five pairs of samples submitted to conventional microbiology or molecular biology laboratories also included auramine stains, culture and AST for mycobacteria. All the results were concordant, showing agreement on two positive samples and three negative samples.
This work demonstrates the feasibility and usefulness of an internal quality assurance program as part of the global quality management in serology, molecular biology and conventional microbiology laboratories. Looking at the level of agreement of the different laboratories, we first observe the high robustness of serological tests, which is partially due to automated systems and commercial kits which are mostly used, enhancing the reproducibility of processes. Moreover, serological tests are achieved on sera, which are by nature homogenous, limiting the variability between different fractions of a given sample.
In molecular biology, the usage of IQA samples shows the common occurrence of discrepancies (10/122 analyses, Fig 1 ). Most discrepant results were due to the stochastic variability of qPCR occurring around the limit of detection, where by definition qPCRs lose part of their sensitivity [ 42 ]. Accordingly, these discrepancies should not be considered as technical errors or insufficiencies. Yet, this variability for weakly positive samples may be overlooked by clinicians who often give too much importance to negative results, even when the negative predictive value is known to be low (i.e. for M . tuberculosis detection and for Listeria PCR in cerebrospinal fluid). While such discrepancies are of limited implication in terms of quality assurance, the single quantitative discrepancy of a 610-folds factor between copies/ml, with almost identical Ct number, was worrisome. Investigations revealed that this difference was the consequence of an error in the manual transcription of the result. In this specific situation, this error did not have clinical impact, since both results would be interpreted as highly positive by clinicians. However, this error highlighted a weak point in our procedures and led us to integrate our automated PCR system with a middleware capable of two directions data transfers from the Applied Biosystems PCR machine to the LIS, in order to definitively abolish any risk of human errors during results retranscription. Made worthy, no false positive seem to have resulted from PCR contaminations. This is particularly remarkable considering the high sensitivity of PCR and likely results from the advanced automatization of our qPCR platform [ 19 ]. Moreover, our IQA process proved itself useful to detect potential errors in this laboratory.
Gram stain reading is well established as a complex procedure that has number of pitfalls and its poor reliability has been described at multiple occasions [ 43 – 47 ]. It is still worrisome to observe that almost 40% of the compared results included at least one discrepancy of more than two relative units, which we categorized as major ( Table 2 ). This rate cannot be directly compared to any other data available in literature, the only similar experiment being the study of Constantine et al. (14), which had only 2% of discrepancies for microscopy, but using a very different definition of discrepancy which allowed much larger differences in the semi-quantitative results. The other studies on the reproducibility of Gram stain use very different methodologies, such as comparison of the concordance between Gram stain and culture results or between different observers reading identical smears, thus providing non-comparable results [ 44 – 47 ].
Observing the results by parameters in Table 2 , the repartition of reported results should be considered. For example, the very high Gram-positive diplococci or yeasts concordance is associated with a very high level of negativity. Kappa score partially helps to compare parameters between them, taking into account the natural proportion of concordance explained by random probability [ 36 ]. According to this score, we conclude that in our laboratory the counts of host cells (leukocytes, erythrocytes) is less reliable than the counts of bacteria. This questions the sense of reporting parameters such as erythrocytes, with such a high level of variability and low clinical impact. Those data also support the need for continuous staff training in this domain and the integration of automatization in Gram reading to improve reproducibility.
In the evaluation of error rates in Gram staining examination, it is important to emphasize that our study included no gold-standard. Thus, our experts (GGR and CDU) could only subjectively assess the most likely origin of the discrepancies. Errors were typically suspected when discrepancies were not compatible with biological variations because they were of very large amplitude or when confusion was suspected (e.g. ++++ Gram-positive cocci versus ++++ erythrocytes). Nevertheless, the factors leading to result heterogeneity and errors are numerous for Gram stains. Indeed, the important heterogeneity of samples such as sputa or fragments and the heterogeneity induced by the staining process can already bring a lot of variability before the reading phase itself, which is a task known as subjective and complex. Moreover, despite the high proportion of discrepant cases, our experts concluded that most of them would have no clinical impact, mainly because they involved host cell quantifications and not detection of bacteria ( Table 1 ). Furthermore, the clinical impact of Gram stains itself is subject to discussion, as for the diagnosis of pneumonia [ 48 , 49 ]. Nevertheless, if the importance of Gram results can be relativized, our work also showed that discrepancies in microscopy results can influence subsequent analytical steps, with significant impact on final culture and AST results transmitted to the clinicians (Tables 3 and 5 ).
Reviewing the comments transmitted with the Gram stain results, an interesting observation was made concerning 4 out of 22 urinary samples that had contradictory comments which concluded once to negativity and once to positivity for the corresponding replicate. These discrepancies were caused by relative quantifications of bacteria that were scored “few” for one sample and “+” or “++” for the other, with an automatic comment announcing positivity in the second case only. Comparing those results with the culture results performed on the same samples, all 4 samples were confirmed positive. This observation questions the usefulness of the “few” category.
The results of culture shown in Table 3 , with 12 significant differences out of 84 cultures pairs (85.7% accuracy), are representative of its complexity. Culture is a dynamic process influenced by many factors, including heterogeneity of samples, Gram stains results, selected culture media, stochastic growth variability and interpretation of the clinical significance of the presence of colonies with choices to identify or not the grown colonies. All these factors contribute to explain the observed discrepancies. Better homogenization of the samples, reconsideration of how Gram results are used to select culture media and training of the personnel concerning analytical choices are all possible ways to improve reliability of the culture steps. Furthermore, regarding the design of an IQA program, longer time between the treatment of the original samples and their replicates was in our data associated with higher level of discrepancy in cultures results. Even this is to be expected, it underlines the importance of shortening the time delay between the inoculation of the original samples and the replicates.
Interestingly, we observed a low rate of significant discrepancies within the AST performed on identical bacteria ( Table 4 , Fig 2 and S2 Table ). On the other hand, steps preceding accomplishment of AST (Gram staining, cultivation, identification and interpretation of culture) often influence significantly the AST transmitted to the clinicians (Tables 3 and 5 ). These observations make us consider that the accuracy of the analytical phase of AST is satisfactory and that we should focus on the pre-analytical factors that are influencing their realization. It could be argued that the clinical information available concerning the IQA samples were different (e.g. the age) or lacking (e.g. the service of hospitalization), influencing the interpretation of the culture. If this may explain some discrepancies in the interpretation of similar culture results, discrepancies originating from diverging interpretation of similar cultures concerned only a minority of samples ( Table 5 ).
Our study, which included only a limited number of samples, provided interesting data on reproducibility. However, samples were not totally chosen in blind but sometimes selected because they addressed a particular analytical question. For instance, cloudy urinary samples were preferably chosen over limpid samples and positive samples are certainly over-represented, increasing the probability of diverging results.
Another limitation of our IQA program concerns specifically the serology laboratory. First, in this facility, blood samples are coming in gel-based serum separating tubes. They are centrifuged and the supernatant is directly transferred into new tubes. This implies that the replicates could only come from already treated samples and that their visual aspect was different from almost all routine samples. The technicians of this facility are routinely checking anterior results from the tested patients to integrate the new results within their serological status. Because we were using a limited pool of anonymous patient profiles, our technicians eventually faced multiple anterior results corresponding to IQA samples processed earlier. Both the visual difference of samples and the anterior results from anonymous patient’s profile led to strong suspicions of the staff working in the serology laboratory about our ongoing experimental IQA process, which finally obliged us to discontinue the IQA in this laboratory. This specific issue did not concern the two other facilities where our process could be completed in secret and on samples that were not pretreated. Despite this limitation, we consider that our process could still have an interest for IQA in serology laboratories, but that it would need to be partly redesigned. A proposition would be to duplicate samples before their arrival in the laboratory. It would solve the problem of visual impact and improve the coverage of pre-analytical steps. It would also be necessary to use more sophisticated fake patient identities, without previous IQA results to prevent their recognition by our coworkers.
Finally, even if our results cover more steps than traditional quality processes, it is still not covering all steps from prescription to interpretation of results by clinicians. Thus, we intend to extend this quality assurance project by collaborating with three other Swiss hospital laboratories. Each laboratory would challenge the others with split samples sent into containers accompanied by etiquettes and identifications used by the receiving laboratories for their external demands. This way, each laboratory would play the role of reference laboratory for one fourth of all samples. This approach would resolve the limitations of our scheme concerning the pre-treatment of the serology samples, the recognition of the samples and the lack of reference results. Such interlaboratory shared quality assurance would also help to provide “external assurance” for some serological and molecular parameters for which there is currently no EQA available (i.e. Tropheryma whipplei ).
In conclusion, this study revealed insightful points to improve and questioned our current practices, such as the use of Gram results to select culture media. Also, this study confirms the great interest of an IQA scheme based on split samples reprocessed in blind. It represents only few hundreds supplementary analyses, on the over 25’000 analyses run every year in our laboratory and this allowed us to generate interesting data on our reproducibility. This design showed itself to be perfectly complementary to other quality management procedures already ongoing in our laboratory and it should be further developed to better delineate the identified limitations and to settle its place in the overall quality assurance program.
Part of this publication was the subject of a master thesis done by Valentin Scherz under the direct supervision of Professor Greub at the School of Medicine of the University of Lausanne. Christian Durussel is the senior laboratory technician, acting at the Institute of Microbiology as the quality officer.
Supporting information
S1 table. categories defining amplitude of discrepancies in conventional microbiology laboratory..
Categories defining the amplitude of the discrepancies observed between A. semi-quantification of parameters pairs of Gram reading; B. semi-quantification of grown colonies; C. Antibiotic susceptibility testing (AST) all performed twice, first as part of the routine diagnostic procedure and then as part of the IQA program.
https://doi.org/10.1371/journal.pone.0187263.s001
S2 Table. Antibiotics sensitivity testing agreement, per antibiotics.
Complete results of per antibiotics analysis of antibiotics sensitivity testing concordances and discrepancies. Minor discrepancies are either sensitive versus intermediate or intermediate versus resistant. Major discrepancies are sensitive versus resistant.
https://doi.org/10.1371/journal.pone.0187263.s002
Acknowledgments
We are grateful to the following laboratory technicians: Sarah Chappuis, Maria Senra-Ortiz and René Brouillet for the replication of the samples. We are also grateful to all technicians for reprocessing the samples and to Tytti Heinonen for her help in correcting this manuscript.
- 1. WHO. Antimicrobial resistance Global Report on Surveillance. Geneva: World Health Organization; 2014. ISBN: 978-92-4-156474-8
- View Article
- PubMed/NCBI
- Google Scholar
- 9. Kohn LT, Corrigan JM, Donaldson MS. To Err Is Human: Building a Safer Health System. Washington, D.C.: National Academy Press; 2000. ISBN: 0-309-51563-7
- 13. Le Conseil fédéral suisse. RS 818.101.32 Ordonnance du 29 avril 2015 sur les laboratoires de microbiologie. Berne: Recueil systrématique du droit fédéral; 2016.
- 14. Laboratory requirements, 42 C.F.R, Sect. 493 (2012).
- 15. International Organization for Standardization (ISO). ISO 15189 Medical laboratories-particular requirements for quality and competence, Geneva: ISO; 2012.
- 29. Eleftheriadou M, Tsimillis KC, editors. Eurachem Guide: Accreditation for Microbiological Laboratories. 2nd editio. 2013. ISBN: 978-91-87017-92-6
- 34. Commission cantonale (VD) d’éthique de la recherche sur l’être humain [Internet]. Soumissions: Premiers pas; c2014 [accessed April 2017]. http://www.cer-vd.ch/soumission/premiers-pas.html
- 35. GraphPad Software [Internet]. GraphPad QuickCalcs: Quantify interrater agreement with kappa; c2016 [accessed November 2016]. http://www.graphpad.com/quickcalcs/kappa1.cfm/
Microbiological Culture Media: A Complete Guide for Pharmaceutical and Healthcare Manufacturers
- December 2017
- Publisher: PDA/DHI
- ISBN: 9781942911159

- The University of Manchester
Discover the world's research
- 25+ million members
- 160+ million publication pages
- 2.3+ billion citations
- Recruit researchers
- Join for free
- Login Email Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google Welcome back! Please log in. Email · Hint Tip: Most researchers use their institutional email address as their ResearchGate login Password Forgot password? Keep me logged in Log in or Continue with Google No account? Sign up
- News & Analysis on Food & Beverage Development & Technology
GUEST ARTICLE
How can microbial quality control help reduce food waste?
29-Sep-2021 - Last updated on 29-Sep-2021 at 15:58 GMT
- Email to a friend

Globally, as much as one-third of the food produced for human consumption is lost or wasted every year. One reason food may be discarded is the presence of microbes that can cause spoilage or illness. This makes proper food safety and chain control in food development and production a key aspect of reducing food waste. Microbiology and Food Safety expert Robyn Eijlander from NIZO discusses how to ensure and control microbial quality when adding novel ingredients or probiotics, or adapting processes.
FoodNavigator: What microbial food safety challenges crop up when developing novel products?
Robyn Eijlander: Consumers constantly demand ‘new’, ‘better’ or 'healthier’ foods. To meet this demand, manufacturers may turn to novel ingredients or adapt their production processes. But this can open the door to new microbial contaminants, which may behave in unexpected ways. Microbes that were never a problem before may now survive and emerge in finished products.
Another consumer trend is for ‘cleaner’, more ‘natural’ foods. But reducing traditional preservatives such as salt or sugar, or replacing them with other ingredients or preservation methods, can affect microbial stability, for instance in shelf-stable foods like sauces.
We also see a desire to add more probiotics to foods. These microbes must not only be safe, but also survive product formulation and ingestion. Probiotic bacterial spore formers can be selected for heat resistance, to ensure stability and robustness during food processing. However, when these strains re-enter the food chain down the line, through plant-based ingredients or raw milk, they could negatively affect food stability and shelf life.
Overall, whenever there is a change in ingredients, formulation or processing, you should look into the potential risks. Spoilage or consumer health risks can damage your brand and reputation, and fixing problems after the fact is more costly than identifying and addressing them early on.
FN: How can food and ingredient companies tackle those challenges?
RE: The first step is to detect potential hazards. If there is a microbial contaminant, you should determine its identity and characteristics: pathogen or spore former, heat- or acid-resistant, biofilm former?
Then, you need to quantify and characterise the risk: predicting the microbe’s growth and inactivation, as well as its potential to produce toxins that might persist even after the bacteria themselves are inactivated, for example. And you want to determine which circumstances could support the presence or growth of the bacteria or toxins.
In the third step, you identify ways to control the contamination, and in the fourth step, you predict, implement and validate your solutions. In step 5, you then monitor and verify your control measures.
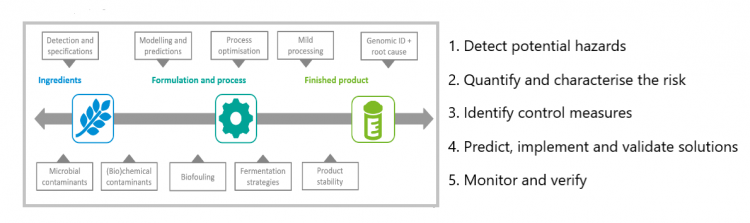
Schematic representation of a step-wise approach to ensure safe and microbially stable food throughout the production chain.
FN: Are all 5 steps needed for each new product or process?
RE: Not necessarily, but by advancing step by step, you cover all the bases. Steps 1 and 2, for example, don’t require a lot of time or resources, but they provide critical insight into microbial safety and quality at an early stage of product development.
And they will tell you whether steps 3 and 4 are necessary: if you are facing a microbial quality or contamination risk, additional characterisation of the contamination and effective control measures are important. After all, it is better to prevent a problem beforehand than to solve it afterwards!
Finally, monitoring the output of your process and verifying the impact of your control measures, is a standard part of food processing, and can either be part of this multi-step process, or a stand-alone action. If you already have your production line set up and you aren’t having any specific issues, monitoring and verifying may be enough. However, identifying, quantifying and characterising potential risks can provide useful insights for optimising this step.
FN: What sorts of models or assays are used during these 5 steps?
RE: The most appropriate approaches will depend on several factors: the type of information you are looking for, the type of product (solid or liquid, dairy or plant-based, etc.), the type of contaminant (microbial or chemical, pathogen or spoiler, etc.). Selecting the best models and assays requires a multidisciplinary approach that includes knowledge of food manufacturing, formulation and processing, as well as microbiology and even bioinformatics.
Let’s take the example of a yoghurt manufacturer who wants to develop a plant-based version, using novel ingredients and processes. They start by identifying the levels of the most common or dangerous contaminants, such as Listeria, Salmonella or Bacillus cereus , or notorious spoilage microorganisms, such as spore formers of other Bacillus species. Classic plating, combined with 16S rRNA gene sequencing and MALDI TOF mass spectrometry, is cost effective and gives good results regarding the presence and identification of these bacterial species.
But it can’t identify low-abundant microbes (<100 CFU/g) or distinguish between strains of bacteria. We know that some pathogenic species have a high strain-to-strain difference in, for instance, toxin-encoding genes. In-depth molecular methods such as PCR gene amplification, whole genome sequencing or metagenomic sequencing, combined with bioinformatics, can provide detailed information on the microbes present. Furthermore, the resulting data can support high-resolution tracking and tracing of notorious microbial contaminants throughout the production chain.
Next, the risk must be quantified and characterised. Predictive computational models or small-scale lab testing are very effective in determining whether the contaminant grows in the product at certain temperatures and how long it takes to reach critical levels, or if the processes adequately inactivate the microbe. Essentially, does anything need to be changed in the product formulation or thermal processes to improve microbial stability of the finished product?
These insights are critical to develop possible control measures and solutions: micro- or lab-scale testing can then identify which is most effective. Challenge tests are also useful for evaluating the product’s shelf life under specific conditions. In step 5, the manufacturer is verifying the impact of the control measures; regular qualitative microbial risk assessments (QMRA) can help ensure microbial quality and safety over time.
FN: What’s in the future for food safety in product development?
RE: Overall, I think there is greater innovation and advancement in food safety and quality when companies consider microbial stability already during research and development. Many pre- and probiotics manufacturers do so, and are also using modern technological advances in molecular technology, sequencing, data management and even artificial intelligence. This holistic approach is not yet as common in general food production, but I hope it will be in the future.
There are some interesting technical innovations that could support this evolution. For instance, handheld devices are making sequencing faster and more accessible for all food manufacturers. You take them on-site, load a sample, and have it sequenced immediately. By combining this technology with decision support system software, informed risk assessments can be completed in hours instead of days or weeks. Futuristic technological applications like these are in reach, and very exciting! They can improve track-and-trace, and open up communication about where contamination is coming from and how to mitigate it.
Related news
- How regenerative farming advances climate goals ADM | Download Product Brochure
- Create a Buzz with Blended Meat ADM | Download Insight Guide
- Download Sweet Trends Report 2024 by Südzucker Südzucker AG | Download White Paper
- Turn up the heat on your culinary creations Givaudan | Download Application Note
- The B2B Marketing Benchmark – Food & Nutrition Barrett Dixon Bell (BDB) | Download Insight Guide
- Study: Natural Preservation Is What Consumers Want Corbion | Download White Paper
Upcoming editorial webinars
- 24 Sep 2024 Tue Webinar Decarbonising Supply Chains at the Source
- 25 Sep 2024 Wed Webinar Eco-friendly Production from Farm to Fork
- 26 Sep 2024 Thu Webinar New Tech and Novel Ingredients for a Sustainable Future
- 08 Oct 2024 Tue Webinar State of the Market
- 15 Oct 2024 Tue Webinar Cognitive Health
- 03 Dec 2024 Tue Webinar Alternative Meat
- 04 Dec 2024 Wed Webinar Alternative Dairy
On-demand webinars
- Unlocking Consumer Emotions: A Comprehensive Framework for Emotional Associations in Food and Beverage Product Development Givaudan

FoodNavigator
- Advertise with us
- Press Releases – Guidelines
- Apply to reuse our content
- Contact the Editor
- Report a technical problem
- Whitelist our newsletters
- Why Register
- Editorial Calendar
- Event Calendar
- Expert Advisory Panel
- BiologyDiscussion.com
- Follow Us On:
- Google Plus
- Publish Now

Essay on Microbiology: Top 6 Essays
ADVERTISEMENTS:
Read this essay to learn about Microbiology. After reading this essay you will learn about: 1. Introduction to Microbiology 2. Eras of Microbiology 3. Some 20th Century Highlights in Microbiology 4. Branches of Microbiology 5. Scope and Future of Microbiology and 6. Modern Microbiology.
Essay on Microbiology Content:
- Essay on the Modern Microbiology
Essay # 1. Meaning of Microbiology:
Microbiology is a branch of biology that deals with microorganisms (often upto a diameter of 1000 µ ) and their effects on other living organisms. It derives its name from the union of three greek words: mikros (= small), bios (= life bearing); and logos (= study).
It ia a multi-disciplinary science of microorganisms and the prefix ‘micro’ generally refers to an object small enough so that microscopic examination is required for detailed visualization. Thus it is the scientific study of microorganisms, including a diverse group of simple life forms like protozoa, algae, molds, bacteria and viruses.
Microbiology is concerned with the structure, function, classification and ways of controlling and using their activities. The earlier works done by Antony Van Leauwenhoek (1632 – 1723), and later in 19th century by Louis Pasteur (1822- 1893) and Robert Koch (1843-1910) etc. laid the foundations of this subject.
Essay # 2. Eras of Microbiology:
1. era of speculation (5000 bc to 1675):.
Prior to Hippocrates (412 B.C.) some Biblical and other historical records often talked of the death tolls that some disease had taken. And Hippocrates seems to have been the first observer to document an Influenza epidemic in the year 412 B.C. Epidemics of plague have been recorded in China since 224 B.C.
Major Plague epidemics occurred in 540 AD in Egypt, reached Constantinopole in 542, and spread to Europe and Asia in the following decade; and was called the Plague of Lustinion named after the emperor of Brezantine Empire from 527 to 565 A.D.
One of the most notorious of the plague epidemics consumed 14 th century Europe. It began in the Volga River Basin in 1345, travelled north through Europe and reached England in 1348, and finally Russia in 1351. It was called as The Great Dying, Magna Mortalis or The Black Death, claimed around one-third to one half the population of Europe and an estimated 40 million deaths worldwide.
It was probably the concept of quarantine (from the Italian quarentina, meaning forty, supposed to be based on the number of days Christ spent in the wilderness) was instituted then. Though the infected individuals were kept isolated from others, the reservoir of the disease causing microbes (Rats) and the rat fleas (the vector) were allowed to roam freely.
The proposed remedies were also interesting and included burning incense sticks, dipping handkerchiefs in aromatic oils, ringing church bells, firing canons, wearing magical figures and bathing in human urine etc. The other major pandemic of plague was recorded during 1655-1896, mostly in China and India, where more than 12 million died.
It was in 1894 that Yersin and Kitasato described the causative agent, now known as Yersinia pestis. And, it wasn’t until 1897 that the mode of transmission was conclusively identified.
During early 1940s Syphilis spread throughout Europe. It was on this ailment Paul Ehrlich, in the first decade of the 20 th century focused his search for the “magic bullet”. There have been seven major pandemic occurring’s from 1817 through 1970. Smallpox alone is estimated to have killed more than 100 million people worldwide.
In the late 18th century the work of Edward Jenner, who developed a vaccine from the similar virus of cowpox, paved the way for the control of this deadly disease. Infact, the term Vaccine was born from the Latin word for cow, vacca, based on Jenner’s work.
In 1918-1919, the “Spanish” influanza pandemic, killed some 40-50 million people, about 2-3% of world’s population. It attacked the very young and very old, taking advantage of their frail immune systems. It also spread across five continents.
Though considerable work was being done in the field of bacteriology, the advances in the field of virology had to wait for the development of the electron microscope by Belgian Physicist Marton (1934).
In the late 17th century a slow yet methodical pursuit began as a new chapter in humanity’s understanding of the hitherto unseen and unknown world. As per WHO report, in 2003 alone, more than 11 million people died of infectious diseases including AIDS, tuberculosis and Malaria and of them half of them in Africa.

2. Era of Observation (1675 to Mid 19 th Century):
The unknown, transmissible substance, Virus (Latin for poison) had always puzzled humanity. Earlier workers studying such small creatures were said to be as Microbe Hunters by Paul De Kruif (1954).
Scientific and methodological understanding of bacteria had already started in this era and the advanced versions of Light and advent of electron microscope started new horizons for the classical microbiology. It is at times seen from 1675 to mid19 th century.
3. Era of Cultivation (Mid 19 th Century to Early 20 th Century):
Although a period of over 150 years of observations and discoveries by many early microbiologists had contributed a lot to the understanding of this discipline, still the cause of infectious diseases was completely unknown till mid 19 th century. In 1857, Pasteur, the chemist with a microscope turned beet sugar into alcohol.
Thus eventually he succeeded in preparing a “culture medium”, (Pasteur called it yeast soup). Infact the lack of suitable means to obtain pure cultures was a major impediment to the development of the “Germ Theory of Disease”. Pasteur thus entered into the field of fermentation and was perhaps the first to associate the growth of specific microbes with the production of specific fermentation products.
It was in the year 1865 that the “Germ Theory of Disease” was inaugurated. After fermentation Pasteur was called to look the cause of silkworm disease in South France, and after observing the diseased eggs and other details he wrote: “If I can be permitted this antithesis, the role of infinitely small being appeared to me infinitely large, either as a cause of various diseases, especially contagious diseases, or as contributors to the decomposition and to the return to the atmosphere of everything which has lived” . (Lechevalier and Solotorovsky, 1965).
Although the prevailing spontaneous generation influenced Pasteur’s work, but he believed that microbes did not arise spontaneously, as the air exposure had failed many of the earlier efforts to demonstrate such things. Later Tyndall, the physicist, and Pasteur postulated that specks of dust carried microbes. This was an experiment which did it all, and is now known as famous swan-neck flask experiment.
In 1877, Fedinand Cohn described the spores of Bacillus subtilis and their heat-resisting properties. Thus the joint efforts of Pasteur, Tyndall and Cohn finally eroded the concept of spontaneous generation. Robert Koch is believed to have used a solid surface (a potato slice) to grow microorganism.
Loeffler and Graffky working in Koch’s lab, devised the streak method whereby they would use a fine wire or needle to streak potato with an organism. Koch, being an amateur photographer, also produced photomicrographs while working on ‘Anthrax’ and used gelatin for this purpose.
But gelatin proved to be a wrong choice as it melted at temperatures above 15°C and thus could not be incubated at the optimal temperatures for the growth of many organisms, they were trying to study. It was in 1882, when Frau Angelina Hesse, the wife of a graduate student in Koch’s lab, suggested agar-agar, came as a final solution.
At 1-1.5%, its properties were well-studied and suited for use in microbiology having a melting point of 100°C and a setting point of less than 45°C. This enabled the incubation of cultures to anything below 100°C.
This technique revolutionized cultural bacteriology and enabled the great advancement of the subject during the last two decades of the 19 th century.
Later the germ theory of disease was advanced by Koch and also demonstrated the association between specific microbes and specific diseases, and showed that there was an external source of the germs and to get the disease, the germs must gain entrance into the victim. Koch was also invited to address the endemic Cholera problem.
He had concluded that the same slightly curved rod in every victim (and not present in Healthier individuals) present in polluted water around huts was the cause of the disease. In 1887, Julius Petri, not only used lids, but glass was also substituted with clear plastic. Also the grid was added to counting chambers.
Chantemesse and Widal, 1887, prepared the first differential medium using glucose and lactose peptone water to differentiate E.coli from Salmonella typhii. After that a Japanese bacteriologist Kitasato developed a test for indole production to further aid in the differentiation of these species. In 1892, Wurtz of Paris introduced the use of indicators in the medium to detect acid production.
Pasteur later developed the preparations from the causative agents of chicken cholera showing the alteration of the virulence and the development of a vaccine produced by laboratory manipulation. This is how Pasteur, ‘Father of Microbiology’ had opened the field of preventive medicine. Pasteur and his colleagues, Emile Roux, and Chamber-land, had further successes using attenuated anthrax bacilli.

Essay # 3. Some 20th Century Highlights in Microbiology :
The 1900’s witnessed tremendous advances on several fronts: microbial metabolism, microbial genetics and molecular biology, antimicrobial therapies, and development of selective and differential culture media and virology, just to name a few.
Also the art of microbial toxonomy moved from a purely morphological basis to one based on metabolic characteistics. The reclassification of species (with molecular methods in use) allowed grouping of organisms according to their genetic make up.
The genetic materials’s role became useful in studies aimed at understanding bacterial variation and antibiotic resistance. The processes of bacterial transformation and mutation were the basis for determining that all genetic information of cells is coded for by DNA.
The year 1928 saw the discovery of Penicillin by Alexander Flemming. Howard Florey associated (driven by the intense need of Second World War), purified it in 1940 and it became commercially available. It also promoted the work and search for other such antimicrobial weapons produced by soil microorganisms.
Later, Streptomycin was discovered and other broad-spectrum antibiotics followed by the development of semisynthetic antibiotics.
The 1920-40s witnessed the development of .many of the selective media used today, as well as techniques for the isolation of anaerobic organisms. In 1953 Watson and Crick, published the double helix model for DNA. In 1983, DNA became a useful tool when Karry Mullis invented the technique of Polymerase Chain Reaction (PCR)
London County Council Laboratory Services in 1950, led to the commercial development of dehydrated media.
The virology progressed due to several reasons made possible by:
(a) The use of fertile eggs as a method for the cultivation of a virus.
(b) The UV-microscopy (developed by Barnard) gave the first view of the elementary bodies of a number of viruses.
(c) The development of tissue culture techniques in the 1920’s and 30’s and
(d) The development of the electron microscope by Marton in 1934.
In 1954, the development of the monolayer technique and that of using cytopathic effect for viral detection led to the availability of live virus vaccines. Since then the development of antiviral chemotherapies has been a thrust area of researches in microbiology. In 1995 the entire genome sequencing was completed for Haemophilus influezae.
Essay # 4. Branches of Microbiology:
Microbiology is a mutidisciplinary subject as the microbes are not only playing useful role in nature through their links in bio-geochemical cycles, symbiotic relationship, in maintaining soil fertility, in bio-remediation, also as plant pathogens, microbes in biotechnology and as amodel organisms for biochemical molecular and genetic processes but are also providing homogenous experimental material relatively quickly. As a result of their link to several branches of science many branches of microbiology can be identified (and are still growing……).
Some of the established branches are as follows:
1. Industrial Microbiology:
It encompasses the uses of a variety of microbes in industrial processes. Initially they were being used for industrial fermentation and waste water treatment. As today industry is linked to biotechnology, several new industrial applications have been found for a variety of microbes.
It is sometimes also studied as microbial biotechnology and is the application of scientific and engineering principles to the processing of materials by microorganisms (such as bacteria, fungi, algae, protozoa and viruses) or plant and animal cells to create useful products or services. Areas of industrial microbiology include quality assurance for the food, pharmaceutical, and chemical industries.
2. Medical Microbiology:
This branch of microbiology deals with the scientific study of pathogenic microbes, the diseases they cause, their mode of survival in environment and their hosts (including life-cycle); their diagnosis, prevention and treatment.
In fact, as early as Varo and Columella in the first century BC had postulated that the diseases were caused by invisible beings (animalia minuta). Von Plenciz (1762) had put forth the idea that each disease was caused by a separate agent.
It covers a variety of topics where microbes are responsible for causing diseases of skin and eye infections, pneumonia (by bacteria), several sexually transmitted diseases (STD’s), minor arthropod diseases, gastrointestinal infections including infections from drinking cow milk harbouing certain pathogenic bacteria and their remedies etc.
3. Agricultural Microbiology:
This branch deals with microbes having an impact on agriculture and food chains. Both, the harmful (microbes causing plant diseases) as well as useful microbes (e.g., N 2 fixing microbes, use of microbes in bio-fertilizers etc.) are studied under this branch.
Certain raminants also carry a mixture of complex bacteria that enable the animal to extract sufficient nutrient from a diet of grasses. Future research in microbial ecology will help to determine in preserving a balance in mirobial communities that favour agriculture.
4. Environmental Microbiology :
In the late 1800’s, and early 19th century Sergei Winogradsky, a Russian Mineralogist, pioneered the field of microbial autotrophs, and initiated the field of Environmental Microbiology. This branch includes the study of composition and physiology of microbial communities of the environment.
It also deals with the activities of microbial entities, their interactions among themselves and with maroorganisms. Adhesion, biofilm formation, global element cycles, biogeochemical processes and microbial life in extremes of environment or unexplored environs all fall in its preview.
5. Food and Dairy Microbiology :
As the microorganisms are ubiquitous (present almost everywhere) food and milk are no exceptions. Hence the microbes are studied from the viewpoint that they (e.g., Bacteria, Yeasts, molds etc.) can either act as spoilage microorganisms or pathogenic microorganisms.
And thus how they can cause spoilage, prevent spoilage through fermentation or can be the cause of human illness, all comes under the realm of this branch of microbiology. It is a thrust area of microbiology these days, as more and more food items are being packaged (including milk and its products) for later use.
6. Biotechnology :
The UN convention on Biological Diversity defined Biotechnology as: any technological application that uses biological systems, living organisms, or derivatives thereof to make or modify products or processes for specific use. Bio-engineering, including recombinant genetic technology of the 21 st century is the science upon which all biotechnological applications are based.
It combines disciplines like genetics, molecular biology, biochemistry, food sciences, mechanical engineering, chemical engineering, microbiology, cell biology and all are interrelated to electronics, information technology and robotics.
7. Bacteriology :
The current science of bacteriology includes the study of both domains of prokaryotic cells (the Bacteria, and Eucarya). But recently due to out-break of molecular techniques applied to phylogeny of life, another group of prokaryotes was defined and informally named ,” archaebacteria (it has now been renamed as Archaea) and included in the study of bacteriology”.
8. Virology :
It is the study of viruses, complexes of nucleic acids and proteins that have the capacity for replication in animal, plant or bacterial cells. To replicate, the viruses use their genomes (DNA or RNA) or of the host cells and cause changes in cells, particularly its antigenicity and may cause several diseases in plants and animals is all covered under this branch.
9. Soil Microbiology :
This branch deals with the biota that inhabits the soil and the processes they mediate. As the soil is a complex environment, colonized by an immense variety of microorganisms, the soil microbiology focuses on soil viruses, bacteria, actinomycetes, fungi and protozoa, but traditionally it has also included investigations of soil animals such as nematodes, mites and other arthropods.
Modem soil microbiology represents an integration of microbiology with the concepts of soil science, chemistry and ecology to understand the functions of microorganism in the soil environment.
10. Sewage Microbiology :
This branch deals with the study of microbial flora of various types of sewage. The sewage may, depending upon source, can contain harmless (E. coli and other coli forms) to potential pathogens including enterococci, Vibrio cholerae, Salmonella typhi, etc. This branch studies their qualitative as well as quantitative details and ways to combat them following various treatment processes.
11. Mycology :
It deals with the study of various fungi. Fungi are eukaryotic organisms and around 300 species are shown to be pathogenic for man. It studies their morphology, taxonomy, biosystematics, distribution, propagation, and several mycotic diseases they cause including hypersensitivity, mycotoxicoses, mycetismus and other infections and their remedies.
12. Phycology :
It is a sub-discipline of botany, and deals with the scientific study of algae. as many species are single celled and microscopic (e.g. phytoplankton and micro algae); yet others are multicellular (some growing) very large as seaweeds such as kelp and sargassum they are also studied in microbiology., it also covers aspects like cyanobacteria (blue-green algae) and other microscopic forms occurring as symbionts in lichens., 13. protozoology :.
Earlier much in use, this branch is the study of protozoa (motile and heterotrophic) protists. Protozoa-despite their small size and unicellularness offer complex and unique biological features. They also serve as experimental models in a variety of cellular, molecular, biochemical and ecological researches.
One of the applied sub-branches of this old branch is medical protozoology (covering protozoa infecting humans). It covers life-cycles, morphological features, host-parasite interactions, geographical distributions, reservoir hosts, method of transmission and control, pathology, immunological aspects, diagnosis and remedies are all included in it.
14. Aquatic Microbiology :
It covers the study of microorganisms and their activities in natural water. As the natural waters include lakes, ponds, streams, rivers, estuaries and oceans, it initially started covering all of them.
But due to the growth of the subject several other branches have also been recognized and are as follows:
(a) Marine microbiology
(b) Estuarine microbiology
(c) Groundwater microbiology, and
(d) Deep-sub-surface microbiology
The aquatic microbiology deals with the variability of aquatic habitat and rapid changes in characteristics and associated microbial component. The microbial activity and biomass measurement studies are performed to follow microbial functions in water ecosystems. Also the balance between N, P, O and H is studied in lakes, eutrophic systems, and streams etc.
15. Marine Microbiology :
As marine environment is the largest part of the biosphere, being about 97-98% of all the water on earth, efforts are being made to study seas, especially deep seas (and their microbial functioning), as 75% of the ocean is below 1000 m depth (and is constantly cold at about 3°C on an average).
The oceanic explorations like Challenger expedition and Galathea expedition were among the initial efforts to critically explore microbial aspects of the deep seas and the nature of psychrophilic bacteria. Barotolerant bacteria are among the unique fauna of deep oceans.
As the most of the earlier work on seas and oceans remains confined to the near-shore and estuarine marine environments, the interest is growing in the off-shore and pelagic ocean microbiology.
16. Estuarine Microbiology :
It deals with the rapid variations in the physicochemical properties in estuaries leading to the establishment of unique microbial communities. This branch also undertakes the study of epiphytic microbial communities of estuarine plants, its biomass, temperature, salinity etc. And as great fluctuations are observed in estuarine environments, so are the varieties of microbial groups including sulphur bacteria, iron bacteria and heterotrophs etc.
17. Groundwater Microbiology :
Earlier the groundwater was assumed to be sterile, as very early studies indicated a decrease in microbes with increasing depths. But the microbiology of groundwater has become an important branch due to increased pressure on groundwater, ever increasing contamination and presence of microbes in deep waters.
18. Deep Sub-Surface microbiology :
This is a recent branch of microbiology and addresses deep aquifers (hundreds of meters below surface). It attempts to study the role of microorganisms in influencing the petroleum, sulphur and other drilling companies and vice-versa.
19. Aeromicrobiology :
It is the study of those invisible microorganisms (less than 1 mm in size) which are present in air. It also covers monitoring, distribution and diversity of airborne microorganisms in indoor and outdoor environments and especially the ones causing breathing trouble and diseases.
20. Geo-Chemical Microbiology :
It covers the role of a variety of microbes in hydrocarbon processing in formation of coal, gases and minerals. It also tells us to employ microbes for recovery of minerals from low grade ores.
21. Bio-Degradation Microbiology :
The microbial degradation of chemicals in the environment is an important route for removal of these compounds. This branch deals with the details of the degradation of hydrocarbons, lignins, cellulose, hemicellulose, polyaromatic hydrocarbons (PAH’s) and pesticides including organophosphates, chlorinated hydrocarbons, dithioates and carbonates.
22. Exomicrobiology :
It deals with the microbes in the outer space (or the extraterrestrial space) but all these microbes were originated on earth. In other words, the space microbiology is called exomicrobiology.
23. Immunology :
It .is the study of our protection from foreign macromolecules or invading microorganisms and our responses to them. These invaders include viruses, bacteria, protozoa or even larger parasites. How our first line of defense (e.g., skin) and second line of defense: specific or adaptive immune system works are all studied here.
24. Parasitology :
Parasitology is the study of parasites, their hosts and the relationship between them. This being a multi-disciplinary branch uses inputs of various techniques from fields such as cell biology, bioinformatics, biochemistry, molecular biology, immunology, genetics, evolution and ecology.
25. Epidemiology :
It is the study of the factors affecting health and illness of populations and use of preventive medicines in order to cure such diseases. It is derived from the term epi = upon, among; demos = people, district, and logos = study meaning “what is upon the people”.
This indicates that it applies to human only. Hippocrates, the famous Greek Physician is sometimes said to have coined the term epidemic (for diseases that are seen at some times but not others); and endemic (for diseases usually found in some places but not in others.)
26. Avian Microbiology :
Microbiology and Veterinary science are complimentary fields in the medical maintenance and treatment of birds. Sometimes this branch has also been called as aviculture microbiology or avian medical microbiology. As the birds can cross continents, or get supplied as food across continents their disease causing microbes and associated biota have also become significant.
27. Veterinary Microbiology :
Veterinary microbiology is concerned with microbial (bacteria, fungal and viral,) diseases of domesticated animals (e.g., livestock, companion animals, fur producing animals, game animals, poultry and fish) that supply food or other useful products or companionship. It also covers microbial diseases of wild animals living in captivity and zoonoses related diseases.
28. Pharmaceutical Microbiology :
Sometimes seen as a part of industrial microbiology, this branch is responsible for screening several drugs (parental and oral drugs) following strict measures for measuring microbiological testing in order to validate certain compounds to standard protocols.
29. Oral Microbiology :
It is a modern branch of microbiology where all the microbes associated with major dental diseases (e.g., caries and periodontal diseases) are studied. It also covers all the resident oral bacteria and host details.
30. Evolutionary Microbiology :
This branch deals with all phases of the systematics of prokaryotes, yeasts and yeast like organisms, including taxonomy, nomenclature, identification, phylogeny, evolution, biodiversity, characterization and culture preservation etc. It also covers all aspects related to the evolution of all micro-organisms, including the protists (as protozoa and algae).
31. Microbial Physiology :
It is an interesting branch of microbiology concerned with the structure, function and relationships of microorganisms, with particular emphasis on how microbes respond in response to changes in environment.
32. Microbial Genetics :
Microbial genetics is a subject area within biotechnology and genetic engineering. It studies the genetics of very small (micro) organisms. It also encompasses the study of the genotype of microbial species and also the expression system in the form of phenotypes.
33. Probiotic and Prebiotic Microbiology:
Probiotics and prebiotics have long been appreciated for their positive influences on gut health. The prebiotics include the non-digestible food ingredients that selectively stimulate the growth and/or activity of beneficial microorganisms in people’s colon. And the probiotics are live microorganisms (in most cases bacteria) that are similar to the microorganisms found in the human gut.
They are also termed as “friendly or good bacteria”. The probiotics are available to consumers mainly in the form of dietary supplements and food. Due to growing consciousness about health, they are nowadays also being used as complementary and alternate medicine (CAM).
One widely used definition of probiotics, is the one given by World Health Organization (WHO) and Food and Agriculture Organization (FAO) of the United Nations; is that, ” Probiotics are live microorganisms, which, when administered in adequate amounts, confer a health benefit on the host.”
Essay # 5. Scope and Future of Microbiology:
The microbes not only need to be understood as causative agents of diseases but are also useful to us as contributors in food production, antibiotic manufacture, vaccine development and environmental management.
Recently the subject had also allowed us to explore and control the following diseases; Foot and mouth disease, Asian Bird Flu, SARS (Severe Acute Respiratory Syndrome), West Nile Virus, and Monkey-pox.
We are now also exploring more Prions related diseases causing a type of amyloid diseases (characterized by normal brain protein being altered and changing into fibres); at times called as: Transmissible Spongiform Encephalopathy (TCS).
Some famous examples include:
Creutzfeldt-Jakob disease in humans (CJD), Bovine Spongiform Encephalopathy (BSE) also known as mad-cow disease, chronic wasting disease (CWD) in deer and elk Kuri in human Feline spongiform Encephalopathy (FSE), and Transmissible mink Encephalopathy (TME). The interest is growing in various zoonotic diseases (of animals that can pass to humans) due to their close proximity of rearing or habitation.
Due to undergoing environmental changes, global warming and related climate changes, various mathematical models are trying to understand the impact of these factors on disease causing microbes of their respective hosts. Immunoparasitology has helped us in the past to understand various pathogens.
But due to a use of wide variety of invasions and immune evasion mechanisms used by various parasites, the nature keeps posing greater challenges for epidemiologists. As a result several genome mapping programmes are underway for disease causing protozoa and helminths (e.g., L. major, P. falciparum, and T. gondii etc.).
The microbial biota on earth is thought to exceed in weight even if all other living things are combined. This is the same as eloquently argued by palaeontologist Stephen Jay Gould (1996), that the 21st century will be the era of the microbes.
As the bacteria alone account for 50% of the biomass of carbon and over 90% of the biomass of Nitrogen and phosphorus combined on our planet, such claims seem to be true. The future microbiology is already an integrative microbiology incorporating inputs from microbial physiology, microbial genetics, microbial ecology and microbial pathogenesis using tools and dialects of various sub disciplines.
The use of microbes in nanotechnology has brought microbiology to engineering and physics. And recent discoveries indicate that microbes also play roles in determining animal behaviour, bringing microbiology to the realm of psychology. Hence the 21st century microbiology is already an integrative science with lots of challenges and benefits to reap.
Essay # 6. Modern Microbiology :
Modern microbiology reaches into many fields of humans including the development of pharmaceutical products, the use of quality-control and dairy product production, the control of disease-causing microorganisms in water, and the industrial applications of microorganisms.
Microorganisms are used to produce vitamins, amino acids, enzymes, and growth supplements. They manufacture many foods including fermented dairy products (sour cream, yogurt, and buttermilk), as well as other fermented foods such as pickles, sauerkraut, breads, and alcoholic beverages.
One of the major areas of applied microbiology is biotechnology. In this discipline microorganisms are used as living factories to produce pharmaceuticals that otherwise could not be manufactured.
These substances include the human hormone insulin, the antiviral substance interferon, numerous blood-clotting factors and clot dissolving enzymes, and a number of bacteria can be engineered to increase plant resistance to insects and frost and biotechnology will represent a major application of microorganisms in the next century.
Related Articles:
- Microbiology and Nursing
- Essay on Microorganisms | Microbiology
- Anybody can ask a question
- Anybody can answer
- The best answers are voted up and rise to the top
Forum Categories
- Animal Kingdom
- Biodiversity
- Biological Classification
- Biology An Introduction 11
- Biology An Introduction
- Biology in Human Welfare 175
- Biomolecules
- Biotechnology 43
- Body Fluids and Circulation
- Breathing and Exchange of Gases
- Cell- Structure and Function
- Chemical Coordination
- Digestion and Absorption
- Diversity in the Living World 125
- Environmental Issues
- Excretory System
- Flowering Plants
- Food Production
- Genetics and Evolution 110
- Human Health and Diseases
- Human Physiology 242
- Human Reproduction
- Immune System
- Living World
- Locomotion and Movement
- Microbes in Human Welfare
- Mineral Nutrition
- Molecualr Basis of Inheritance
- Neural Coordination
- Organisms and Population
- Photosynthesis
- Plant Growth and Development
- Plant Kingdom
- Plant Physiology 261
- Principles and Processes
- Principles of Inheritance and Variation
- Reproduction 245
- Reproduction in Animals
- Reproduction in Flowering Plants
- Reproduction in Organisms
- Reproductive Health
- Respiration
- Structural Organisation in Animals
- Transport in Plants
- Trending 14
Privacy Overview
| Cookie | Duration | Description |
|---|---|---|
| cookielawinfo-checkbox-analytics | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Analytics". |
| cookielawinfo-checkbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-others | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Other. |
| cookielawinfo-checkbox-performance | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Performance". |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
- Resume Builder
- Resume Templates
- Resume Formats
- Resume Examples
- Cover Letter Builder
- Cover Letter Templates
- Cover Letter Formats
- Cover Letter Examples
- Career Advice
- Interview Questions
- Resume Skills
- Resume Objectives
- Job Description
- Job Responsibilities
- FAQ’s
Quality Control Microbiologist Cover Letter Example
Writing a cover letter for a Quality Control Microbiologist position can be a challenge. It is important to have a well-written and effective letter to get the best chance of being considered for the job. Whether you are new to the job market or an experienced professional, this guide provides helpful tips to craft an impressive cover letter and stand out from the competition. With an example cover letter included, you can ensure that your application is ready to submit and be in with a chance of success.
If you didn’t find what you were looking for, be sure to check out our complete library of cover letter examples .

Download the Cover Letter Sample in Word Document – Click Below

Start building your dream career today!
Create your professional cover letter in just 5 minutes with our easy-to-use cover letter builder!
Quality Control Microbiologist Cover Letter Sample
Dear [Recruiter],
I am writing to apply for the position of Quality Control Microbiologist at [Company Name]. With my Bachelor’s degree in Microbiology and six years of experience in the biotechnology industry, I have the technical expertise and knowledge to ensure the quality and safety of products at your organization.
In my current role as Quality Control Microbiologist at [Company Name], I have been responsible for ensuring that incoming ingredients and materials meet the quality standards set by the organization. I have conducted tests to detect the presence of pathogens and contaminants, as well as identifying any issues that may be present in a batch of ingredients. Additionally, I have developed and implemented quality systems and processes to ensure that all products meet the standards set by regulatory agencies.
My experience in the biotechnology industry has also provided me with the ability to use modern laboratory techniques such as DNA sequencing and PCR. I have also developed a strong attention to detail and the ability to work under pressure while ensuring accuracy when performing tests.
I am confident that I have the necessary skills and expertise to excel in this position and I look forward to discussing further how my qualifications can be of benefit to [Company Name]. Thank you for your time and consideration.
[Your Name]
Create My Cover Letter
Build a profession cover letter in just minutes for free.
Looking to improve your resume? Our resume examples with writing guide and tips offers extensive assistance.
What should a Quality Control Microbiologist cover letter include?
A Quality Control Microbiologist cover letter should include the following:
- A brief introduction that explains your interest in the Quality Control Microbiologist role and why you feel your qualifications and experience make you the ideal candidate for the job.
- A summary of your relevant experience in the field of Quality Control Microbiology, including any specific qualifications or formal training you may have.
- A discussion of your technical capabilities and accomplishments, including any lab testing, sample preparation, and data analysis you have conducted.
- A description of your ability to work collaboratively and effectively with other team members.
- Examples of the different ways in which you have contributed to successful projects in the field of Quality Control Microbiology.
- A closing statement that expresses your enthusiasm and commitment to join the team and contribute to the success of the organization.
Quality Control Microbiologist Cover Letter Writing Tips
Writing a cover letter for a Quality Control Microbiologist position can be a challenge. To make sure you stand out from the competition, here are some tips to help you create a winning cover letter.
- Research the company: Before writing your cover letter, research the company to gain a thorough understanding of their products and services. This will help you tailor your letter and demonstrate that you’ve taken the time to understand the role you’re applying for.
- Highlight your skills: Be sure to highlight the skills and qualifications that make you the perfect fit for the position. Don’t forget to mention any accomplishments and awards you have related to the position, as well as any certifications or training that make you a standout candidate.
- Keep it brief and to- the- point: Quality Control Microbiologists are in high demand, so employers don’t have time to read long, drawn- out cover letters. Keep your letter succinct and straightforward, and make sure you provide the information that the employer needs to make a decision.
- Emphasize your passion: Employers want to hire passionate candidates who can bring enthusiasm to the role. Make sure your cover letter reflects your enthusiasm and dedication to the field, and explain why you’re the perfect candidate.
- Proofread: Finally, be sure to proofread your cover letter before submitting it. Even a few mistakes can make the difference between getting an interview and being passed over.
By following these tips, you’ll be well on your way to creating a winning cover letter for a Quality Control Microbiologist position. Good luck!
Common mistakes to avoid when writing Quality Control Microbiologist Cover letter
Writing a quality control microbiologist cover letter is an important part of getting an interview. A well- crafted one can help you stand out from the competition and give you the best chance at securing the job. However, there are some common mistakes that can easily be avoided.
- Not customizing your cover letter: A generic one- size- fits- all cover letter will not cut it. You should customize your cover letter to include specific details about your experience, qualifications, and skills that make you an excellent fit for the job.
- Failing to explain why you are a great fit: While you may have an impressive resume and qualifications, you need to explain in your cover letter why you are the best candidate for the job. Highlight the reasons why you would be the best choice and how your skills and experience make you a great asset to the company.
- Not showing enthusiasm: Showing enthusiasm in your cover letter is key. Express your excitement for the job and how you are looking forward to the opportunity to use your skills and experience to help the company reach its goals.
- Not addressing the hiring manager directly: It is important to address the hiring manager directly in your cover letter. Take the time to research who the hiring manager is and address them by name.
- Not proofreading: Before submitting your cover letter, make sure you read it over and proofread it. Check for any grammar and spelling errors, typos, and awkward phrasing. Doing this can help you make a great impression and avoid any potential red flags.
Key takeaways
The quality control microbiologist role is a highly detailed and specialized position, and prospective employees should demonstrate their experience and expertise in the field in their cover letter. Crafting a quality control microbiologist cover letter that stands out can help you make a strong impression with potential employers. Here are some key takeaways for writing an impressive quality control microbiologist cover letter:
- Highlight your experience and qualifications as they relate to the role: Make sure to emphasize your experience and qualifications that are relevant to the quality control microbiologist role. Include specific details about your knowledge of laboratory techniques, relevant training and certifications, and any experience in similar roles.
- Showcase your attention to detail: Quality control microbiologists need to have strong attention to detail and be able to think critically. Showcase your ability to do this by providing examples of how you have caught errors or improved processes in your past roles.
- Demonstrate your knowledge of the industry: Showcase your knowledge of the company and the industry as a whole in your cover letter. In addition to understanding the duties of the role, it will be important to understand the company’s mission, goals, and values.
- Show your enthusiasm for the role: The cover letter is an opportunity to demonstrate your enthusiasm for the role and the company. Talk about why you are excited to be considered for the quality control microbiologist role and what you can contribute to the team.
- Keep it organized and professional: Make sure to keep your cover letter organized and professional. Use bullet points to highlight your qualifications and make it easy to read. Proofread your cover letter to make sure there are no errors and that it is written in a clear and concise manner.
Frequently Asked Questions
1.how do i write a cover letter for an quality control microbiologist job with no experience.
Writing a cover letter for a Quality Control Microbiologist job with no experience can be daunting, but there are ways to highlight your unique skills and abilities to make a great impression on the hiring manager. Start by introducing yourself and explaining why you’re interested in the position. Include your educational background and any relevant work experience, such as laboratory research or experience working with microorganisms. Focus on your strengths and training, such as your attention to detail, strong analytical skills, and data- driven approach to problem- solving. Finally, emphasize your enthusiasm for the job and eagerness to contribute to the organization.
2.How do I write a cover letter for an Quality Control Microbiologist job experience?
When writing a cover letter for a Quality Control Microbiologist job with experience, focus on the specific duties, experiences, and accomplishments that make you the ideal candidate for the position. Start by highlighting your most recent experience as a Quality Control Microbiologist, such as conducting analysis, diagnosing problems, and developing testing protocols. Describe how your background in microbiology and lab work has prepared you to excel in the job. Additionally, draw upon your experience to show the hiring manager your ability to think critically, solve problems, and collaborate with other teams.
3.How can I highlight my accomplishments in Quality Control Microbiologist cover letter?
When writing a Quality Control Microbiologist cover letter, it’s important to highlight your accomplishments in order to stand out to the hiring manager. Start by reflecting on prior successful projects that you’ve completed in the job. Include specific details about how you implemented best practices when conducting analysis, identified and corrected errors, or developed creative solutions to complex problems.
In addition to this, be sure to check out our cover letter templates , cover letter formats , cover letter examples , job description , and career advice pages for more helpful tips and advice.
Let us help you build your Cover Letter!
Make your cover letter more organized and attractive with our Cover Letter Builder

Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
New quality productivity and industrial structure in china: the moderating effect of environmental regulation.

1. Introduction
2. literature review, 2.1. literature on new quality productivity and industrial structure, 2.2. literature on environmental regulation and industrial structure, 3. theoretical analysis and hypothesis development, 3.1. the connotation of new quality productivity, 3.2. analysis of the mechanism between new quality productivity and industrial structure, 3.3. analysis of the mechanism of new quality productivity, environmental regulation and industrial structure, 4. methods and data, 4.1. construction and measurement of new quality productivity index system, 4.1.1. construction of new quality productivity index system, 4.1.2. measurement of new quality productivity index system, 4.2. variable selection specification, 4.3. econometric modeling, 5. results and discussion, 5.1. analysis of the results of the neoplasm productivity measurements, 5.1.1. overall analysis of new quality productivity measurement results, 5.1.2. analysis of new quality productivity measurement results in different dimensions, 5.2. benchmark regression and robustness test results, 5.3. endogeneity issues and robustness tests, 5.4. moderating effects test of environmental regulation, 6. conclusions and prospect, 6.1. conclusions, 6.2. prospect, author contributions, institutional review board statement, informed consent statement, data availability statement, acknowledgments, conflicts of interest.
- Hu, W.; Wang, D. How does environmental regulation influence China’s carbon productivity? An empirical analysis based on the spatial spillover effect. J. Clean. Prod. 2020 , 257 , 120484. [ Google Scholar ] [ CrossRef ]
- Bai, B.L.; Wang, S.L. Analysis on Socialist Productivity Theory with Chinese Characteristics: The Enrichment and Development of Marxist Political Economy. Shanghai J. Econ. 2017 , 3–9+33. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Zhou, M.; Guo, J.H.; Wang, W.H. Research on Digital Industry Empowering the Modern Industrial System under the Guidance of New Quality Productivity: Based on the Perspective of Supplementing Nodes, Establishing Links and Fixing Networks. Manag. World 2024 , 40 , 1–26. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Qi, C.S. How to Understand that “New Quality Productivity Itself is Green Productivity”. Economist 2024 , 7 , 15–23. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Gu, H.L. From “Overall Jump in the Level of Social Productivity” to New Quality Productivity--Research on the Theoretical Innovation of Xi Jin-ping's Economic Thought on Social Productivity in the New Era. Economist 2024 , 6 , 5–15. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Zhou, W.; Xu, L.Y. On New Quality Productivity: Connotative Characteristics and Important Focus. Reform 2023 , 10 , 1–13. (In Chinese) [ Google Scholar ]
- Ling, X.X.; Xie, H.; Tuo, L.; Jin, Z. The Three Dimensions of New Quality Productivity: Temporal-Spatial, Structural and Technological Dimensions. J. Xinjiang Norm. Univ. (Philos. Soc. Sci.) 2024 , 45 , 67–76. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Shen, K.; Jin, T.; Zhao, Q. To Energize High-quality Development by New-quality Productivity. Nanjing J. Soc. Sci. 2024 , 1 , 37–42. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Xu, Z.; Zhang, J. New Quality Productive Forces to Promote the Transformation and Upgrading of Manufacturing Industry: Value Orientation, Logic Mechanism and Key Measures. J. Soc. Sci. Hunan Norm. Univ. 2024 , 53 , 104–113. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Pan, J.; Tao, H. To understand the three dimensions of the new quality productivity. J. Xi’an Jiaotong Univ. (Soc. Sci.) 2024 , 44 , 12–19. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Ma, L.; Li, X.; Pan, Y. Global Industrial Chain Resilience Research: Theory and Measurement. Systems 2023 , 11 , 466. [ Google Scholar ] [ CrossRef ]
- Guo, C.; Chen, X.; Peng, L. Research on new quality productivity promoting modern industrial system construction. J. Xi’an Jiaotong Univ. (Soc. Sci.) 2024 , 44 , 1–11. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Li, J.A.H. Effects of time-dependent environmental regulations on air pollution: Evidence from the Changsha-Zhuzhou-Xiangtan region, China. World Dev. 2021 , 138 , 105267. [ Google Scholar ] [ CrossRef ]
- Antweiler, W.; Copeland, B.R.; Taylor, M.S. Is Free Trade Good for the Environment? Am. Econ. Rev. 2001 , 91 , 877–908. [ Google Scholar ] [ CrossRef ]
- Lanoie, P.; Patry, M.; Lajeunesse, R. Environmental regulation and productivity: Testing the porter hypothesis. J. Prod. Anal. 2008 , 30 , 121–128. [ Google Scholar ] [ CrossRef ]
- Chakraborty, D.; Mukherjee, S. How do trade and investment flows affect environmental sustainability? Evidence from panel data. Environ. Dev. 2013 , 6 , 34–47. [ Google Scholar ] [ CrossRef ]
- Wu, T.; Wen, L.; Yi, M. Balancing growth targets and environmental regulations: An empirical analysis of dual policy impact on corporate environmental responsibility-insights from China. J. Environ. Manag. 2024 , 355 , 120500. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Huiping, W.; Runjie, Z. Effects of environmental regulation on CO 2 emissions: An empirical analysis of 282 cities in China. Sustain. Prod. Consum. 2022 , 29 , 259–272. [ Google Scholar ] [ CrossRef ]
- Rubayyat, H.; Khorshed, A. Dynamic relationship among environmental regulation, innovation, CO 2 emissions, population, and economic growth in OECD countries: A panel investigation. J. Clean. Prod. 2019 , 231 , 1100–1109. [ Google Scholar ] [ CrossRef ]
- Cai, X.; Zhu, B.; Zhang, H. Can direct environmental regulation promote green technology innovation in heavily polluting industries? Evidence from Chinese listed companies. Sci. Total Environ. 2020 , 746 , 140810. [ Google Scholar ] [ CrossRef ]
- Chen, X.; Qian, W. Effect of marine environmental regulation on the industrial structure adjustment of manufacturing industry: An empirical analysis of China’s eleven coastal provinces. Mar. Policy 2020 , 113 , 103797. [ Google Scholar ] [ CrossRef ]
- Zhang, G.; Zhang, P.; Zhang, Z.G. Impact of Environmental Regulations on Industrial Structure Upgrading: An Empirical Study on Beijing-Tianjin-Hebei Region in China. J. Clean. Prod. 2019 , 238 , 117848. [ Google Scholar ] [ CrossRef ]
- Porter, M.E.; Linde, C.V.D. Toward a New Conception of the Environment-Competitiveness Relationship. J. Econ. Perspect. 1995 , 9 , 97–118. [ Google Scholar ] [ CrossRef ]
- Du, K.; Cheng, Y.; Yao, X. Environmental regulation, green technology innovation, and industrial structure upgrading: The road to the green transformation of Chinese cities. Energ. Econ. 2021 , 98 , 105247. [ Google Scholar ] [ CrossRef ]
- Qiu, S.; Wang, Z.; Geng, S. How do environmental regulation and foreign investment behavior affect green productivity growth in the industrial sector? An empirical test based on Chinese provincial panel data. J. Environ. Manag. 2021 , 287 , 112282. [ Google Scholar ] [ CrossRef ]
- Wang, J. New Productive Forces: A Theoretical Frame and Index System. J. Northwest. Univ. (Philos. Soc. Sci. Ed.) 2024 , 54 , 35–44. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Song, J.; Zhang, J.; Pan, Y. Research on the Impact of ESG Development on New Quality Productive Forces of Enterprises Empirical Evidence from Chinese A-share Listed Companies. Cont. Econ. Manag. 2024 , 46 , 1–11. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Wang, J.; Wang, R. New Quality Productivity: Index Construction and Spatiotemporal Evolution. J. Xi’an Univ. Financ. Econ. 2024 , 37 , 31–47. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Gong, X.; Yan, Y. The Basic Meaning, Realization Mechanism and Practice Path of New Quality Productivity. He Soc. Sci. 2024 , 32 , 15–22. (In Chinese) [ Google Scholar ]
- Wenjie, Z.; Ning, X.; Chengyu, L. Impact of digital input on enterprise green productivity: Micro evidence from the Chinese manufacturing industry. J. Clean. Prod. 2023 , 414 , 137272. [ Google Scholar ] [ CrossRef ]
- Huaping, S.; Kofi, B.E.; Kwaku, A.K. Energy efficiency: The role of technological innovation and knowledge spillover. Technol. Forecast. Soc. 2021 , 167 , 120659. [ Google Scholar ] [ CrossRef ]
- Lei, W.; Yangyang, C.; Stephen, T.R. Will researching digital technology really empower green development? Technol. Soc. 2021 , 66 , 101638. [ Google Scholar ] [ CrossRef ]
- Zhang, W.; Xuan, Y. How to improve the regional energy efficiency via intelligence? Empirical analysis based on provincial panel data in China. Bus. Manag. J. 2022 , 44 , 27–46. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Peng, J.S.; Li, J.H.; Zheng, Z.H. The Interactions between Producer Services and Manufacturing: An Empirical Analysis Based on Input-Output Subsystem Model. Appl. Mech. Mater. 2013 , 2279 , 3041–3045. [ Google Scholar ] [ CrossRef ]
- Zhu, X.; Zuo, X.; Li, H. The dual effects of heterogeneous environmental regulation on the technological innovation of Chinese steel enterprises—Based on a high-dimensional fixed effects model. Ecol. Econ. 2021 , 188 , 107113. [ Google Scholar ] [ CrossRef ]
- Chen, Y.P.; Zhuo, Z.; Huang, Z.; Li, W. Environmental regulation and ESG of SMEs in China: Porter hypothesis re-tested. Sci. Total Environ. 2022 , 850 , 157967. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Maoliang, B.; Zhenzi, Q.; Beibei, L. Voluntary environmental regulation and firm innovation in China. Econ. Model. 2020 , 89 , 10–18. [ Google Scholar ] [ CrossRef ]
- Lin, B.; Tan, R. Economic Agglomeration and Green Economy Efficiency in China. Econ. Res. J. 2019 , 54 , 119–132. (In Chinese) [ Google Scholar ]
- Zhao, T.; Zhang, Z.; Liang, S. Digital Economy, Entrepreneurship, and High-Quality Economic Development: Empirical Evidence from Urban China. Manag. World 2020 , 36 , 65–76. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Guo, F.; Wang, J.; Wang, F.; Kong, T.; Zhang, X.; Cheng, Z. Measuring China’s Digital Financial Inclusion: Index Compilation and Spatial Characteristics. China Econ. Q. 2020 , 19 , 1401–1418. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Hwang, C.L.; Yoon, K.P. Multiple Attribute Decision Making. Methods and Applications. A State-of-the-Art Survey ; Springer: Berlin/Heidelberg, Germany, 1981. [ Google Scholar ] [ CrossRef ]
- Gan, C.; Zheng, R.; Yu, D. An Empirical Study on the Effects of Industrial Structure on Economic Growth and Fluctuations in China. Econ. Res. J. 2011 , 46 , 4–16+31. (In Chinese) [ Google Scholar ]
- Wang, Y.; Wu, M. Has China Entered the Era of Servitization? Judgment Standards and Present Stage of China. Econ. Probl. 2016 , 2 , 62–68. (In Chinese) [ Google Scholar ] [ CrossRef ]
- Jaffe, A.B.; Newell, R.G.; Stavins, R.N. Environmental Policy and Technological Change. Environ. Resour. Econ. 2002 , 22 , 41–70. [ Google Scholar ] [ CrossRef ]
- Liu, Y.; Yang, Y.; Li, H. Digital Economy Development, Industrial Structure Upgrading and Green Total Factor Productivity: Empirical Evidence from China’s Cities. Int. J. Environ. Res. Public Health 2022 , 19 , 2414. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Arellano, M.; Bond, S. Some Tests of Specification for Panel Data: Monte Carlo Evidence and an Application to Employment Equations. Rev. Econ. Stud. 1991 , 58 , 277–297. [ Google Scholar ] [ CrossRef ]
- Blundell, R.; Bond, S. Initial conditions and moment restrictions in dynamic panel data model. J. Econom. 1998 , 87 , 115–143. [ Google Scholar ] [ CrossRef ]
- Bond, S.R. Dynamic panel data models: A guide to micro data methods and practice. Port. Econ. J. 2002 , 1 , 141–162. [ Google Scholar ] [ CrossRef ]
- Shuke, F.; Zhuo, M.; Bing, N. Research on the spatial differences of pollution-intensive industry transfer under the environmental regulation in China. Ecol. Indic. 2021 , 129 , 107921. [ Google Scholar ] [ CrossRef ]
Click here to enlarge figure
| Indicator Dimension | Primary Indicator | Secondary Indicator | Measurement Method | Direction of Effect |
|---|---|---|---|---|
| Innovation driving force | Innovation input | Scientific research fund | Internal expenditure on R&D/GDP | + |
| Scientific manpower | R&D full-time personnel | + | ||
| The optimization of the labor market | Number of students in colleges and universities/total employment population | + | ||
| Innovation output | Patent output | Number of domestic patent applications granted | + | |
| High-tech industry output | High-tech industry new product sales revenue/GDP | + | ||
| Labor productivity | GDP/total employment population | + | ||
| Green driving force | Resource consumption | Energy intensity | Energy consumption/GDP | − |
| Land resources | Population density | − | ||
| Atmospheric resources | SO emissions | − | ||
| Green and environmental protection | Greening rate | Forest coverage rate | + | |
| Urban environmental protection | Investment in the urban environment | + | ||
| Greenhouse effect | CO emissions | − | ||
| Terminal pollution control | Domestic garbage disposal capacity | Domestic garbage harmless treatment rate | + | |
| Solid waste treatment capacity | Common industrial solid wastes utilized/common industrial solid wastes generated | + | ||
| Wastewater Treatment capacity | Daily treatment capacity of Wastewater | + | ||
| productivity driving force | Traditional infrastructure | Transportation resources | (Highway Miles + Railroad Miles)/Jurisdictional Area | + |
| Educational resources | Number of colleges and universities per 10,000 people | + | ||
| Medical resources | Number of beds in medical and health institutions | + | ||
| Digital economy development | Internet-related output | Total telecommunications business per capita | + | |
| Digital economy employment level | Number of employees in the information transmission, software, and information technology services industry/employed population in urban organizations | + | ||
| Internet Penetration Rate | The number of Internet users per 100 people | + | ||
| Digital Inclusive Finance | Digital Inclusive Finance index | + |
| (1) | (2) | (3) | (4) | (5) | (6) | (7) | (8) |
|---|---|---|---|---|---|---|---|
| Types | Variables | Indicators | Obs | Mean | SD | Min | Max |
| Explained variables | rat | Rationalization of industries | 330 | 0.1524 | 0.0937 | 0.0082 | 0.4515 |
| upg | Ppgrading of industries | 330 | 1.3415 | 0.7320 | 0.5271 | 5.2440 | |
| Core explanatory variables | nqp | New quality productivity | 330 | 0.3883 | 0.0775 | 0.2288 | 0.6418 |
| ino | Innovation driving force | 330 | 0.2406 | 0.1254 | 0.0275 | 0.6757 | |
| gre | Green Driving Force | 330 | 0.4776 | 0.0814 | 0.2977 | 0.7161 | |
| pro | Production Driving Force | 330 | 0.3359 | 0.0993 | 0.1133 | 0.6121 | |
| Moderating variable | evi | Environmental Regulation | 330 | 11.3379 | 12.0869 | 0.0860 | 110.3389 |
| Control variables | cos | Consumption level | 330 | 0.3801 | 0.0683 | 0.2220 | 0.5384 |
| fdi | Foreign investment | 330 | 0.8381 | 0.8092 | 0.0003 | 3.5760 | |
| tra | Foreign trade | 330 | 0.2653 | 0.2908 | 0.0076 | 1.5482 | |
| gov | Government intervention | 330 | 0.2487 | 0.1025 | 0.1066 | 0.6430 | |
| tax | Tax burden level | 330 | 0.0819 | 0.0293 | 0.0443 | 0.1997 |
| Province | District | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | 2021 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beijing | 1 | 0.4444 | 0.4734 | 0.4946 | 0.5188 | 0.5202 | 0.5322 | 0.5375 | 0.5503 | 0.5648 | 0.5556 | 0.5691 |
| Tianjin | 1 | 0.3986 | 0.4093 | 0.4180 | 0.4232 | 0.4308 | 0.4326 | 0.4336 | 0.4416 | 0.4498 | 0.4801 | 0.4988 |
| Hebei | 1 | 0.2549 | 0.2585 | 0.2683 | 0.2781 | 0.2951 | 0.3209 | 0.3427 | 0.3515 | 0.3732 | 0.4070 | 0.4226 |
| Shanxi | 2 | 0.2288 | 0.2566 | 0.2579 | 0.2629 | 0.2650 | 0.2830 | 0.3032 | 0.3210 | 0.3352 | 0.3520 | 0.3640 |
| Inner Mongolia | 3 | 0.2671 | 0.2722 | 0.2926 | 0.2989 | 0.3021 | 0.3140 | 0.3257 | 0.3272 | 0.3360 | 0.3507 | 0.3671 |
| Liaoning | 4 | 0.3145 | 0.3329 | 0.3422 | 0.3408 | 0.3422 | 0.3694 | 0.3813 | 0.3967 | 0.4033 | 0.4235 | 0.4448 |
| Jilin | 4 | 0.3079 | 0.3199 | 0.3416 | 0.3452 | 0.3477 | 0.3651 | 0.3735 | 0.3930 | 0.4089 | 0.4234 | 0.4378 |
| Heilongjiang | 4 | 0.2977 | 0.3159 | 0.3334 | 0.3347 | 0.3369 | 0.3499 | 0.3476 | 0.3648 | 0.3770 | 0.3924 | 0.4047 |
| Shanghai | 1 | 0.3857 | 0.3986 | 0.4140 | 0.4167 | 0.4249 | 0.4248 | 0.4400 | 0.4640 | 0.4789 | 0.4984 | 0.5115 |
| Jiangsu | 1 | 0.4024 | 0.4282 | 0.4501 | 0.4583 | 0.4657 | 0.4690 | 0.4806 | 0.5137 | 0.5268 | 0.5684 | 0.5825 |
| Zhejiang | 1 | 0.4079 | 0.4358 | 0.4549 | 0.4640 | 0.4798 | 0.4867 | 0.4986 | 0.5310 | 0.5498 | 0.5809 | 0.5933 |
| Anhui | 2 | 0.3357 | 0.3524 | 0.3697 | 0.3755 | 0.3898 | 0.3938 | 0.4130 | 0.4275 | 0.4405 | 0.4755 | 0.4960 |
| Fujian | 1 | 0.3695 | 0.3960 | 0.4092 | 0.4125 | 0.4161 | 0.4201 | 0.4298 | 0.4524 | 0.4600 | 0.4802 | 0.4991 |
| Jiangxi | 2 | 0.3225 | 0.3372 | 0.3421 | 0.3508 | 0.3599 | 0.3629 | 0.3826 | 0.4101 | 0.4397 | 0.4517 | 0.4748 |
| Shandong | 1 | 0.3516 | 0.3715 | 0.3969 | 0.4028 | 0.4101 | 0.4257 | 0.4480 | 0.4649 | 0.4671 | 0.4991 | 0.5218 |
| Henan | 2 | 0.2672 | 0.2878 | 0.3101 | 0.3201 | 0.3357 | 0.3564 | 0.3965 | 0.4151 | 0.4343 | 0.4706 | 0.4960 |
| Hubei | 2 | 0.3468 | 0.3598 | 0.3745 | 0.3890 | 0.3938 | 0.4189 | 0.4255 | 0.4471 | 0.4679 | 0.4819 | 0.5010 |
| Hunan | 2 | 0.3271 | 0.3415 | 0.3545 | 0.3651 | 0.3785 | 0.3874 | 0.4052 | 0.4293 | 0.4455 | 0.4715 | 0.4839 |
| Guangdong | 1 | 0.4061 | 0.4235 | 0.4427 | 0.4566 | 0.4736 | 0.4833 | 0.5078 | 0.5572 | 0.5811 | 0.6211 | 0.6418 |
| Guangxi | 3 | 0.3288 | 0.3427 | 0.3559 | 0.3600 | 0.3678 | 0.3731 | 0.3828 | 0.3970 | 0.4104 | 0.4350 | 0.4380 |
| Hainan | 1 | 0.3342 | 0.3537 | 0.3647 | 0.3641 | 0.3744 | 0.3779 | 0.3794 | 0.3930 | 0.4093 | 0.4227 | 0.4352 |
| Chongqing | 3 | 0.3407 | 0.3529 | 0.3730 | 0.3826 | 0.3974 | 0.4049 | 0.4173 | 0.4380 | 0.4534 | 0.4785 | 0.4937 |
| Sichuan | 3 | 0.3007 | 0.3180 | 0.3357 | 0.3493 | 0.3768 | 0.3868 | 0.4010 | 0.4339 | 0.4480 | 0.4732 | 0.4956 |
| Guizhou | 3 | 0.2420 | 0.2617 | 0.2746 | 0.3087 | 0.3243 | 0.3419 | 0.3545 | 0.3802 | 0.4015 | 0.4279 | 0.4437 |
| Yunnan | 3 | 0.2879 | 0.2962 | 0.3254 | 0.3338 | 0.3460 | 0.3524 | 0.3673 | 0.3862 | 0.4057 | 0.4233 | 0.4386 |
| Shaanxi | 3 | 0.2992 | 0.3165 | 0.3341 | 0.3460 | 0.3648 | 0.3852 | 0.3814 | 0.3945 | 0.4016 | 0.4305 | 0.4244 |
| Gansu | 3 | 0.2516 | 0.2611 | 0.2774 | 0.2828 | 0.2903 | 0.3074 | 0.3171 | 0.3379 | 0.3667 | 0.3719 | 0.3814 |
| Qinghai | 3 | 0.2742 | 0.2779 | 0.2810 | 0.2927 | 0.2933 | 0.3047 | 0.3165 | 0.3338 | 0.3461 | 0.3524 | 0.3677 |
| Ningxia | 3 | 0.2873 | 0.2996 | 0.3114 | 0.3257 | 0.3194 | 0.3209 | 0.3294 | 0.3457 | 0.3371 | 0.3555 | 0.3713 |
| Xinjiang | 3 | 0.2372 | 0.2455 | 0.2507 | 0.2524 | 0.2779 | 0.2867 | 0.3037 | 0.3165 | 0.3178 | 0.3406 | 0.3494 |
| Province | New Quality Productivity Index | Ranking | Innovation Driving Force Index | Ranking | Green Driving Force Index | Ranking | Production Driving Force Score | Ranking |
|---|---|---|---|---|---|---|---|---|
| Beijign | 0.5237 | 1 | 0.3726 | 5 | 0.6062 | 2 | 0.5203 | 1 |
| Tianjin | 0.4379 | 6 | 0.4242 | 4 | 0.4840 | 15 | 0.3752 | 8 |
| Hebei | 0.3248 | 25 | 0.1864 | 19 | 0.3838 | 27 | 0.3281 | 15 |
| Shanxi | 0.2936 | 29 | 0.1853 | 20 | 0.3384 | 30 | 0.3029 | 20 |
| Inner Mongolia | 0.3140 | 26 | 0.1642 | 23 | 0.3650 | 28 | 0.3307 | 14 |
| Liaoning | 0.3719 | 17 | 0.2491 | 12 | 0.4429 | 20 | 0.3544 | 12 |
| Jilin | 0.3694 | 20 | 0.1994 | 17 | 0.4792 | 16 | 0.2935 | 24 |
| Heilongjiang | 0.3505 | 22 | 0.1950 | 18 | 0.4332 | 22 | 0.3213 | 17 |
| Shanghai | 0.4416 | 5 | 0.3687 | 6 | 0.5082 | 11 | 0.3847 | 5 |
| Jiangsu | 0.4860 | 4 | 0.4965 | 1 | 0.5473 | 5 | 0.3767 | 7 |
| Zhejiang | 0.4984 | 3 | 0.4508 | 3 | 0.6169 | 1 | 0.3580 | 11 |
| Anhui | 0.4063 | 11 | 0.2400 | 13 | 0.5314 | 8 | 0.3189 | 18 |
| Fujian | 0.4314 | 8 | 0.2777 | 8 | 0.5570 | 4 | 0.3148 | 19 |
| Jiangxi | 0.3849 | 14 | 0.2247 | 14 | 0.4951 | 13 | 0.2943 | 23 |
| Shandong | 0.4327 | 7 | 0.3463 | 7 | 0.4935 | 14 | 0.4016 | 4 |
| Henan | 0.3718 | 18 | 0.2191 | 15 | 0.4332 | 23 | 0.3815 | 6 |
| Hubei | 0.4188 | 9 | 0.2530 | 10 | 0.5261 | 9 | 0.3750 | 9 |
| Hunan | 0.3990 | 12 | 0.2141 | 16 | 0.5063 | 12 | 0.3595 | 10 |
| Guangdong | 0.5086 | 2 | 0.4767 | 2 | 0.6012 | 3 | 0.4063 | 3 |
| Guangxi | 0.3810 | 16 | 0.1316 | 26 | 0.5352 | 6 | 0.2580 | 28 |
| Hainan | 0.3826 | 15 | 0.1379 | 25 | 0.5247 | 10 | 0.2497 | 29 |
| Chongqing | 0.4120 | 10 | 0.2634 | 9 | 0.5336 | 7 | 0.2977 | 21 |
| Sichuan | 0.3926 | 13 | 0.1765 | 21 | 0.4679 | 18 | 0.4177 | 2 |
| Guizhou | 0.3419 | 23 | 0.1274 | 27 | 0.4496 | 19 | 0.2921 | 25 |
| Yunnan | 0.3603 | 21 | 0.0877 | 29 | 0.4727 | 17 | 0.3270 | 16 |
| Shaanxi | 0.3707 | 19 | 0.2519 | 11 | 0.4384 | 21 | 0.3474 | 13 |
| Gansu | 0.3132 | 27 | 0.1484 | 24 | 0.3980 | 26 | 0.2623 | 27 |
| Qinghai | 0.3128 | 28 | 0.0816 | 30 | 0.4046 | 24 | 0.2485 | 30 |
| Ningxia | 0.3276 | 24 | 0.1696 | 22 | 0.4012 | 25 | 0.2814 | 26 |
| Xinjiang | 0.2889 | 30 | 0.1010 | 28 | 0.3523 | 29 | 0.2961 | 22 |
| (1) | (2) | (3) | (4) | |
|---|---|---|---|---|
| Rat | Ino | Gre | Pro | |
| nqp | −0.6228 *** | |||
| (0.0884) | ||||
| ino | −0.4507 *** | |||
| (0.0706) | ||||
| gre | −0.7016 *** | |||
| (0.0929) | ||||
| pro | −0.4172 *** | |||
| (0.0518) | ||||
| cos | −0.2001 ** | −0.2640 ** | −0.1745 ** | −0.1560 * |
| (0.0868) | (0.0979) | (0.0803) | (0.0936) | |
| fdi | −0.0031 | −0.0012 | −0.0096 | −0.0073 |
| (0.0079) | (0.0085) | (0.0077) | (0.0071) | |
| tra | −0.0830 *** | −0.0724 *** | −0.0888 *** | −0.0882 *** |
| (0.0272) | (0.0241) | (0.0268) | (0.0331) | |
| gov | −0.3002 | −0.4197 * | −0.2931 * | −0.1048 |
| (0.2139) | (0.2130) | (0.1730) | (0.1527) | |
| tax | 0.7976 ** | 1.0145 ** | 0.9029 ** | 0.2676 |
| (0.3363) | (0.3760) | (0.3740) | (0.2652) | |
| evi | 0.0003 | 0.0004 | 0.0008** | 0.0002 |
| (0.0003) | (0.0004) | (0.0003) | (0.0003) | |
| _cons | 0.5009 *** | 0.3984 *** | 0.5756 *** | 0.3836 *** |
| (0.0530) | (0.0452) | (0.0614) | (0.0389) | |
| Hausman | 14.35 | 14.70 | −148.48 | 6.23 |
| p-Value | 0.0259 | 0.0401 | - | 0.5126 |
| R | 0.650 | 0.580 | 0.586 | 0.665 |
| N | 330 | 330 | 330 | 330 |
| (1) | (2) | (3) | (4) | |
|---|---|---|---|---|
| Upg | Ino | Gre | Pro | |
| nqp | 2.5179 *** | |||
| (0.4119) | ||||
| ino | 1.4677 *** | |||
| (0.4265) | ||||
| gre | 2.5862 *** | |||
| (0.4729) | ||||
| pro | 1.7481 *** | |||
| (0.2264) | ||||
| cos | 0.4155 | 0.7269 ** | 0.4388 | 0.2349 |
| (0.3237) | (0.3295) | (0.3400) | (0.3142) | |
| fdi | −0.0066 | −0.0027 | 0.0080 | −0.0029 |
| (0.0493) | (0.0674) | (0.0546) | (0.0464) | |
| tra | −0.8545 *** | −0.9306 *** | −0.8993 *** | −0.8746 *** |
| (0.2848) | (0.3059) | (0.2951) | (0.2339) | |
| gov | 3.7884 *** | 4.4497 *** | 4.3143 *** | 3.1945 *** |
| (0.8260) | (0.786) | (0.8920) | (0.7256) | |
| tax | −5.8453 ** | −7.5855 *** | −7.8955 *** | −4.0714 ** |
| (2.1639) | (2.3196) | (2.1473) | (1.8121) | |
| evi | 0.0007 | −0.0001 | −0.0008 | 0.0018 ** |
| (0.0009) | (0.0014) | (0.0011) | (0.0008) | |
| _cons | −0.0336 | 0.4769 *** | −0.2454 | 0.4177 *** |
| (0.1572) | (0.1430) | (0.2065) | (0.1196) | |
| Hausman | 294.44 | 178.05 | 459.34 | 3428.56 |
| p-Value | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| R | 0.740 | 0.669 | 0.705 | 0.776 |
| N | 330 | 330 | 330 | 330 |
| (1) | (2) | (3) | (4) | |
|---|---|---|---|---|
| Sys-GMM | Diff-GMM | |||
| Rat | Upg | Rat | Upg | |
| L.rat | 0.7200 *** | 0.7552 *** | ||
| (0.0204) | (0.0187) | |||
| L.upg | 0.9922 *** | 0.6629 *** | ||
| (0.0399) | (0.0390) | |||
| nqp | −0.1121 *** | 0.7525 *** | −0.0439 *** | 0.3572 ** |
| (0.0187) | (0.1096) | (0.0161) | (0.1772) | |
| cos | −0.0121 | 0.4463 *** | −0.0130 * | 0.2589 *** |
| (0.0087) | (0.0779) | (0.0067) | (0.0392) | |
| fdi | −0.0120 *** | −0.0889 *** | −0.0051 *** | −0.0206 ** |
| (0.0036) | (0.0212) | (0.0014) | (0.0084) | |
| tra | −0.0740 *** | −0.1777 *** | −0.0001 | −0.6128 *** |
| (0.0194) | (0.0393) | (0.0077) | (0.1028) | |
| gov | −0.3254 *** | 1.3933 *** | −0.3838 *** | 2.2454 *** |
| (0.0288) | (0.3295) | (0.0234) | (0.1483) | |
| tax | 0.3911 *** | −0.3031 | 0.7349 *** | −2.6063 *** |
| (0.1013) | (0.5166) | (0.0933) | (0.4328) | |
| evi | −0.0001 *** | −0.0001 | −0.0001 *** | 0.0005 |
| (0.0000) | (0.0004) | (0.0000) | (0.0003) | |
| _cons | 0.1501 *** | −0.2537 *** | 0.0862 *** | 0.0341 |
| (0.0102) | (0.0974) | (0.0100) | (0.0636) | |
| AR(1)-P | 0.0135 | 0.0431 | 0.0176 | 0.1144 |
| AR(2)-P | 0.0564 | 0.1158 | 0.1069 | 0.2759 |
| Sargan | 28.0614 | 26.0649 | 26.9213 | 23.6456 |
| p-Value | 0.7911 | 0.9807 | 0.3598 | 0.8570 |
| N | 263 | 263 | 223 | 223 |
| (1) | (2) | (3) | (4) | (5) | (6) | |
|---|---|---|---|---|---|---|
| Benchmark Regression | Sys-GMM | Diff-GMM | ||||
| Rat | Upg | Rat | Upg | Rat | Upg | |
| L.rat | 0.7125 *** | 0.7287 *** | ||||
| (0.0205) | (0.0211) | |||||
| L.upg | 0.9823 *** | 0.6250 *** | ||||
| (0.0379) | (0.0360) | |||||
| nqp | −0.5742 *** | 2.6335 *** | −0.1051 *** | 0.7579 *** | −0.0550 *** | 0.4162 ** |
| (0.0767) | (0.4859) | (0.0300) | (0.1588) | (0.0186) | (0.1982) | |
| evi | 0.0065 *** | 0.0090 | 0.0015 *** | −0.0131 *** | 0.0013 *** | −0.0049 ** |
| (0.0020) | (0.0095) | (0.0003) | (0.0026) | (0.0003) | (0.0025) | |
| evnqp | −0.0194 *** | −0.0260 | −0.0051 *** | 0.0380 *** | −0.0045 *** | 0.0167 ** |
| (0.0062) | (0.0301) | (0.0009) | (0.0084) | (0.0009) | (0.0074) | |
| cos | −0.1557 ** | 0.4530 | 0.0187 ** | 0.3827 *** | −0.0108 | 0.3762 *** |
| (0.0776) | (0.3144) | (0.0087) | (0.0917) | (0.0097) | (0.0643) | |
| fdi | −0.0026 | −0.0047 | −0.0111** | −0.0546** | −0.0030 | −0.0231 |
| (0.0083) | (0.0468) | (0.0053) | (0.0265) | (0.0018) | (0.0150) | |
| tra | −0.0943 *** | −0.8557 *** | −0.0806 *** | −0.1629 *** | −0.0001 | −0.5476 *** |
| (0.0276) | (0.2899) | (0.0203) | (0.0560) | (0.0084) | (0.1183) | |
| gov | −0.1855 | 3.8440 *** | −0.3345 *** | 1.4725 *** | −0.3579 *** | 2.3434 *** |
| (0.1589) | (0.8234) | (0.0402) | (0.3534) | (0.0327) | (0.1488) | |
| tax | 0.7018 ** | −5.6357 ** | 0.5115 *** | 0.1629 | 0.7793 *** | −3.3264 *** |
| (0.2858) | (2.2077) | (0.0860) | (0.7969) | (0.0980) | (0.4858) | |
| _cons | 0.4536 *** | −0.11657 | 0.1423 *** | −0.2424 * | 0.0849 *** | 0.0267 |
| (0.0444) | (0.1971) | (0.0167) | (0.1279) | (0.0155) | (0.1119) | |
| Hausman | 12.34 | 316.71 | ||||
| p-Value | 0.0900 | 0.0000 | ||||
| R | 0.683 | 0.742 | ||||
| AR(1)-P | 0.0115 | 0.0461 | 0.0157 | 0.1737 | ||
| AR(2)-P | 0.0760 | 0.1498 | 0.1446 | 0.2314 | ||
| Sargan | 28.0984 | 23.4695 | 27.5126 | 22.5209 | ||
| p-Value | 0.9615 | 0.9933 | 0.3308 | 0.6055 | ||
| N | 330 | 330 | 263 | 263 | 223 | 223 |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Shao, C.; Dong, H.; Gao, Y. New Quality Productivity and Industrial Structure in China: The Moderating Effect of Environmental Regulation. Sustainability 2024 , 16 , 6796. https://doi.org/10.3390/su16166796
Shao C, Dong H, Gao Y. New Quality Productivity and Industrial Structure in China: The Moderating Effect of Environmental Regulation. Sustainability . 2024; 16(16):6796. https://doi.org/10.3390/su16166796
Shao, Changhua, Han Dong, and Yuan Gao. 2024. "New Quality Productivity and Industrial Structure in China: The Moderating Effect of Environmental Regulation" Sustainability 16, no. 16: 6796. https://doi.org/10.3390/su16166796
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals

COMMENTS
Quality control is an important aspect in producing products or services, in order to ensure quality in accordance with company standards so that it can meet consumer expectations. 2.1.3. Quality control in medical microbiology The issue of quality control (QC) in the medical microbiology laboratory is complex.
Clinical microbiology laboratories perform quality control (QC) for their analytical test methods, which is the initial level of controlling a procedure. ... Follow and enforce the policies and procedures that govern the criteria for appropriate choice and use of microbiology services: Write, promote, and enforce the policies and procedures ...
David Roesti, PhD, works at Novartis Pharma AG in Stein, Switzerland, and is responsible for defining the microbial control strategy at the site and is a global subject matter expert in microbiology for the Novartis group. He is also is an elected member of the General Chapters Microbiology Expert Committee of the Unites States Pharmacopoeia 2015 2020 revision cycle.
Abstract. Given the importance of microbiology results on patient care, high quality standards are expected. Internal quality assurance (IQA) could mitigate the limitations of internal quality control, competency assessment and external quality assurance, adding a longitudinal insight, including pre- and post-analytical steps.
Quality Control of Stains. Microbiology laboratories use a wide variety of stains such as simple stains (methylene blue), negative staining (India ink), differential stains (gram stain, Z-N stain for acid fast bacilli, Giemsa stain), and other stains such as auramine- rhodamine stain for acid fast bacilli, lactophenol cotton blue, and so on.
Abstract. The quality control (QC) practice in a microbiology laboratory is a bit different than what we have covered till now. This chapter provides an overview of quality control specific to microbiology. Pre-analytical and analytical phases emphasis on QC in culture media, stains, processing of samples for culture, and various ...
The quality principles are organized under three main categories: quality infrastructure, laboratory operations, and quality assurance and continual improvement. The roles and responsibilities to establish and sustain a QMS are outlined for microbiology laboratory staff, laboratory management personnel, and the institution's leadership.
Thus this book is welcome as an excellent practical guide to the institution of quality assurance (QA) in microbiology. Since it was first published in 1991 under the title Quality Control: Principles and Practice in the Microbiology Laboratory there has been considerable revision, with the addition of several sections and many new contributors ...
Abstract. Quality assurance (QA) is the total process whereby the quality of laboratory reports can be guaranteed. The term quality control covers that part of QA, which primarily concerns the control of errors in the performance of tests and verification of test results. All materials, equipment and procedures must be adequately controlled.
Quality control in microbiology, with the proliferation and widespread usage of commercial identification systems, has largely passed into the hands of the manufacturer. In this review the author attempts to provide a spectrum of information concerning the validity, precision, and specificity of commercially available systems and individual methods for the identification of bacteria, of ...
The Handbook of Microbiological Quality Control provides guidance on safe microbiological practices, including laboratory design and sampling techniques. The design storage, use and quality ...
Overall, to ensure product safety, pharmaceutical companies must be versed in the important role of microbiological testing in product research and development, process validation, manufacturing, and quality control. ATCC supports the maintenance of product integrity, reputation, and safety by providing top-quality, fully characterized strains ...
Given the importance of microbiology results on patient care, high quality standards are expected. Internal quality assurance (IQA) could mitigate the limitations of internal quality control, competency assessment and external quality assurance, adding a longitudinal insight, including pre- and post-analytical steps. Here, we implemented an IQA program in our clinical microbiology facilities ...
Taking into account that 90 percent of quality control microbiology remains reliant upon culture based methods, this unique text focuses on microbiological culture media as applied to ...
A comprehensive microbial control program should allow to identify the risk of contamination to the product as well as the different mitigation steps to control this risk. The systems used to control the environment of the facility are qualified prior to running operations and are part of the facilities' life cycle.
Quality control in microbiology, with the proliferation and widespread usage of commercial identification systems, has largely passed into the hands of the manufacturer. In this review the author attempts to provide a spectrum of information concerning the validity, precision, and specificity of commercially available systems and individual ...
Initial processing and incubation of clinical specimens for bacteriology are complex and involve many decision‐making steps depending on the culture type and specimen submitted. Many of the decision‐making steps can be standardized and automated…. Back to Table of Contents. ClinMicroNow Books. Cases in Medical Microbiology and Infectious ...
In the third step, you identify ways to control the contamination, and in the fourth step, you predict, implement and validate your solutions. In step 5, you then monitor and verify your control measures. Schematic representation of a step-wise approach to ensure safe and microbially stable food throughout the production chain. Image source: NIZO.
Essay # 6. Modern Microbiology: Modern microbiology reaches into many fields of humans including the development of pharmaceutical products, the use of quality-control and dairy product production, the control of disease-causing microorganisms in water, and the industrial applications of microorganisms.
A Quality Control Microbiologist cover letter should include the following: A brief introduction that explains your interest in the Quality Control Microbiologist role and why you feel your qualifications and experience make you the ideal candidate for the job. A summary of your relevant experience in the field of Quality Control Microbiology ...
Academia.edu no longer supports Internet Explorer. To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your ...
To explore the connotation and development level of China's new quality productivity, this paper constructs an index system based on innovation, greenness, and productivity. This system is used to describe the development level of China's new quality productivity. Using relevant data from 30 provincial administrative regions in China from 2011 to 2021, the entropy weight-TOPSIS method was ...