METHODS article
Using deep learning for image-based plant disease detection.

- 1 Digital Epidemiology Lab, EPFL, Geneva, Switzerland
- 2 School of Life Sciences, EPFL, Lausanne, Switzerland
- 3 School of Computer and Communication Sciences, EPFL, Lausanne, Switzerland
- 4 Department of Entomology, College of Agricultural Sciences, Penn State University, State College, PA, USA
- 5 Department of Biology, Eberly College of Sciences, Penn State University, State College, PA, USA
- 6 Center for Infectious Disease Dynamics, Huck Institutes of Life Sciences, Penn State University, State College, PA, USA
Crop diseases are a major threat to food security, but their rapid identification remains difficult in many parts of the world due to the lack of the necessary infrastructure. The combination of increasing global smartphone penetration and recent advances in computer vision made possible by deep learning has paved the way for smartphone-assisted disease diagnosis. Using a public dataset of 54,306 images of diseased and healthy plant leaves collected under controlled conditions, we train a deep convolutional neural network to identify 14 crop species and 26 diseases (or absence thereof). The trained model achieves an accuracy of 99.35% on a held-out test set, demonstrating the feasibility of this approach. Overall, the approach of training deep learning models on increasingly large and publicly available image datasets presents a clear path toward smartphone-assisted crop disease diagnosis on a massive global scale.

Introduction
Modern technologies have given human society the ability to produce enough food to meet the demand of more than 7 billion people. However, food security remains threatened by a number of factors including climate change ( Tai et al., 2014 ), the decline in pollinators ( Report of the Plenary of the Intergovernmental Science-PolicyPlatform on Biodiversity Ecosystem and Services on the work of its fourth session, 2016 ), plant diseases ( Strange and Scott, 2005 ), and others. Plant diseases are not only a threat to food security at the global scale, but can also have disastrous consequences for smallholder farmers whose livelihoods depend on healthy crops. In the developing world, more than 80 percent of the agricultural production is generated by smallholder farmers ( UNEP, 2013 ), and reports of yield loss of more than 50% due to pests and diseases are common ( Harvey et al., 2014 ). Furthermore, the largest fraction of hungry people (50%) live in smallholder farming households ( Sanchez and Swaminathan, 2005 ), making smallholder farmers a group that's particularly vulnerable to pathogen-derived disruptions in food supply.
Various efforts have been developed to prevent crop loss due to diseases. Historical approaches of widespread application of pesticides have in the past decade increasingly been supplemented by integrated pest management (IPM) approaches ( Ehler, 2006 ). Independent of the approach, identifying a disease correctly when it first appears is a crucial step for efficient disease management. Historically, disease identification has been supported by agricultural extension organizations or other institutions, such as local plant clinics. In more recent times, such efforts have additionally been supported by providing information for disease diagnosis online, leveraging the increasing Internet penetration worldwide. Even more recently, tools based on mobile phones have proliferated, taking advantage of the historically unparalleled rapid uptake of mobile phone technology in all parts of the world ( ITU, 2015 ).
Smartphones in particular offer very novel approaches to help identify diseases because of their computing power, high-resolution displays, and extensive built-in sets of accessories, such as advanced HD cameras. It is widely estimated that there will be between 5 and 6 billion smartphones on the globe by 2020. At the end of 2015, already 69% of the world's population had access to mobile broadband coverage, and mobile broadband penetration reached 47% in 2015, a 12-fold increase since 2007 ( ITU, 2015 ). The combined factors of widespread smartphone penetration, HD cameras, and high performance processors in mobile devices lead to a situation where disease diagnosis based on automated image recognition, if technically feasible, can be made available at an unprecedented scale. Here, we demonstrate the technical feasibility using a deep learning approach utilizing 54,306 images of 14 crop species with 26 diseases (or healthy) made openly available through the project PlantVillage ( Hughes and Salathé, 2015 ). An example of each crop—disease pair can be seen in Figure 1 .

Figure 1. Example of leaf images from the PlantVillage dataset, representing every crop-disease pair used. (1) Apple Scab, Venturia inaequalis (2) Apple Black Rot, Botryosphaeria obtusa (3) Apple Cedar Rust, Gymnosporangium juniperi-virginianae (4) Apple healthy (5) Blueberry healthy (6) Cherry healthy (7) Cherry Powdery Mildew, Podoshaera clandestine (8) Corn Gray Leaf Spot, Cercospora zeae-maydis (9) Corn Common Rust, Puccinia sorghi (10) Corn healthy (11) Corn Northern Leaf Blight, Exserohilum turcicum (12) Grape Black Rot, Guignardia bidwellii , (13) Grape Black Measles (Esca), Phaeomoniella aleophilum, Phaeomoniella chlamydospora (14) Grape Healthy (15) Grape Leaf Blight, Pseudocercospora vitis (16) Orange Huanglongbing (Citrus Greening), Candidatus Liberibacter spp. (17) Peach Bacterial Spot, Xanthomonas campestris (18) Peach healthy (19) Bell Pepper Bacterial Spot, Xanthomonas campestris (20) Bell Pepper healthy (21) Potato Early Blight, Alternaria solani (22) Potato healthy (23) Potato Late Blight, Phytophthora infestans (24) Raspberry healthy (25) Soybean healthy (26) Squash Powdery Mildew, Erysiphe cichoracearum (27) Strawberry Healthy (28) Strawberry Leaf Scorch, Diplocarpon earlianum (29) Tomato Bacterial Spot, Xanthomonas campestris pv. vesicatoria (30) Tomato Early Blight, Alternaria solani (31) Tomato Late Blight, Phytophthora infestans (32) Tomato Leaf Mold, Passalora fulva (33) Tomato Septoria Leaf Spot, Septoria lycopersici (34) Tomato Two Spotted Spider Mite, Tetranychus urticae (35) Tomato Target Spot, Corynespora cassiicola (36) Tomato Mosaic Virus (37) Tomato Yellow Leaf Curl Virus (38) Tomato healthy.
Computer vision, and object recognition in particular, has made tremendous advances in the past few years. The PASCAL VOC Challenge ( Everingham et al., 2010 ), and more recently the Large Scale Visual Recognition Challenge (ILSVRC) ( Russakovsky et al., 2015 ) based on the ImageNet dataset ( Deng et al., 2009 ) have been widely used as benchmarks for numerous visualization-related problems in computer vision, including object classification. In 2012, a large, deep convolutional neural network achieved a top-5 error of 16.4% for the classification of images into 1000 possible categories ( Krizhevsky et al., 2012 ). In the following 3 years, various advances in deep convolutional neural networks lowered the error rate to 3.57% ( Krizhevsky et al., 2012 ; Simonyan and Zisserman, 2014 ; Zeiler and Fergus, 2014 ; He et al., 2015 ; Szegedy et al., 2015 ). While training large neural networks can be very time-consuming, the trained models can classify images very quickly, which makes them also suitable for consumer applications on smartphones.
Deep neural networks have recently been successfully applied in many diverse domains as examples of end to end learning. Neural networks provide a mapping between an input—such as an image of a diseased plant—to an output—such as a crop~disease pair. The nodes in a neural network are mathematical functions that take numerical inputs from the incoming edges, and provide a numerical output as an outgoing edge. Deep neural networks are simply mapping the input layer to the output layer over a series of stacked layers of nodes. The challenge is to create a deep network in such a way that both the structure of the network as well as the functions (nodes) and edge weights correctly map the input to the output. Deep neural networks are trained by tuning the network parameters in such a way that the mapping improves during the training process. This process is computationally challenging and has in recent times been improved dramatically by a number of both conceptual and engineering breakthroughs ( LeCun et al., 2015 ; Schmidhuber, 2015 ).
In order to develop accurate image classifiers for the purposes of plant disease diagnosis, we needed a large, verified dataset of images of diseased and healthy plants. Until very recently, such a dataset did not exist, and even smaller datasets were not freely available. To address this problem, the PlantVillage project has begun collecting tens of thousands of images of healthy and diseased crop plants ( Hughes and Salathé, 2015 ), and has made them openly and freely available. Here, we report on the classification of 26 diseases in 14 crop species using 54,306 images with a convolutional neural network approach. We measure the performance of our models based on their ability to predict the correct crop-diseases pair, given 38 possible classes. The best performing model achieves a mean F 1 score of 0.9934 (overall accuracy of 99.35%), hence demonstrating the technical feasibility of our approach. Our results are a first step toward a smartphone-assisted plant disease diagnosis system.
Dataset Description
We analyze 54,306 images of plant leaves, which have a spread of 38 class labels assigned to them. Each class label is a crop-disease pair, and we make an attempt to predict the crop-disease pair given just the image of the plant leaf. Figure 1 shows one example each from every crop-disease pair from the PlantVillage dataset. In all the approaches described in this paper, we resize the images to 256 × 256 pixels, and we perform both the model optimization and predictions on these downscaled images.
Across all our experiments, we use three different versions of the whole PlantVillage dataset. We start with the PlantVillage dataset as it is, in color; then we experiment with a gray-scaled version of the PlantVillage dataset, and finally we run all the experiments on a version of the PlantVillage dataset where the leaves were segmented, hence removing all the extra background information which might have the potential to introduce some inherent bias in the dataset due to the regularized process of data collection in case of PlantVillage dataset. Segmentation was automated by the means of a script tuned to perform well on our particular dataset. We chose a technique based on a set of masks generated by analysis of the color, lightness and saturation components of different parts of the images in several color spaces (Lab and HSB). One of the steps of that processing also allowed us to easily fix color casts, which happened to be very strong in some of the subsets of the dataset, thus removing another potential bias.
This set of experiments was designed to understand if the neural network actually learns the “notion” of plant diseases, or if it is just learning the inherent biases in the dataset. Figure 2 shows the different versions of the same leaf for a randomly selected set of leaves.
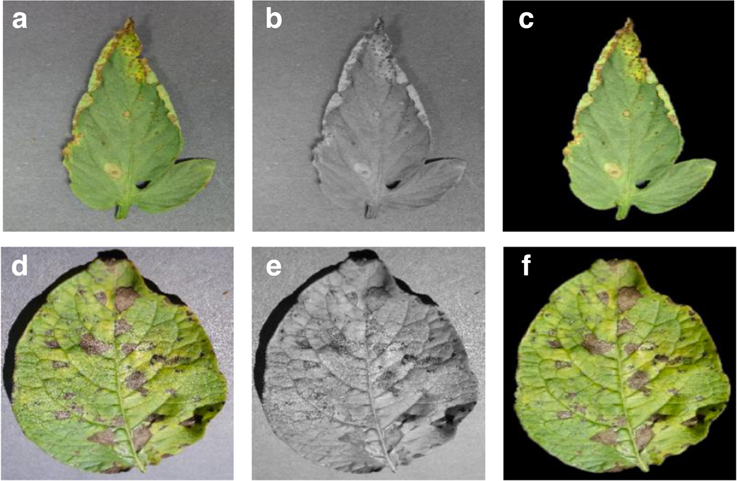
Figure 2. Sample images from the three different versions of the PlantVillage dataset used in various experimental configurations. (A) Leaf 1 color, (B) Leaf 1 grayscale, (C) Leaf 1 segmented, (D) Leaf 2 color, (E) Leaf 2 gray-scale, (F) Leaf 2 segmented.
Measurement of Performance
To get a sense of how our approaches will perform on new unseen data, and also to keep a track of if any of our approaches are overfitting, we run all our experiments across a whole range of train-test set splits, namely 80–20 (80% of the whole dataset used for training, and 20% for testing), 60–40 (60% of the whole dataset used for training, and 40% for testing), 50–50 (50% of the whole dataset used for training, and 50% for testing), 40–60 (40% of the whole dataset used for training, and 60% for testing) and finally 20–80 (20% of the whole dataset used for training, and 80% for testing). It must be noted that in many cases, the PlantVillage dataset has multiple images of the same leaf (taken from different orientations), and we have the mappings of such cases for 41,112 images out of the 54,306 images; and during all these test-train splits, we make sure all the images of the same leaf goes either in the training set or the testing set. Further, for every experiment, we compute the mean precision, mean recall, mean F 1 score, along with the overall accuracy over the whole period of training at regular intervals (at the end of every epoch). We use the final mean F 1 score for the comparison of results across all of the different experimental configurations.
We evaluate the applicability of deep convolutional neural networks for the classification problem described above. We focus on two popular architectures, namely AlexNet ( Krizhevsky et al., 2012 ), and GoogLeNet ( Szegedy et al., 2015 ), which were designed in the context of the “Large Scale Visual Recognition Challenge” (ILSVRC) ( Russakovsky et al., 2015 ) for the ImageNet dataset ( Deng et al., 2009 ).
The AlexNet architecture (see Figure S2) follows the same design pattern as the LeNet-5 ( LeCun et al., 1989 ) architecture from the 1990s. The LeNet-5 architecture variants are usually a set of stacked convolution layers followed by one or more fully connected layers. The convolution layers optionally may have a normalization layer and a pooling layer right after them, and all the layers in the network usually have ReLu non-linear activation units associated with them. AlexNet consists of 5 convolution layers, followed by 3 fully connected layers, and finally ending with a softMax layer. The first two convolution layers (conv{1, 2}) are each followed by a normalization and a pooling layer, and the last convolution layer (conv5) is followed by a single pooling layer. The final fully connected layer (fc8) has 38 outputs in our adapted version of AlexNet (equaling the total number of classes in our dataset), which feeds the softMax layer. The softMax layer finally exponentially normalizes the input that it gets from (fc8), thereby producing a distribution of values across the 38 classes that add up to 1. These values can be interpreted as the confidences of the network that a given input image is represented by the corresponding classes. All of the first 7 layers of AlexNet have a ReLu non-linearity activation unit associated with them, and the first two fully connected layers (fc{6, 7}) have a dropout layer associated with them, with a dropout ratio of 0.5.
The GoogleNet architecture on the other hand is a much deeper and wider architecture with 22 layers, while still having considerably lower number of parameters (5 million parameters) in the network than AlexNet (60 million parameters). An application of the “network in network” architecture ( Lin et al., 2013 ) in the form of the inception modules is a key feature of the GoogleNet architecture. The inception module uses parallel 1 × 1, 3 × 3, and 5 × 5 convolutions along with a max-pooling layer in parallel, hence enabling it to capture a variety of features in parallel. In terms of practicality of the implementation, the amount of associated computation needs to be kept in check, which is why 1 × 1 convolutions before the above mentioned 3 × 3, 5 × 5 convolutions (and also after the max-pooling layer) are added for dimensionality reduction. Finally, a filter concatenation layer simply concatenates the outputs of all these parallel layers. While this forms a single inception module, a total of 9 inception modules is used in the version of the GoogLeNet architecture that we use in our experiments. A more detailed overview of this architecture can be found for reference in ( Szegedy et al., 2015 ).
We analyze the performance of both these architectures on the PlantVillage dataset by training the model from scratch in one case, and then by adapting already trained models (trained on the ImageNet dataset) using transfer learning. In case of transfer learning, we re-initialize the weights of layer fc8 in case of AlexNet, and of the loss {1,2,3}/classifier layers in case of GoogLeNet. Then, when training the model, we do not limit the learning of any of the layers, as is sometimes done for transfer learning. In other words, the key difference between these two learning approaches (transfer vs. training from scratch) is in the initial state of weights of a few layers, which lets the transfer learning approach exploit the large amount of visual knowledge already learned by the pre-trained AlexNet and GoogleNet models extracted from ImageNet ( Russakovsky et al., 2015 ).
To summarize, we have a total of 60 experimental configurations, which vary on the following parameters:
1. Choice of deep learning architecture:
2. Choice of training mechanism:
Transfer Learning,
Training from Scratch.
3. Choice of dataset type:
Gray scale,
Leaf Segmented.
4. Choice of training-testing set distribution:
Train: 80%, Test: 20%,
Train: 60%, Test: 40%,
Train: 50%, Test: 50%,
Train: 40%, Test: 60%,
Train: 20%, Test: 80%.
Throughout this paper, we have used the notation of Architecture:TrainingMechanism:DatasetType:Train-Test-Set-Distribution to refer to particular experiments. For instance, to refer to the experiment using the GoogLeNet architecture, which was trained using transfer learning on the gray-scaled PlantVillage dataset on a train—test set distribution of 60–40, we will use the notation GoogLeNet:TransferLearning:GrayScale:60–40 .
Each of these 60 experiments runs for a total of 30 epochs, where one epoch is defined as the number of training iterations in which the particular neural network has completed a full pass of the whole training set. The choice of 30 epochs was made based on the empirical observation that in all of these experiments, the learning always converged well within 30 epochs (as is evident from the aggregated plots (Figure 3 ) across all the experiments).
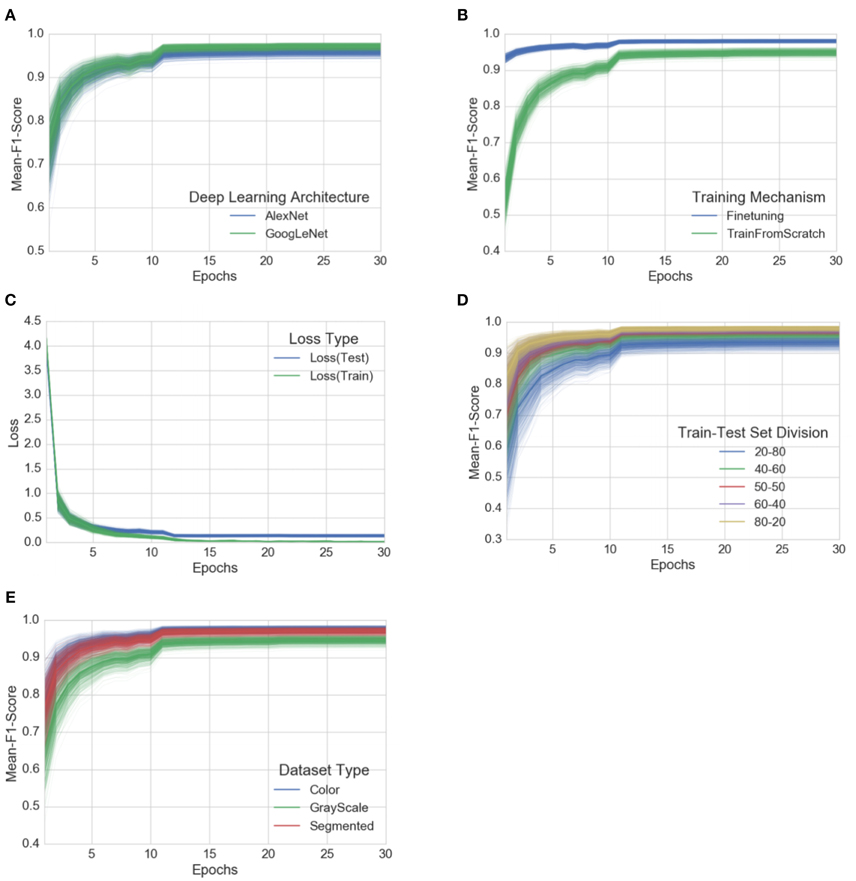
Figure 3. Progression of mean F 1 score and loss through the training period of 30 epochs across all experiments, grouped by experimental configuration parameters . The intensity of a particular class at any point is proportional to the corresponding uncertainty across all experiments with the particular configurations. (A) Comparison of progression of mean F 1 score across all experiments, grouped by deep learning architecture, (B) Comparison of progression of mean F 1 score across all experiments, grouped by training mechanism, (C) Comparison of progression of train-loss and test-loss across all experiments, (D) Comparison of progression of mean F 1 score across all experiments, grouped by train-test set splits, (E) Comparison of progression of mean F 1 score across all experiments, grouped by dataset type. A similar plot of all the observations, as it is, across all the experimental configurations can be found in the Supplementary Material.
To enable a fair comparison between the results of all the experimental configurations, we also tried to standardize the hyper-parameters across all the experiments, and we used the following hyper-parameters in all of the experiments:
• Solver type: Stochastic Gradient Descent,
• Base learning rate: 0.005,
• Learning rate policy: Step (decreases by a factor of 10 every 30/3 epochs),
• Momentum: 0.9,
• Weight decay: 0.0005,
• Gamma: 0.1,
• Batch size: 24 (in case of GoogLeNet), 100 (in case of AlexNet).
All the above experiments were conducted using our own fork of Caffe ( Jia et al., 2014 ), which is a fast, open source framework for deep learning. The basic results, such as the overall accuracy can also be replicated using a standard instance of caffe.
At the outset, we note that on a dataset with 38 class labels, random guessing will only achieve an overall accuracy of 2.63% on average. Across all our experimental configurations, which include three visual representations of the image data (see Figure 2 ), the overall accuracy we obtained on the PlantVillage dataset varied from 85.53% (in case of AlexNet::TrainingFromScratch::GrayScale::80–20 ) to 99.34% (in case of GoogLeNet::TransferLearning::Color::80–20 ), hence showing strong promise of the deep learning approach for similar prediction problems. Table 1 shows the mean F 1 score, mean precision, mean recall, and overall accuracy across all our experimental configurations. All the experimental configurations run for a total of 30 epochs each, and they almost consistently converge after the first step down in the learning rate.
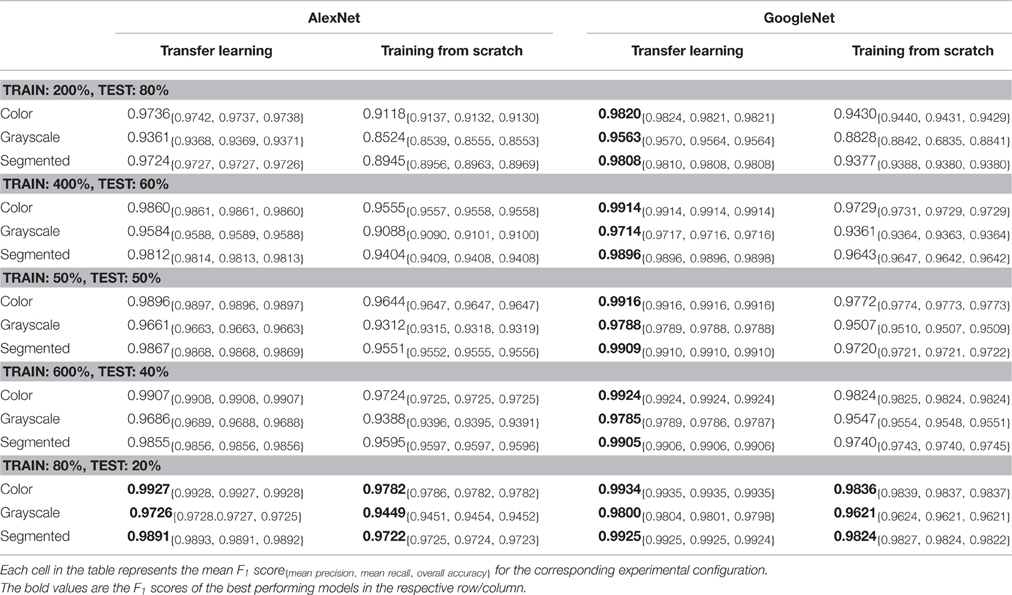
Table 1. Mean F 1 score across various experimental configurations at the end of 30 epochs .
To address the issue of over-fitting, we vary the test set to train set ratio and observe that even in the extreme case of training on only 20% of the data and testing the trained model on the rest 80% of the data, the model achieves an overall accuracy of 98.21% (mean F 1 score of 0.9820) in the case of GoogLeNet::TransferLearning::Color::20–80 . As expected, the overall performance of both AlexNet and GoogLeNet do degrade if we keep increasing the test set to train set ratio (see Figure 3D ), but the decrease in performance is not as drastic as we would expect if the model was indeed over-fitting. Figure 3C also shows that there is no divergence between the validation loss and the training loss, confirming that over-fitting is not a contributor to the results we obtain across all our experiments.
Among the AlexNet and GoogLeNet architectures, GoogLeNet consistently performs better than AlexNet (Figure 3A ), and based on the method of training, transfer learning always yields better results (Figure 3B ), both of which were expected.
The three versions of the dataset (color, gray-scale, and segmented) show a characteristic variation in performance across all the experiments when we keep the rest of the experimental configuration constant. The models perform the best in case of the colored version of the dataset. When designing the experiments, we were concerned that the neural networks might only learn to pick up the inherent biases associated with the lighting conditions, the method and apparatus of collection of the data. We therefore experimented with the gray-scaled version of the same dataset to test the model's adaptability in the absence of color information, and its ability to learn higher level structural patterns typical to particular crops and diseases. As expected, the performance did decrease when compared to the experiments on the colored version of the dataset, but even in the case of the worst performance, the observed mean F 1 score was 0.8524 (overall accuracy of 85.53%). The segmented versions of the whole dataset was also prepared to investigate the role of the background of the images in overall performance, and as shown in Figure 3E , the performance of the model using segmented images is consistently better than that of the model using gray-scaled images, but slightly lower than that of the model using the colored version of the images.
While these approaches yield excellent results on the PlantVillage dataset which was collected in a controlled environment, we also assessed the model's performance on images sampled from trusted online sources, such as academic agriculture extension services. Such images are not available in large numbers, and using a combination of automated download from Bing Image Search and IPM Images with a visual verification step, we obtained two small, verified datasets of 121 (dataset 1) and 119 images (dataset 2), respectively (see Supplementary Material for a detailed description of the process). Using the best model on these datasets, we obtained an overall accuracy of 31.40% in dataset 1, and 31.69% in dataset 2, in successfully predicting the correct class label (i.e., crop and disease information) from among 38 possible class labels. We note that a random classifier will obtain an average accuracy of only 2.63%. Across all images, the correct class was in the top-5 predictions in 52.89% of the cases in dataset 1, and in 65.61% of the cases in dataset 2. The best models for the two datasets were GoogLeNet:Segmented:TransferLearning:80–20 for dataset 1, and GoogLeNet:Color:TransferLearning:80–20 for dataset 2. An example image from theses datasets, along with its visualization of activations in the initial layers of an AlexNet architecture, can be seen in Figure 4 .
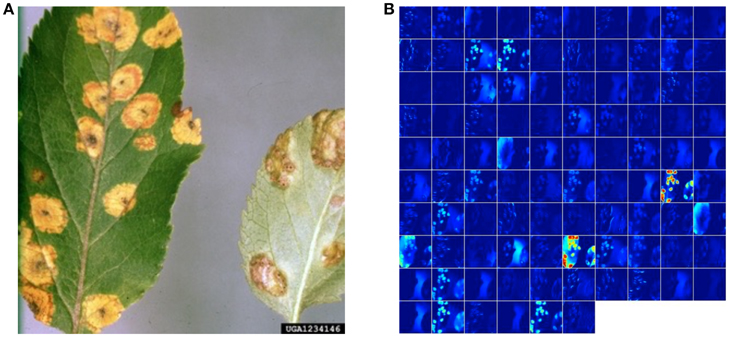
Figure 4. Visualization of activations in the initial layers of an AlexNet architecture demonstrating that the model has learnt to efficiently activate against the diseased spots on the example leaf. (A) Example image of a leaf suffering from Apple Cedar Rust, selected from the top-20 images returned by Bing Image search for the keywords “Apple Cedar Rust Leaves” on April 4th, 2016. Image Reference: Clemson University - USDA Cooperative Extension Slide Series, Bugwood. org. (B) Visualization of activations in the first convolution layer(conv1) of an AlexNet architecture trained using AlexNet:Color:TrainFromScratch:80–20 when doing a forward pass on the image in shown in panel b.
So far, all results have been reported under the assumption that the model needs to detect both the crop species and the disease status. We can limit the challenge to a more realistic scenario where the crop species is provided, as it can be expected to be known by those growing the crops. To assess this the performance of the model under this scenario, we limit ourselves to crops where we have at least n > = 2 (to avoid trivial classification) or n > = 3 classes per crop. In the n > = 2 case, dataset 1 contains 33 classes distributed among 9 crops. Random guessing in such a dataset would achieve an accuracy of 0.225, while our model has an accuracy of 0.478. In the n > = 3 case, the dataset contains 25 classes distributed among 5 crops. Random guessing in such a dataset would achieve an accuracy of 0.179, while our model has an accuracy of 0.411.
Similarly, in the n > = 2 case, dataset 2 contains 13 classes distributed among 4 crops. Random guessing in such a dataset would achieve an accuracy of 0.314, while our model has an accuracy of 0.545. In the n > = 3 case, the dataset contains 11 classes distributed among 3 crops. Random guessing in such a dataset would achieve an accuracy of 0.288, while our model has an accuracy of 0.485.
The performance of convolutional neural networks in object recognition and image classification has made tremendous progress in the past few years. ( Krizhevsky et al., 2012 ; Simonyan and Zisserman, 2014 ; Zeiler and Fergus, 2014 ; He et al., 2015 ; Szegedy et al., 2015 ). Previously, the traditional approach for image classification tasks has been based on hand-engineered features, such as SIFT ( Lowe, 2004 ), HoG ( Dalal and Triggs, 2005 ), SURF ( Bay et al., 2008 ), etc., and then to use some form of learning algorithm in these feature spaces. The performance of these approaches thus depended heavily on the underlying predefined features. Feature engineering itself is a complex and tedious process which needs to be revisited every time the problem at hand or the associated dataset changes considerably. This problem occurs in all traditional attempts to detect plant diseases using computer vision as they lean heavily on hand-engineered features, image enhancement techniques, and a host of other complex and labor-intensive methodologies.
In addition, traditional approaches to disease classification via machine learning typically focus on a small number of classes usually within a single crop. Examples include a feature extraction and classification pipeline using thermal and stereo images in order to classify tomato powdery mildew against healthy tomato leaves ( Raza et al., 2015 ); the detection of powdery mildew in uncontrolled environments using RGB images ( Hernández-Rabadán et al., 2014 ); the use of RGBD images for detection of apple scab ( Chéné et al., 2012 ) the use of fluorescence imaging spectroscopy for detection of citrus huanglongbing ( Wetterich et al., 2012 ) the detection of citrus huanglongbing using near infrared spectral patterns ( Sankaran et al., 2011 ) and aircraft-based sensors ( Garcia-Ruiz et al., 2013 ) the detection of tomato yellow leaf curl virus by using a set of classic feature extraction steps, followed by classification using a support vector machines pipeline ( Mokhtar et al., 2015 ), and many others. A very recent review on the use of machine learning on plant phenotyping ( Singh et al., 2015 ) extensively discusses the work in this domain. While neural networks have been used before in plant disease identification ( Huang, 2007 ) (for the classification and detection of Phalaenopsis seedling disease like bacterial soft rot, bacterial brown spot, and Phytophthora black rot), the approach required representing the images using a carefully selected list of texture features before the neural network could classify them.
Our approach is based on recent work Krizhevsky et al. (2012) which showed for the first time that end-to-end supervised training using a deep convolutional neural network architecture is a practical possibility even for image classification problems with a very large number of classes, beating the traditional approaches using hand-engineered features by a substantial margin in standard benchmarks. The absence of the labor-intensive phase of feature engineering and the generalizability of the solution makes them a very promising candidate for a practical and scaleable approach for computational inference of plant diseases.
Using the deep convolutional neural network architecture, we trained a model on images of plant leaves with the goal of classifying both crop species and the presence and identity of disease on images that the model had not seen before. Within the PlantVillage data set of 54,306 images containing 38 classes of 14 crop species and 26 diseases (or absence thereof), this goal has been achieved as demonstrated by the top accuracy of 99.35%. Thus, without any feature engineering, the model correctly classifies crop and disease from 38 possible classes in 993 out of 1000 images. Importantly, while the training of the model takes a lot of time (multiple hours on a high performance GPU cluster computer), the classification itself is very fast (less than a second on a CPU), and can thus easily be implemented on a smartphone. This presents a clear path toward smartphone-assisted crop disease diagnosis on a massive global scale.
However, there are a number of limitations at the current stage that need to be addressed in future work. First, when tested on a set of images taken under conditions different from the images used for training, the model's accuracy is reduced substantially, to just above 31%. It's important to note that this accuracy is much higher than the one based on random selection of 38 classes (2.6%), but nevertheless, a more diverse set of training data is needed to improve the accuracy. Our current results indicate that more (and more variable) data alone will be sufficient to substantially increase the accuracy, and corresponding data collection efforts are underway.
The second limitation is that we are currently constrained to the classification of single leaves, facing up, on a homogeneous background. While these are straightforward conditions, a real world application should be able to classify images of a disease as it presents itself directly on the plant. Indeed, many diseases don't present themselves on the upper side of leaves only (or at all), but on many different parts of the plant. Thus, new image collection efforts should try to obtain images from many different perspectives, and ideally from settings that are as realistic as possible.
At the same time, by using 38 classes that contain both crop species and disease status, we have made the challenge harder than ultimately necessary from a practical perspective, as growers are expected to know which crops they are growing. Given the very high accuracy on the PlantVillage dataset, limiting the classification challenge to the disease status won't have a measurable effect. However, on the real world datasets, we can measure noticeable improvements in accuracy. Overall, the presented approach works reasonably well with many different crop species and diseases, and is expected to improve considerably with more training data.
Finally, it's worth noting that the approach presented here is not intended to replace existing solutions for disease diagnosis, but rather to supplement them. Laboratory tests are ultimately always more reliable than diagnoses based on visual symptoms alone, and oftentimes early-stage diagnosis via visual inspection alone is challenging. Nevertheless, given the expectation of more than 5 Billion smartphones in the world by 2020—of which almost a Billion in Africa ( GSMA Intelligence, 2016 )—we do believe that the approach represents a viable additional method to help prevent yield loss. What's more, in the future, image data from a smartphone may be supplemented with location and time information for additional improvements in accuracy. Last but not least, it would be prudent to keep in mind the stunning pace at which mobile technology has developed in the past few years, and will continue to do so. With ever improving number and quality of sensors on mobiles devices, we consider it likely that highly accurate diagnoses via the smartphone are only a question of time.
Author Contributions
MS, DH, and SM conceived the study and wrote the paper. SM implemented the algorithm described.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Boris Conforty for help with the segmentation. We thank Kelsee Baranowski, Ryan Bringenberg, and Megan Wilkerson for taking the images and Kelsee Baranowski for image curation. We thank Anna Sostarecz, Kaity Gonzalez, Ashtyn Goodreau, Kalley Veit, Ethan Keller, Parand Jalili, Emma Volk, Nooeree Samdani, Kelsey Pryze for additional help with image curation. We thank EPFL, and the Huck Institutes at Penn State University for support. We are particularly grateful for access to EPFL GPU cluster computing resources.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01419
The data and the code used in this paper are available at the following locations:
Data: https://github.com/salathegroup/plantvillage_deeplearning_paper_dataset
Code: https://github.com/salathegroup/plantvillage_deeplearning_paper_analysis
More image data can be found at https://www.plantvillage.org/en/plant_images
Bay, H., Ess, A., Tuytelaars, T., and Van Gool, L. (2008). Speeded-up robust features (surf). Comput. Vis. Image Underst. 110, 346–359. doi: 10.1016/j.cviu.2007.09.014
CrossRef Full Text | Google Scholar
Chéné, Y., Rousseau, D., Lucidarme, P., Bertheloot, J., Caffier, V., Morel, P., et al. (2012). On the use of depth camera for 3d phenotyping of entire plants. Comput. Electron. Agric. 82, 122–127. doi: 10.1016/j.compag.2011.12.007
Dalal, N., and Triggs, B. (2005). “Histograms of oriented gradients for human detection,” in Computer Vision and Pattern Recognition, 2005. CVPR 2005. IEEE Computer Society Conference on. (IEEE) (Washington, DC).
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei L. (2009). “Imagenet: A large-scale hierarchical image database,” in Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference on. (IEEE).
Google Scholar
Ehler, L. E. (2006). Integrated pest management (ipm): definition, historical development and implementation, and the other ipm. Pest Manag. Sci. 62, 787–789. doi: 10.1002/ps.1247
PubMed Abstract | CrossRef Full Text | Google Scholar
Everingham, M., Van Gool, L., Williams, C. K., Winn, J., and Zisserman, A. (2010). The pascal visual object classes (voc) challenge. Int. J. Comput. Vis. 88, 303–338. doi: 10.1007/s11263-009-0275-4
Garcia-Ruiz, F., Sankaran, S., Maja, J. M., Lee, W. S., Rasmussen, J., and Ehsani R. (2013). Comparison of two aerial imaging platforms for identification of huanglongbing-infected citrus trees. Comput. Electron. Agric. 91, 106–115. doi: 10.1016/j.compag.2012.12.002
GSMA Intelligence (2016). The Mobile Economy- Africa 2016 . London: GSMA.
Harvey, C. A., Rakotobe, Z. L., Rao, N. S., Dave, R., Razafimahatratra, H., Rabarijohn, R. H., et al. (2014). Extreme vulnerability of smallholder farmers to agricultural risks and climate change in madagascar. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130089. doi: 10.1098/rstb.2013.008
He, K., Zhang, X., Ren, S., and Sun, J. (2015). Deep residual learning for image recognition. arXiv:1512.03385.
PubMed Abstract | Google Scholar
Hernández-Rabadán, D. L., Ramos-Quintana, F., and Guerrero Juk, J. (2014). Integrating soms and a bayesian classifier for segmenting diseased plants in uncontrolled environments. Sci. World J. 2014:214674. doi: 10.1155/2014/214674
Huang, K. Y. (2007). Application of artificial neural network for detecting phalaenopsis seedling diseases using color and texture features. Comput. Electron. Agric. 57, 3–11. doi: 10.1016/j.compag.2007.01.015
Hughes, D. P., and Salathé, M. (2015). An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv:1511.08060
ITU (2015). ICT Facts and Figures – the World in 2015. Geneva: International Telecommunication Union.
Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J., Girshick, R., et al. (2014). Caffe: Convolutional architecture for fast feature embedding. arXiv:1408.5093.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). “Imagenet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems , eds F. Pereira, C. J. C. Burges, L. Bottou, and K. Q. Weinberger (Curran Associates, Inc.), 1097–1105.
LeCun, Y., Boser, B., Denker, J. S., Henderson, D., Howard, R. E., Hubbard, W., et al. (1989). Backpropagation applied to handwritten zip code recognition. Neural Comput. 1, 541–551. doi: 10.1162/neco.1989.1.4.541
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539
Lin, M., Chen, Q., and Yan, S. (2013). Network in network. arXiv:1312.4400.
Lowe, D. G. (2004). Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60, 91–110. doi: 10.1023/B:VISI.0000029664.99615.94
Mokhtar, U., Ali, M. A., Hassanien, A. E., and Hefny, H. (2015). “Identifying two of tomatoes leaf viruses using support vector machine,” in Information Systems Design and Intelligent Applications , eds J. K. Mandal, S. C. Satapathy, M. K. Sanyal, P. P. Sarkar, A. Mukhopadhyay (Springer), 771–782.
Raza, S.-A., Prince, G., Clarkson, J. P., Rajpoot, N. M., et al. (2015). Automatic detection of diseased tomato plants using thermal and stereo visible light images. PLoS ONE 10:e0123262. doi: 10.1371/journal.pone.0123262. Available online at: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0123262
Report of the Plenary of the Intergovernmental Science-PolicyPlatform on Biodiversity Ecosystem Services on the work of its fourth session (2016). Plenary of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services Fourth session . Kuala Lumpur. Available online at: http://www.ipbes.net/sites/default/files/downloads/pdf/IPBES-4-4-19-Amended-Advance.pdf
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., et al. (2015). ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252. doi: 10.1007/s11263-015-0816-y
Sanchez, P. A., and Swaminathan, M. S. (2005). Cutting world hunger in half. Science 307, 357–359. doi: 10.1126/science.1109057
Sankaran, S., Mishra, A., Maja, J. M., and Ehsani, R. (2011). Visible-near infrared spectroscopy for detection of huanglongbing in citrus orchards. Comput. Electron. Agric. 77, 127–134. doi: 10.1016/j.compag.2011.03.004
Schmidhuber, J. (2015). Deep learning in neural networks: an overview. Neural Netw. 61, 85–117. doi: 10.1016/j.neunet.2014.09.003
Simonyan, K., and Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556.
Singh, A., Ganapathysubramanian, B., Singh, A. K., and Sarkar, S. (2015). Machine learning for highthroughput stress phenotyping in plants. Trends Plant Sci. 21, 110–124 doi: 10.1016/j.tplants.2015.10.015
PubMed Abstract | CrossRef Full Text
Strange, R. N., and Scott, P. R. (2005). Plant disease: a threat to global food security. Phytopathology 43, 83–116. doi: 10.1146/annurev.phyto.43.113004.133839
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., et al. (2015). “Going deeper with convolutions,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition .
Tai, A. P., Martin, M. V., and Heald, C. L. (2014). Threat to future global food security from climate change and ozone air pollution. Nat. Clim. Chang 4, 817–821. doi: 10.1038/nclimate2317
UNEP (2013). Smallholders, Food Security, and the Environment . Rome : International Fund for Agricultural Development (IFAD). Available online at: https://www.ifad.org/documents/10180/666cac2414b643c2876d9c2d1f01d5dd
Wetterich, C. B., Kumar, R., Sankaran, S., Junior, J. B., Ehsani, R., and Marcassa, L. G. (2012). A comparative study on application of computer vision and fluorescence imaging spectroscopy for detection of huanglongbing citrus disease in the usa and brazil. J. Spectrosc. 2013:841738. doi: 10.1155/2013/841738
Zeiler, M. D., and Fergus, R. (2014). “Visualizing and understanding convolutional networks,” in Computer Vision–ECCV 2014 , eds D. Fleet, T. Pajdla, B. Schiele, and T. Tuytelaars (Springer), 818–833.
Keywords: crop diseases, machine learning, deep learning, digital epidemiology
Citation: Mohanty SP, Hughes DP and Salathé M (2016) Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 7:1419. doi: 10.3389/fpls.2016.01419
Received: 19 June 2016; Accepted: 06 September 2016; Published: 22 September 2016.
Reviewed by:
Copyright © 2016 Mohanty, Hughes and Salathé. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcel Salathé, [email protected]
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
- Open access
- Published: 24 February 2021
Plant diseases and pests detection based on deep learning: a review
- Jun Liu ORCID: orcid.org/0000-0001-8769-5981 1 &
- Xuewei Wang 1
Plant Methods volume 17 , Article number: 22 ( 2021 ) Cite this article
127k Accesses
337 Citations
17 Altmetric
Metrics details
Plant diseases and pests are important factors determining the yield and quality of plants. Plant diseases and pests identification can be carried out by means of digital image processing. In recent years, deep learning has made breakthroughs in the field of digital image processing, far superior to traditional methods. How to use deep learning technology to study plant diseases and pests identification has become a research issue of great concern to researchers. This review provides a definition of plant diseases and pests detection problem, puts forward a comparison with traditional plant diseases and pests detection methods. According to the difference of network structure, this study outlines the research on plant diseases and pests detection based on deep learning in recent years from three aspects of classification network, detection network and segmentation network, and the advantages and disadvantages of each method are summarized. Common datasets are introduced, and the performance of existing studies is compared. On this basis, this study discusses possible challenges in practical applications of plant diseases and pests detection based on deep learning. In addition, possible solutions and research ideas are proposed for the challenges, and several suggestions are given. Finally, this study gives the analysis and prospect of the future trend of plant diseases and pests detection based on deep learning.
Plant diseases and pests detection is a very important research content in the field of machine vision. It is a technology that uses machine vision equipment to acquire images to judge whether there are diseases and pests in the collected plant images [ 1 ]. At present, machine vision-based plant diseases and pests detection equipment has been initially applied in agriculture and has replaced the traditional naked eye identification to some extent.
For traditional machine vision-based plant diseases and pests detection method, conventional image processing algorithms or manual design of features plus classifiers are often used [ 2 ]. This kind of method usually makes use of the different properties of plant diseases and pests to design the imaging scheme and chooses appropriate light source and shooting angle, which is helpful to obtain images with uniform illumination. Although carefully constructed imaging schemes can greatly reduce the difficulty of classical algorithm design, but also increase the application cost. At the same time, under natural environment, it is often unrealistic to expect the classical algorithms designed to completely eliminate the impact of scene changes on the recognition results [ 3 ]. In real complex natural environment, plant diseases and pests detection is faced with many challenges, such as small difference between the lesion area and the background, low contrast, large variations in the scale of the lesion area and various types, and a lot of noise in the lesion image. Also, there are a lot of disturbances when collecting plant diseases and pests images under natural light conditions. At this time, the traditional classical methods often appear helpless, and it is difficult to achieve better detection results.
In recent years, with the successful application of deep learning model represented by convolutional neural network (CNN) in many fields of computer vision (CV, computer-vision), for example, traffic detection [ 4 ], medical Image Recognition [ 5 ], Scenario text detection [ 6 ], expression recognition [ 7 ], face Recognition [ 8 ], etc. Several plant diseases and pests detection methods based on deep learning are applied in real agricultural practice, and some domestic and foreign companies have developed a variety of deep learning-based plant diseases and pests detection Wechat applet and photo recognition APP software. Therefore, plant diseases and pests detection method based on deep learning not only has important academic research value, but also has a very broad market application prospect.
In view of the lack of comprehensive and detailed discussion on plant diseases and pests detection methods based on deep learning, this study summarizes and combs the relevant literatures from 2014 to 2020, aiming to help researchers quickly and systematically understand the relevant methods and technologies in this field. The content of this study is arranged as follows: “ Definition of plant diseases and pests detection problem ” section gives the definition of plant diseases and pests detection problem; “ Image recognition technology based on deep learning ” section focuses on the detailed introduction of image recognition technology based on deep learning; “ Plant diseases and pests detection methods based on deep learning ” section analyses the three kinds of plant diseases and pests detection methods based on deep learning according to network structure, including classification, detection and segmentation network; “ Dataset and performance comparison ” section introduces some datasets of plant diseases and pests detection and compares the performance of the existing studies; “ Challenges ” section puts forward the challenges of plant diseases and pests detection based on deep learning; “ Conclusions and future directions ” section prospects the possible research focus and development direction in the future.
Definition of plant diseases and pests detection problem
Definition of plant diseases and pests
Plant diseases and pests is one kind of natural disasters that affect the normal growth of plants and even cause plant death during the whole growth process of plants from seed development to seedling and to seedling growth. In machine vision tasks, plant diseases and pests tend to be the concepts of human experience rather than a purely mathematical definition.
Definition of plant diseases and pests detection
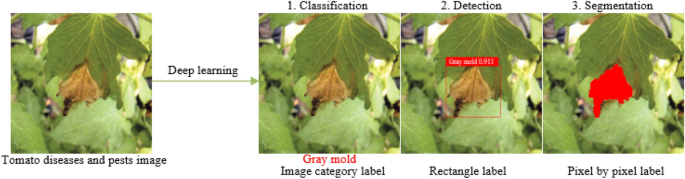
Compared with the definite classification, detection and segmentation tasks in computer vision [ 9 ], the requirements of plant diseases and pests detection is very general. In fact, its requirements can be divided into three different levels: what, where and how [ 10 ]. In the first stage, “what” corresponds to the classification task in computer vision. As shown in Fig. 1 , the label of the category to which it belongs is given. The task in this stage can be called classification and only gives the category information of the image. In the second stage, “where” corresponds to the location task in computer vision, and the positioning of this stage is the rigorous sense of detection. This stage not only acquires what types of diseases and pests exist in the image, but also gives their specific locations. As shown in Fig. 1 , the plaque area of gray mold is marked with a rectangular box. In the third stage, “how” corresponds to the segmentation task in computer vision. As shown in Fig. 1 , the lesions of gray mold are separated from the background pixel by pixel, and a series of information such as the length, area, location of the lesions of gray mold can be further obtained, which can assist the higher-level severity level evaluation of plant diseases and pests. Classification describes the image globally through feature expression, and then determines whether there is a certain kind of object in the image by means of classification operation; while object detection focuses on local description, that is, answering what object exists in what position in an image, so in addition to feature expression, object structure is the most obvious feature that object detection differs from object classification. That is, feature expression is the main research line of object classification, while structure learning is the research focus of object detection. Although the function requirements and objectives of the three stages of plant diseases and pests detection are different, yet in fact, the three stages are mutually inclusive and can be converted. For example, the “where” in the second stage contains the process of “what” in the first stage, and the “how” in the third stage can finish the task of “where” in the second stage. Also, the “what” in the first stage can achieve the goal of the second and the third stages through some methods. Therefore, the problem in this study is collectively referred to as plant diseases and pests detection as conventions in the following text, and the terminology differentiates only when different network structures and functions are adopted.
Comparison with traditional plant diseases and pests detection methods
To better illustrate the characteristics of plant diseases and pests detection methods based on deep learning, according to existing references [ 11 , 12 , 13 , 14 , 15 ], a comparison with traditional plant diseases and pests detection methods is given from four aspects including essence, method, required conditions and applicable scenarios. Detailed comparison results are shown in Table 1 .
Image recognition technology based on deep learning
Compared with other image recognition methods, the image recognition technology based on deep learning does not need to extract specific features, and only through iterative learning can find appropriate features, which can acquire global and contextual features of images, and has strong robustness and higher recognition accuracy.
Deep learning theory
The concept of Deep Learning (DL) originated from a paper published in Science by Hinton et al. [ 16 ] in 2006. The basic idea of deep learning is: using neural network for data analysis and feature learning, data features are extracted by multiple hidden layers, each hidden layer can be regarded as a perceptron, the perceptron is used to extract low-level features, and then combine low-level features to obtain abstract high-level features, which can significantly alleviate the problem of local minimum. Deep learning overcomes the disadvantage that traditional algorithms rely on artificially designed features and has attracted more and more researchers’ attention. It has now been successfully applied in computer vision, pattern recognition, speech recognition, natural language processing and recommendation systems [ 17 ].
Traditional image classification and recognition methods of manual design features can only extract the underlying features, and it is difficult to extract the deep and complex image feature information [ 18 ]. And deep learning method can solve this bottleneck. It can directly conduct unsupervised learning from the original image to obtain multi-level image feature information such as low-level features, intermediate features and high-level semantic features. Traditional plant diseases and pests detection algorithms mainly adopt the image recognition method of manual designed features, which is difficult and depends on experience and luck, and cannot automatically learn and extract features from the original image. On the contrary, deep learning can automatically learn features from large data without manual manipulation. The model is composed of multiple layers, which has good autonomous learning ability and feature expression ability, and can automatically extract image features for image classification and recognition. Therefore, deep learning can play a great role in the field of plant diseases and pests image recognition. At present, deep learning methods have developed many well-known deep neural network models, including deep belief network (DBN), deep Boltzmann machine (DBM), stack de-noising autoencoder (SDAE) and deep convolutional neural network (CNN) [ 19 ]. In the area of image recognition, the use of these deep neural network models to realize automate feature extraction from high-dimensional feature space offers significant advantages over traditional manual design feature extraction methods. In addition, as the number of training samples grows and the computational power increases, the characterization power of deep neural networks is being further improved. Nowadays, the boom of deep learning is sweeping both industry and academia, and the performance of deep neural network models are all significantly ahead of traditional models. In recent years, the most popular deep learning framework is deep convolutional neural network.
- Convolutional neural network
Convolutional Neural Networks, abbreviated as CNN, has a complex network structure and can perform convolution operations. As shown in Fig. 2 , the convolutional neural network model is composed of input layer, convolution layer, pooling layer, full connection layer and output layer. In one model, the convolution layer and the pooling layer alternate several times, and when the neurons of the convolution layer are connected to the neurons of the pooling layer, no full connection is required. CNN is a popular model in the field of deep learning. The reason lies in the huge model capacity and complex information brought about by the basic structural characteristics of CNN, which enables CNN to play an advantage in image recognition. At the same time, the successes of CNN in computer vision tasks have boosted the growing popularity of deep learning.
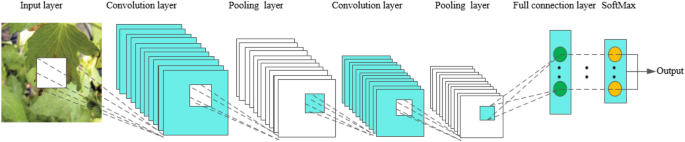
The basic structure of CNN
In the convolution layer, a convolution core is defined first. The convolution core can be considered as a local receptive field, and the local receptive field is the greatest advantage of the convolution neural network. When processing data information, the convolution core slides on the feature map to extract part of the feature information. After the feature extraction of the convolution layer, the neurons are input into the pooling layer to extract the feature again. At present, the commonly used methods of pooling include calculating the mean, maximum and random values of all values in the local receptive field [ 20 , 21 ]. After the data entering several convolution layers and pooling layers, they enter the full-connection layer, and the neurons in the full-connection layer are fully connected with the neurons in the upper layer. Finally, the data in the full-connection layer can be classified by the softmax method, and then the values are transmitted to the output layer for output results.
Open source tools for deep learning
The commonly used third-party open source tools for deep learning are Tensorflow [ 22 ], Torch/PyTorch [ 23 ], Caffe [ 24 ], Theano [ 25 ]. The different characteristics of each open source tool are shown in Table 2 .
The four commonly used deep learning third-party open source tools all support cross-platform operation, and the platforms that can be run include Linux, Windows, iOS, Android, etc. Torch/PyTorch and Tensorflow have good scalability and support a large number of third-party libraries and deep network structures, and have the fastest training speed when training large CNN networks on GPU.
Plant diseases and pests detection methods based on deep learning
This section gives a summary overview of plant diseases and pests detection methods based on deep learning. Since the goal achieved is completely consistent with the computer vision task, plant diseases and pests detection methods based on deep learning can be seen as an application of relevant classical networks in the field of agriculture. As shown in Fig. 3 , the network can be further subdivided into classification network, detection network and segmentation network according to the different network structures. As can be seen from Fig. 3 , this paper is subdivided into several different sub-methods according to the processing characteristics of each type of methods.
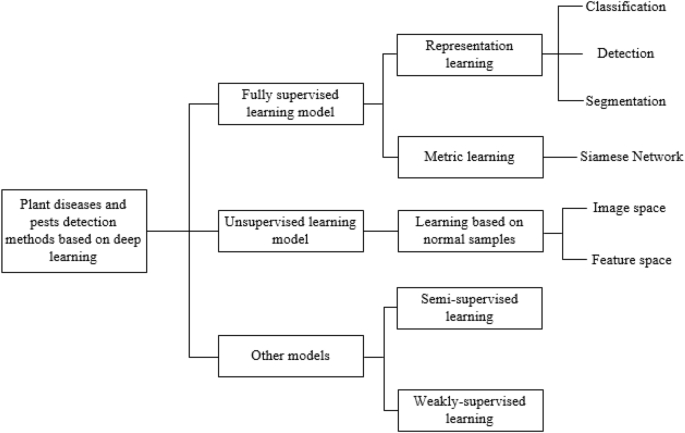
Framework of plant diseases and pests detection methods based on deep learning
Classification network
In real natural environment, the great differences in shape, size, texture, color, background, layout and imaging illumination of plant diseases and pests make the recognition a difficult task. Due to the strong feature extraction capability of CNN, the adoption of CNN-based classification network has become the most commonly used pattern in plant diseases and pests classification. Generally, the feature extraction part of CNN classification network consists of cascaded convolution layer + pooling layer, followed by full connection layer (or average pooling layer) + softmax structure for classification. Existing plant diseases and pests classification network mostly use the muture network structures in computer vision, including AlexNet [ 26 ], GoogleLeNet [ 27 ], VGGNet [ 28 ], ResNet [ 29 ], Inception V4 [ 30 ], DenseNets [ 31 ], MobileNet [ 32 ] and SqueezeNet [ 33 ]. There are also some studies which have designed network structures based on practical problems [ 34 , 35 , 36 , 37 ]. By inputting a test image into the classification network, the network analyses the input image and returns a label that classifies the image. According to the difference of tasks achieved by the classification network method, it can be subdivided into three subcategories: using the network as a feature extractor, using the network for classification directly and using the network for lesions location.
Using network as feature extractor
In the early studies on plant diseases and pests classification methods based on deep learning, many researchers took advantage of the powerful feature extraction capability of CNN, and the methods were combined with traditional classifiers [ 38 ]. First, the images are input into a pretrained CNN network to obtain image characterization features, and the acquired features are then input into a conventional machine learning classifier (e.g., SVM) for classification. Yalcin et al. [ 39 ] proposed a convolutional neural network architecture to extract the features of images while performing experiments using SVM classifiers with different kernels and feature descriptors such as LBP and GIST, the experimental results confirmed the effectiveness of the approach. Fuentes et al. [ 40 ] put forward the idea of CNN based meta architecture with different feature extractors, and the input images included healthy and infected plants, which were identified as their respective classes after going through the meta architecture. Hasan et al. [ 41 ] identified and classified nine different types of rice diseases by using the features extracted from DCNN model and input into SVM, and the accuracy achieved 97.5%.
Using network for classification directly
Directly using classification network to classify lesions is the earliest common means of CNN applied in plant diseases and pests detection. According to the characteristics of existing research work, it can be further subdivided into original image classification, classification after locating Region of Interest (ROI) and multi-category classification.
Original image classification. That is, directly put the collected complete plant diseases and pests image into the network for learning and training. Thenmozhi et al. [ 42 ] proposed an effective deep CNN model, and transfer learning is used to fine-tune the pre-training model. Insect species were classified on three public insect datasets with accuracy of 96.75%, 97.47% and 95.97%, respectively. Fang et al. [ 43 ] used ResNet50 in plant diseases and pests detection. The focus loss function was used instead of the standard cross-entropy loss function, and the Adam optimization method was used to identify the leaf disease grade, and the accuracy achieved 95.61%.
Classification after locating ROI. For the whole image acquired, we should focus on whether there is a lesion in a fixed area, so we often obtain the region of interest (ROI) in advance, and then input the ROI into the network to judge the category of diseases and pests. Nagasubramanian et al. [ 44 ] used a new three-dimensional deep convolution neural network (DCNN) and salience map visualization method to identify healthy and infected samples of soybean stem rot, and the classification accuracy achieved 95.73%.
Multi-category classification. When the number of plant diseases and pests class to be classified exceed 2 class, the conventional plant diseases and pests classification network is the same as the original image classification method, that is, the output nodes of the network are the number of plant diseases and pests class + 1 (including normal class). However, multi-category classification methods often use a basic network to classify lesions and normal samples, and then share feature extraction parts on the same network to modify or increase the classification branches of lesion categories. This approach is equivalent to preparing a pre-training weight parameter for subsequent multi-objective plant diseases and pests classification network, which is obtained by binary training between normal samples and plant diseases and pests samples. Picon et al. [ 45 ] proposed a CNN architecture to identify 17 diseases in 5 crops, which seamlessly integrates context metadata, allowing training of a single multi-crop model. The model can achieve the following goals: (a) obtains richer and more robust shared visual features than the corresponding single crop; (b) is not affected by different diseases in which different crops have similar symptoms; (c) seamlessly integrates context to perform crop conditional disease classification. Experiments show that the proposed model alleviates the problem of data imbalance, and the average balanced accuracy is 0.98, which is superior to other methods and eliminates 71% of classifier errors.
Using network for lesions location
Generally, the classification network can only complete the classification of image label level. In fact, it can also achieve the location of lesions and the pixel-by-pixel classification by combining different techniques and methods. According to the different means used, it can be further divided into three forms: sliding window, heatmap and multi-task learning network.
Sliding window. This is the simplest and intuitive method to achieve the location of lesion coarsely. The image in the sliding window is input into the classification network for plant diseases and pests detection by redundant sliding on the original image through a smaller size window. Finally, all sliding windows are connected to obtain the results of the location of lesion. Chen et al. [ 46 ] used CNN classification network based on sliding window to build a framework for characteristics automatic learning, feature fusion, recognition and location regression calculation of plant diseases and pests species, and the recognition rate of 38 common symptoms in the field was 50–90%.
Heatmap. This is an image that reflects the importance of each region in the image, the darker the color represents the more important. In the field of plant diseases and pests detection, the darker the color in the heatmap represents the greater the probability that it is the lesion. In 2017, Dechant et al. [ 47 ] trained CNN to make heatmap to show the probability of infection in each region in maize disease images, and these heatmaps were used to classify the complete images, dividing each image into containing or not containing infected leaves. At runtime, it takes about 2 min to generate a heatmap for an image (1.6 GB of memory) and less than one second to classify a set of three heatmaps (800 MB of memory). Experiments show that the accuracy is 96.7% on the test dataset. In 2019, Wiesner-Hanks et al. [ 48 ] used heatmap method to obtain accurate contour areas of maize diseases, the model can accurately depict lesions as low as millimeter scale from the images collected by UAVs, with an accuracy rate of 99.79%, which is the best scale of aerial plant disease detection achieved so far.
Multi-task learning network. If the pure classified network does not add any other skills, it could only realize the image level classification. Therefore, to accurately locate the location of plant diseases and pests, the designed network should often add an extra branch, and the two branches would share the results of the feature extracting. In this way, the network generally had the classification and segmentation output of the plant diseases and pests, forming a multi-task learning network. It takes into account the characteristics of both network. For segmentation network branches, each pixel in the image can be used as a training sample to train the network. Therefore, the multi-task learning network not only uses the segmentation branches to output the specific segmentation results of the lesions, but also greatly reduces the requirements of the classification network for samples. Ren et al. [ 49 ] constructed a Deconvolution-Guided VGNet (DGVGNet) model to identify plant leaf diseases which were easily disturbed by shadows, occlusions and light intensity. The deconvolution was used to guide the CNN classifier to focus on the real lesion sites. The test results show that the accuracy of disease class identification is 99.19%, the pixel accuracy of lesion segmentation is 94.66%, and the model has good robustness in occlusion, low light and other environments.
To sum up, the method based on classification network is widely used in practice, and many scholars have carried out application research on the classification of plant diseases and pests [ 50 , 51 , 52 , 53 ]. At the same time, different sub-methods have their own advantages and disadvantages, as shown in Table 3 .
Detection network
Object positioning is one of the most basic tasks in the field of computer vision. It is also the closest task to plant diseases and pests detections in the traditional sense. Its purpose is to obtain accurate location and category information of the object. At present, object detection methods based on deep learning emerge endlessly. Generally speaking, plant diseases and pests detection network based on deep learning can be divided into: two stage network represented by Faster R-CNN [ 54 ]; one stage network represented by SSD [ 55 ] and YOLO [ 56 , 57 , 58 ]. The main difference between the two networks is that the two-stage network needs to first generate a candidate box (proposal) that may contain the lesions, and then further execute the object detection process. In contrast, the one-stage network directly uses the features extracted in the network to predict the location and class of the lesions.
Plant diseases and pests detection based on two stages network
The basic process of two-stage detection network (Faster R-CNN) is to obtain the feature map of the input image through the backbone network first, then calculate the anchor box confidence using RPN and get the proposal. Then, input the feature map of the proposal area after ROIpooling to the network, fine-tune the initial detection results, and finally get the location and classification results of the lesions. Therefore, according to the characteristics of plant diseases and pests detection, common methods often improve on the backbone structure or its feature map, anchor ratio, ROIpooling and loss function. In 2017, Fuentes et al. [ 59 ] first used Faster R-CNN to locate tomato diseases and pests directly, combined with deep feature extractors such as VGG-Net and ResNet, the mAP value reached 85.98% in a dataset containing 5000 tomato diseases and pests of 9 categories. In 2019, Ozguven et al. [ 60 ] proposed a Faster R-CNN structure for automatic detection of beet leaf spot disease by changing the parameters of CNN model. 155 images were trained and tested. The results show that the overall correct classification rate of this method is 95.48%. Zhou et al. [ 61 ] presented a fast rice disease detection method based on the fusion of FCM-KM and Faster R-CNN. The application results of 3010 images showed that: the detection accuracy and time of rice blast, bacterial blight, and sheath blight are 96.71%/0.65 s, 97.53%/0.82 s and 98.26%/0.53 s respectively. Xie et al. [ 62 ] proposed a Faster DR-IACNN model based on the self-built grape leaf disease dataset (GLDD) and Faster R-CNN detection algorithm, the Inception-v1 module, Inception-ResNet-v2 module and SE are introduced. The proposed model achieved higher feature extraction ability, the mAP accuracy was 81.1% and the detection speed was 15.01FPS. The two-stage detection network has been devoted to improving the detection speed to improve the real-time and practicability of the detection system, but compared with the single-stage detection network, it is still not concise enough, and the inference speed is still not fast enough.
Plant diseases and pests detection based on one stage network
The one-stage object detection algorithm has eliminated the region proposal stage, but directly adds the detection head to the backbone network for classification and regression, thus greatly improving the inference speed of the detection network. The single-stage detection network is divided into two types, SSD and YOLO, both of which use the whole image as the input of the network, and directly return the position of the bounding box and the category to which it belongs at the output layer.
Compared with the traditional convolutional neural network, the SSD selects VGG16 as the trunk of the network, and adds a feature pyramid network to obtain features from different layers and make predictions. Singh et al. [ 63 ] built the PlantDoc dataset for plant disease detection. Considering that the application should predict in mobile CPU in real time, an application based on MobileNets and SSD was established to simplify the detection of model parameters. Sun et al. [ 64 ] presented an instance detection method of multi-scale feature fusion based on convolutional neural network, which is improved on the basis of SSD to detect maize leaf blight under complex background. The proposed method combined data preprocessing, feature fusion, feature sharing, disease detection and other steps. The mAP of the new model is higher (from 71.80 to 91.83%) than that of the original SSD model. The FPS of the new model has also improved (from 24 to 28.4), reaching the standard of real-time detection.
YOLO considers the detection task as a regression problem, and uses global information to directly predict the bounding box and category of the object to achieve end-to-end detection of a single CNN network. YOLO can achieve global optimization and greatly improve the detection speed while satisfying higher accuracy. Prakruti et al. [ 65 ] presented a method to detect pests and diseases on images captured under uncontrolled conditions in tea gardens. YOLOv3 was used to detect pests and diseases. While ensuring real-time availability of the system, about 86% mAP was achieved with 50% IOU. Zhang et al. [ 66 ] combined the pooling of spatial pyramids with the improved YOLOv3, deconvolution is implemented by using the combination of up-sampling and convolution operation, which enables the algorithm to effectively detect small size crop pest samples in the image and reduces the problem of relatively low recognition accuracy due to the diversity of crop pest attitudes and scales. The average recognition accuracy can reach 88.07% by testing 20 class of pests collected in real scene.
In addition, there are many studies on using detection network to identify diseases and pests [ 47 , 67 , 68 , 69 , 70 , 71 , 72 , 73 ]. With the development of object detection network in computer vision, it is believed that more and more new detection models will be applied in plant diseases and pests detection in the future. In summary, in the field of plant diseases and pests detection which emphasizes detection accuracy at this stage, more models based on two-stage are used, and in the field of plant diseases and pests detection which pursue detection speed more models based on one-stage are used.
Can detection network replace classification network? The task of detection network is to solve the location problem of plant diseases and pests. The task of classification network is to judge the class of plant diseases and pests. Visually, the hidden information of detection network includes the category information, that is, the category information of plant diseases and pests that need to be located needs to be known beforehand, and the corresponding annotation information should be given in advance to judge the location of plant diseases and pests. From this point of view, the detection network seems to include the steps of the classification network, that is, the detection network can answer “what kind of plant diseases and pests are in what place”. But there is a misconception, in which “what kind of plant diseases and pests” is given a priori, that is, what is labelled during training is not necessarily the real result. In the case of strong model differentiation, that is, when the detection network can give accurate results, the detection network can answer “what kind of plant diseases and pests are in what place” to a certain extent. However, in the real world, in many cases, it cannot uniquely reflect the uniqueness of plant diseases and pests categories, only can answer “what kind of plant diseases and pests may be in what place”, then the involvement of the classification network is necessary. Thus, the detection network cannot replace the classification network.
Segmentation network
Segmentation network converts the plant diseases and pests detection task to semantic and even instance segmentation of lesions and normal areas. It not only finely divides the lesion area, but also obtains the location, category and corresponding geometric properties (including length, width, area, outline, center, etc.). It can be roughly divided into: Fully Convolutional Networks (FCN) [ 74 ] and Mask R-CNN [ 75 ].
Full convolution neural network (FCN) is the basis of image semantics segmentation. At present, almost all semantics segmentation models are based on FCN. FCN first extracts and codes the features of the input image using convolution, then gradually restores the feature image to the size of the input image by deconvolution or up sampling. Based on the differences in FCN network structure, the plant diseases and pests segmentation methods can be divided into conventional FCN, U-net [ 76 ] and SegNet [ 77 ].
Conventional FCN. Wang et al. [ 78 ] presented a new method of maize leaf disease segmentation based on full convolution neural network to solve the problem that traditional computer vision is susceptible to different illumination and complex background, and the segmentation accuracy reached 96.26. Wang et al. [ 79 ] proposed a plant diseases and pests segmentation method based on improved FCN. In this method, a convolution layer was used to extract multi-layer feature information from the input maize leaf lesion image, and the size and resolution of the input image were restored by deconvolution operation. Compared with the original FCN method, not only the integrity of the lesion was guaranteed, but also the segmentation of small lesion area was highlighted, and the accuracy rate reached 95.87%.
U-net. U-net is not only a classical FCN structure, but also a typical encoder-decoder structure. It is characterized by introducing a layer-hopping connection, fusing the feature map in the coding stage with that in the decoding stage, which is beneficial to the recovery of segmentation details. Lin et al. [ 80 ] used U-net based convolutional neural network to segment 50 cucumber powdery mildew leaves collected in natural environment. Compared with the original U-net, a batch normalization layer was added behind each convolution layer, making the neural network insensitive to weight initialization. The experiment shows that the convolutional neural network based on U-net can accurately segment powdery mildew on cucumber leaves at the pixel level with an average pixel accuracy of 96.08%, which is superior to the existing K-means, Random-forest and GBDT methods. The U-net method can segment the lesion area in a complex background, and still has good segmentation accuracy and segmentation speed with fewer samples.
SegNet. It is also a classical encoder–decoder structure. Its feature is that the up-sampling operation in the decoder takes advantage of the index of the largest pooling operation in the encoder. Kerkech et al. [ 81 ] presented an image segmentation method for unmanned aerial vehicles. Visible and infrared images (480 samples from each range) were segmented using SegNet to identify four categories: shadows, ground, healthy and symptomatic grape vines. The detection rates of the proposed method on grape vines and leaves were 92% and 87%, respectively.
Mask R-CNN is one of the most commonly used image instance segmentation methods at present. It can be considered as a multitask learning method based on detection and segmentation network. When multiple lesions of the same type have adhesion or overlap, instance segmentation can separate individual lesions and further count the number of lesions. However, semantic segmentation often treats multiple lesions of the same type as a whole. Stewart et al. [ 82 ] trained a Mask R-CNN model to segment maize northern leaf blight (NLB) lesions in an unmanned aerial vehicle image. The trained model can accurately detect and segment a single lesion. At the IOU threshold of 0.50, the IOU between the baseline true value and the predicted lesion was 0.73, and the average accuracy was 0.96. Also, some studies combine the Mask R-CNN framework with object detection networks for plant diseases and pests detection. Wang et al. [ 83 ] used two different models, Faster R-CNN and ask R-CNN, in which Faster R-CNN was used to identify the class of tomato diseases and Mask R-CNN was used to detect and segment the location and shape of the infected area. The results showed that the proposed model can quickly and accurately identify 11 class of tomato diseases, and divide the location and shape of infected areas. Mask R-CNN reached a high detection rate of 99.64% for all class of tomato diseases.
Compared with the classification and detection network methods, the segmentation method has advantages in obtaining the lesion information. However, like the detection network, it requires a lot of annotation data, and its annotation information is pixel by pixel, which often takes a lot of effort and cost.
Dataset and performance comparison
This section first gives a brief introduction to the plant diseases and pests related datasets and the evaluation index of deep learning model, then compares and analyses the related models of plant diseases and pests detection based on deep learning in recent years.
Datasets for plant diseases and pests detection
Plant diseases and pests detection datasets are the basis for research work. Compared with ImageNet, PASCAL-VOC2007/2012 and COCO in computer vision tasks, there is not a large and unified dataset for plant diseases and pests detection. The plant diseases and pests dataset can be acquired by self-collection, network collection and use of public datasets. Among them, self-collection of image dataset is often obtained by unmanned aerial remote sensing, ground camera photography, Internet of Things monitoring video or video recording, aerial photography of unmanned aerial vehicle with camera, hyperspectral imager, near-infrared spectrometer, and so on. Public datasets typically come from PlantVillage, an existing well-known public standard library. Relatively, self-collected datasets of plant diseases and pests in real natural environment are more practical. Although more and more researchers have opened up the images collected in the field, it is difficult to compare them uniformly based on different class of diseases under different detection objects and scenarios. This section provides links to a variety of plant diseases and pests detection datasets in conjunction with existing studies. As shown in Table 4 .
Evaluation indices
Evaluation indices can vary depending on the focus of the study. Common evaluation indices include \(Precision\) , \(Recall\) , mean Average Precision (mAP) and the harmonic Mean F1 score based on \(Precision\) and \(Recall\) .
\(Precision\) and \(Recall\) are defined as:
In Formula ( 1 ) and Formula ( 2 ), TP (True Positive) is true-positive, predicted to be 1 and actually 1, indicating the number of lesions correctly identified by the algorithm. FP (False Positive) is false-positive, predicted to be 1 and actually 0, indicating the number of lesions incorrectly identified by the algorithm. FN (False Negative) is false-negative, predicted to be 0 and actually 1, indicating the number of unrecognized lesions.
Detection accuracy is usually assessed using mAP. The average accuracy of each category in the dataset needs to be calculated first:
In the above-mentioned formula, \(N\left( {class} \right)\) represents the number of all categories, \(Precision\left( j \right)\) and \(Recall\left( j \right)\) represents the precision and recall of class j respectively.
Average accuracy for each category is defined as mAP:
The greater the value of \(mAP\) , the higher the recognition accuracy of the algorithm; conversely, the lower the accuracy of the algorithm.
F1 score is also introduced to measure the accuracy of the model. F1 score takes into account both the accuracy and recall of the model. The formula is
Frames per second (FPS) is used to evaluate the recognition speed. The more frames per second, the faster the algorithm recognition speed; conversely, the slower the algorithm recognition speed.
Performance comparison of existing algorithms
At present, the research on plant diseases and pests based on deep learning involves a wide range of crops, including all kinds of vegetables, fruits and food crops. The tasks completed include not only the basic tasks of classification, detection and segmentation, but also more complex tasks such as the judgment of infection degree.
At present, most of the current deep learning-based methods for plant diseases and pests detection are applied on specific datasets, many datasets are not publicly available, there is still no single publicly available and comprehensive dataset that will allow all algorithms to be uniformly compared. With the continuous development of deep learning, the application performance of some typical algorithms on different datasets has been gradually improved, and the mAP, F1 score and FPS of the algorithms have all been increased.
The breakthroughs achieved in the existing studies are amazing, but due to the fact that there is still a certain gap between the complexity of the infectious diseases and pests images in the existing studies and the real-time field diseases and pests detection based on mobile devices. Subsequent studies will need to find breakthroughs in larger, more complex, and more realistic datasets.
Small dataset size problem
At present, deep learning methods are widely used in various computer vision tasks, plant diseases and pests detection is generally regarded as specific application in the field of agriculture. There are too few agricultural plant diseases and pests samples available. Compared with open standard libraries, self-collected data sets are small in size and laborious in labeling data. Compared with more than 14 million sample data in ImageNet datasets, the most critical problem facing plant diseases and pests detection is the problem of small samples. In practice, some plant diseases have low incidence and high cost of disease image acquisition, resulting in only a few or dozen training data collected, which limits the application of deep learning methods in the field of plant diseases and pests identification. In fact, for the problem of small samples, there are currently three different solutions.
Data amplification, synthesis and generation
Data amplification is a key component of training deep learning models. An optimized data amplification strategy can effectively improve the plant diseases and pests detection effect. The most common method of plant diseases and pests image expansion is to acquire more samples using image processing operations such as mirroring, rotating, shifting, warping, filtering, contrast adjustment, and so on for the original plant diseases and pests samples. In addition, Generative Adversarial Networks (GANs) [ 93 ] and Variational automatic encoder (VAE) [ 94 ] can generate more diverse samples to enrich limited datasets.
Transfer learning and fine-tuning classical network model
Transfer learning (TL) transfers knowledge learned from generic large datasets to specialized areas with relatively small amounts of data. When transfer learning develops a model for newly collected unlabeled samples, it can start with a training model by a similar known dataset. After fine-tuning parameters or modifying components, it can be applied to localized plant disease and pest detection, which can reduce the cost of model training and enable the convolution neural network to adapt to small sample data. Oppenheim et al. [ 95 ] collected infected potato images of different sizes, hues and shapes under natural light and classified by fine-tuning the VGG network. The results showed that, the transfer learning and training of new networks were effective. Too et al. [ 96 ] evaluated various classical networks by fine-tuning and contrast. The experimental results showed that the accuracy of Dense-Nets improved with the number of iterations. Chen et al. [ 97 ] used transfer learning and fine-tuning to identify rice disease images under complex background conditions and achieved an average accuracy of 92.00%, which proves that the performance of transfer learning is better than training from scratch.
Reasonable network structure design
By designing a reasonable network structure, the sample requirements can be greatly reduced. Zhang et al. [ 98 ] constructed a three-channel convolution neural network model for plant leaf disease recognition by combining three color components. Each channel TCCNN component is composed of three color RGB leaf disease images. Liu et al. [ 99 ] presented an improved CNN method for identifying grape leaf diseases. The model used a depth-separable convolution instead of a standard convolution to alleviate overfitting and reduce the number of parameters. For the different size of grape leaf lesions, the initial structure was applied to the model to improve the ability of multi-scale feature extraction. Compared with the standard ResNet and GoogLeNet structures, this model has faster convergence speed and higher accuracy during training. The recognition accuracy of this algorithm was 97.22%.
Fine-grained identification of small-size lesions in early identification
Small-size lesions in early identification.
Accurate early detection of plant diseases is essential to maximize the yield [ 36 ]. In the actual early identification of plant diseases and pests, due to the small size of the lesion object itself, multiple down sampling processes in the deep feature extraction network tend to cause small-scale objects to be ignored. Moreover, due to the background noise problem on the collected images, large-scale complex background may lead to more false detection, especially on low-resolution images. In view of the shortage of existing algorithms, the improvement direction of small object detection algorithm is analyzed, and several strategies such as attention mechanism are proposed to improve the performance of small target detection.
The use of attention mechanism makes resources allocated more rationally. The essence of attention mechanism is to quickly find region of interest and ignore unimportant information. By learning the characteristics of plant diseases and pests images, features can be separated using weighted sum method with weighted coefficient, and the background noise in the image can be suppressed. Specifically, the attention mechanism module can get a salient image, and seclude the object from the background, and the Softmax function can be used to manipulate the feature image, and combine it with the original feature image to obtain new fusion features for noise reduction purposes. In future studies on early recognition of plant diseases and pests, attention mechanisms can be used to effectively select information and allocate more resources to region of interest to achieve more accurate detection. Karthik et al. [ 100 ] applied attention mechanism on the residual network and experiments were carried out using the plantVillage dataset, which achieved 98% overall accuracy.
Fine-grained identification
First, there is a large difference within the class, that is, the visual characteristics of plant diseases and pests belonging to the same class are quite different. The reason is that the aforementioned external factors such as uneven illumination, dense occlusion, blurred equipment dithering and other interferences, resulting in different image samples belonging to the same kind of diseases and pests differ greatly. Plant diseases and pests detection in complex scenarios is a very challenging task of fine-grained recognition [ 101 ]. The existence of growth variations of diseases and pests results in distinct differences in the characterization of the same diseases and pests at different stages, forming the “intra-class difference” fine-grained characteristics.
Secondly, there is fuzziness between classes, that is, objects of different classes have some similarity. There are many detailed classifications of biological subspecies and subclasses of different kinds of diseases and pests, and there are some similarities of biological morphology and life habits among the subclasses, which lead to the problem of fine-grained identification of “inter-class similarity”. Barbedo believed that similar symptoms could be produced, which even phytopathologists could not correctly distinguish [ 102 ].
Thirdly, background disturbance makes it impossible for plant diseases and pests to appear in a very clean background in the real world. Background can be very complex and interfere with objects of interest, which makes plant diseases and pests detection more difficult. Some literature often ignores this issue because images are captured under controlled conditions [ 103 ].
Relying on the existing deep learning methods can not effectively identify the fine-grained characteristics of diseases and pests that exist naturally in the application of the above actual agricultural scenarios, resulting in technical difficulties such as low identification accuracy and generalization robustness, which has long restricted the performance improvement of decision-making management of diseases and pests by the Intelligent Agricultural Internet of Things [ 104 ]. The existing research is only suitable for fine-grained identification of fewer class of diseases and pests, can not solve the problem of large-scale, large-category, accurate and efficient identification of diseases and pests, and is difficult to deploy directly to the mobile terminals of smart agriculture.
Detection performance under the influence of illumination and occlusion
Lighting problems.
Previous studies have collected images of plant diseases and pests mostly in indoor light boxes [ 105 ]. Although this method can effectively eliminate the influence of external light to simplify image processing, it is quite different from the images collected under real natural light. Because natural light changes very dynamically, and the range in which the camera can accept dynamic light sources is limited, it is easy to cause image color distortion when above or below this limit. In addition, due to the difference of view angle and distance during image collection, the apparent characteristics of plant diseases and pests change greatly, which brings great difficulties to the visual recognition algorithm.
Occlusion problem
At present, most researchers intentionally avoid the recognition of plant diseases and pests in complex environments. They only focus on a single background. They use the method of directly intercepting the area of interest to the collected images, but seldom consider the occlusion problem. As a result, the recognition accuracy under occlusion is low and the practicability is greatly reduced. Occlusion problems are common in real natural environments, including blade occlusion caused by changes in blade posture, branch occlusion, light occlusion caused by external lighting, and mixed occlusion caused by different types of occlusion. The difficulties of plant diseases and pests identification under occlusion are the lack of features and noise overlap caused by occlusion. Different occlusion conditions have different degrees of impact on the recognition algorithm, resulting in false detection or even missed detection. In recent years, with the maturity of deep learning algorithms under restricted conditions, some researchers have gradually challenged the identification of plant diseases and pests under occluded conditions [ 106 , 107 ], and significant progress has been made, which lays a good foundation for the application of plant diseases and pests identification in real-world scenarios. However, occlusion is random and complex. The training of the basic framework is difficult and the dependence on the performance of hardware devices still exists, we should strengthen the innovation and optimization of the basic framework, including the design of lightweight network architecture. The exploration of GAN and other aspects should be enhanced, while ensuring the accuracy of detection, the difficulty of model training should be reduced. GAN has prominent advantages in dealing with posture changes and chaotic background, but its design is not yet mature, and it is easy to crash in learning and cause model uncontrollable problems during training. We should strengthen the exploration of network performance to make it easier to quantify the quality of the model.
Detection speed problem
Compared with traditional methods, deep learning algorithms have better results, but their computational complexity is also higher. If the detection accuracy is guaranteed, the model needs to fully learn the characteristics of the image and increase the computational load, which will inevitably lead to slow detection speed and can not meet the needs of real-time. In order to ensure the detection speed, it is usually necessary to reduce the amount of calculation. However, this will cause insufficient training and result in false or missed detection. Therefore, it is important to design an efficient algorithm with both detection accuracy and detection speed.
Plant diseases and pests detection methods based on deep learning include three main links in agricultural applications: data labeling, model training and model inference. In real-time agricultural applications, more attention is paid to model inference. Currently, most plant diseases and pests detection methods focus on the accuracy of recognition. Little attention is paid to the efficiency of model inference. In reference [ 108 ], to improve the efficiency of the model calculation process to meet the actual agricultural needs, a deep separable convolution structure model for plant leaf disease detection was introduced. Several models were trained and tested. The classification accuracy of Reduced MobileNet was 98.34%, the parameters were 29 times less than VGG, and 6 times less than MobileNet. This shows an effective compromise between delay and accuracy, which is suitable for real-time crop diseases diagnosis on resource-constrained mobile devices.
Conclusions and future directions
Compared with traditional image processing methods, which deal with plant diseases and pests detection tasks in several steps and links, plant diseases and pests detection methods based on deep learning unify them into end-to-end feature extraction, which has a broad development prospects and great potential. Although plant diseases and pests detection technology is developing rapidly, it has been moving from academic research to agricultural application, there is still a certain distance from the mature application in the real natural environment, and there are still some problems to be solved.
Plant diseases and pests detection dataset
Deep learning technology has made some achievements in the identification of plant diseases and pests. Various image recognition algorithms have also been further developed and extended, which provides a theoretical basis for the identification of specific diseases and pests. However, the collection of image samples in previous studies mostly come from the characterization of disease spots, insect appearance characteristics or the characterization of insect pests and leaves. Most of the research results are limited to the laboratory environment and are applicable only to the plant diseases and pests images obtained at the time. The main reason for this is that the growth of plants is cyclical, continuous, seasonal and regional. Similarly, the characteristics of the same disease or pest at different growing stages of crops are different. Images of different plant species vary from region to region. As a result, most of the existing research results are not universal. Even with a high recognition rate in a single trial, the validity of the data obtained at other times cannot be guaranteed.
Most of the existing studies are based on the images generated in the visible range, but the electromagnetic wave outside the visible range also contains a lot of information, so the comprehensive information such as visible light, near infrared, multi-spectral should be fused to achieve the acquisition of plant diseases and pests dataset. Future research should focus on multi-information fusion method to obtain and identify plant diseases and pests information.
In addition, image databases of different kinds of plant diseases and pests in real natural environments are still in the blank stage. Future research should make full use of the data information acquisition platform such as portable field spore auto-capture instrument, unmanned aerial vehicle aerial photography system, agricultural internet of things monitoring equipment, which performs large-area and coverage identification of farmland and makes up for the lack of randomness of image samples in previous studies. Also, it can ensures the comprehensiveness and accuracy of dataset, and improves the generality of the algorithm.
Early recognition of plant diseases and pests
In the application of plant diseases and pests identification, the manifestation symptoms are not obvious, so early diagnosis is very difficult whether it is by visual observation or computer interpretation. However, the research significance and demand of early diagnosis are greater, which is more conducive to the prevention and control of plant diseases and pests and prevent their spread and development. The best image quality can be obtained when the sunlight is sufficient, and taking pictures in cloudy weather will increase the complexity of image preprocessing and reduce the recognition effect. In addition, in the early stage of plant diseases and pests occurrence, even high-resolution images are difficult to analyze. It is necessary to combine meteorological and plant protection data such as temperature and humidity to realize the recognition and prediction of diseases and pests. By consulting the existing research literatures, there are few reports on the early diagnosis of plant diseases and pests.
Network training and learning
When plant diseases and pests are visually identified manually, it is difficult to collect samples of all plant diseases and pests types, and many times only healthy data (positive samples) are available. However, most of the current plant diseases and pests detection methods based on deep learning are supervised learning based on a large number of diseases and pests samples, so manual collection of labelled datasets requires a lot of manpower, so unsupervised learning needs to be explored. Deep learning is a black box, which requires a large number of labelled training samples for end-to-end learning and has poor interpretability. Therefore, how to use the prior knowledge of brain-inspired computing and human-like visual cognitive model to guide the training and learning of the network is also a direction worthy of studying. At the same time, deep models need a large amount of memory and are extremely time-consuming during testing, which makes them unsuitable for deployment on mobile platforms with limited resources. It is important to study how to reduce complexity and obtain fast-executing models without losing accuracy. Finally, the selection of appropriate hyper-parameters has always been a major obstacle to the application of deep learning model to new tasks, such as learning rate, filter size, step size and number, these hyper-parameters have a strong internal dependence, any small adjustment may have a greater impact on the final training results.
Interdisciplinary research
Only by more closely integrating empirical data with theories such as agronomic plant protection, can we establish a field diagnosis model that is more in line with the rules of crop growth, and will further improve the effectiveness and accuracy of plant diseases and pests identification. In the future, it is necessary to go from image analysis at the surface level to identification of the occurrence mechanism of diseases and pests, and transition from simple experimental environment to practical application research that comprehensively considers crop growth law, environmental factors, etc.
In summary, with the development of artificial intelligence technology, the research focus of plant diseases and pests detection based on machine vision has shifted from classical image processing and machine learning methods to deep learning methods, which solved the difficult problems that could not be solved by traditional methods. There is still a long distance from the popularization of practical production and application, but this technology has great development potential and application value. To fully explore the potential of this technology, the joint efforts of experts from relevant disciplines are needed to effectively integrate the experience knowledge of agriculture and plant protection with deep learning algorithms and models, so as to make plant diseases and pests detection based on deep learning mature. Also, the research results should be integrated into agricultural machinery equipment to truly land the corresponding theoretical results.
Availability of data and materials
For relevant data and codes, please contact the corresponding author of this manuscript.
Lee SH, Chan CS, Mayo SJ, Remagnino P. How deep learning extracts and learns leaf features for plant classification. Pattern Recogn. 2017;71:1–13.
Article Google Scholar
Tsaftaris SA, Minervini M, Scharr H. Machine learning for plant phenotyping needs image processing. Trends Plant Sci. 2016;21(12):989–91.
Article CAS PubMed Google Scholar
Fuentes A, Yoon S, Park DS. Deep learning-based techniques for plant diseases recognition in real-field scenarios. In: Advanced concepts for intelligent vision systems. Cham: Springer; 2020.
Google Scholar
Yang D, Li S, Peng Z, Wang P, Wang J, Yang H. MF-CNN: traffic flow prediction using convolutional neural network and multi-features fusion. IEICE Trans Inf Syst. 2019;102(8):1526–36.
Sundararajan SK, Sankaragomathi B, Priya DS. Deep belief cnn feature representation based content based image retrieval for medical images. J Med Syst. 2019;43(6):1–9.
Melnyk P, You Z, Li K. A high-performance CNN method for offline handwritten chinese character recognition and visualization. Soft Comput. 2019;24:7977–87.
Li J, Mi Y, Li G, Ju Z. CNN-based facial expression recognition from annotated rgb-d images for human–robot interaction. Int J Humanoid Robot. 2019;16(04):504–5.
Kumar S, Singh SK. Occluded thermal face recognition using bag of CNN(BoCNN). IEEE Signal Process Lett. 2020;27:975–9.
Wang X. Deep learning in object recognition, detection, and segmentation. Found Trends Signal Process. 2016;8(4):217–382.
Article CAS Google Scholar
Boulent J, Foucher S, Théau J, St-Charles PL. Convolutional neural networks for the automatic identification of plant diseases. Front Plant Sci. 2019;10:941.
Article PubMed PubMed Central Google Scholar
Kumar S, Kaur R. Plant disease detection using image processing—a review. Int J Comput Appl. 2015;124(2):6–9.
Martineau M, Conte D, Raveaux R, Arnault I, Munier D, Venturini G. A survey on image-based insect classification. Pattern Recogn. 2016;65:273–84.
Jayme GAB, Luciano VK, Bernardo HV, Rodrigo VC, Katia LN, Claudia VG, et al. Annotated plant pathology databases for image-based detection and recognition of diseases. IEEE Latin Am Trans. 2018;16(6):1749–57.
Kaur S, Pandey S, Goel S. Plants disease identification and classification through leaf images: a survey. Arch Comput Methods Eng. 2018;26(4):1–24.
CAS Google Scholar
Shekhawat RS, Sinha A. Review of image processing approaches for detecting plant diseases. IET Image Process. 2020;14(8):1427–39.
Hinton GE, Salakhutdinov R. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7.
Liu W, Wang Z, Liu X, et al. A survey of deep neural network architectures and their applications. Neurocomputing. 2017;234:11–26.
Fergus R. Deep learning methods for vision. CVPR 2012 Tutorial; 2012.
Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35(8):1798–828.
Article PubMed Google Scholar
Boureau YL, Le Roux N, Bach F, Ponce J, Lecun Y. [IEEE 2011 IEEE international conference on computer vision (ICCV)—Barcelona, Spain (2011.11.6–2011.11.13)] 2011 international conference on computer vision—ask the locals: multi-way local pooling for image recognition; 2011. p. 2651–8.
Zeiler MD, Fergus R. Stochastic pooling for regularization of deep convolutional neural networks. Eprint Arxiv. arXiv:1301.3557 . 2013.
TensorFlow. https://www.tensorflow.org/ .
Torch/PyTorch. https://pytorch.org/ .
Caffe. http://caffe.berkeleyvision.org/ .
Theano. http://deeplearning.net/software/theano/ .
Krizhenvshky A, Sutskever I, Hinton G. Imagenet classification with deep convolutional networks. In: Proceedings of the conference neural information processing systems (NIPS), Lake Tahoe, NV, USA, 3–8 December; 2012. p. 1097–105.
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions. In: Proceedings of the 2015 IEEE conference on computer vision and pattern recognition, Boston, MA, USA, 7–12 June; 2015. p. 1–9.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. arXiv:1409.1556 . 2014.
Xie S, Girshick R, Dollár P, Tu Z, He K. Aggregated residual transformations for deep neural networks. arXiv. arXiv:1611.05431 . 2017.
Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence. 2016.
Huang G, Lrj Z, Maaten LVD, et al. Densely connected convolutional networks. In: IEEE conference on computer vision and pattern recognition. 2017. p. 2261–9.
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H. MobileNets: efficient convolutional neural networks for mobile vision applications. arXiv. arXiv:1704.04861 . 2017.
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50 × fewer parameters and < 0.5 MB model size. arXiv. arXiv:1602.07360 . 2016.
Priyadharshini RA, Arivazhagan S, Arun M, Mirnalini A. Maize leaf disease classification using deep convolutional neural networks. Neural Comput Appl. 2019;31(12):8887–95.
Wen J, Shi Y, Zhou X, Xue Y. Crop disease classification on inadequate low-resolution target images. Sensors. 2020;20(16):4601.
Article PubMed Central Google Scholar
Thangaraj R, Anandamurugan S, Kaliappan VK. Automated tomato leaf disease classification using transfer learning-based deep convolution neural network. J Plant Dis Prot. 2020. https://doi.org/10.1007/s41348-020-00403-0 .
Atila M, Uar M, Akyol K, Uar E. Plant leaf disease classification using efficientnet deep learning model. Ecol Inform. 2021;61:101182.
Sabrol H, Kumar S. Recent studies of image and soft computing techniques for plant disease recognition and classification. Int J Comput Appl. 2015;126(1):44–55.
Yalcin H, Razavi S. Plant classification using convolutional neural networks. In: 2016 5th international conference on agro-geoinformatics (agro-geoinformatics). New York: IEEE; 2016.
Fuentes A, Lee J, Lee Y, Yoon S, Park DS. Anomaly detection of plant diseases and insects using convolutional neural networks. In: ELSEVIER conference ISEM 2017—The International Society for Ecological Modelling Global Conference, 2017. 2017.
Hasan MJ, Mahbub S, Alom MS, Nasim MA. Rice disease identification and classification by integrating support vector machine with deep convolutional neural network. In: 2019 1st international conference on advances in science, engineering and robotics technology (ICASERT). 2019.
Thenmozhi K, Reddy US. Crop pest classification based on deep convolutional neural network and transfer learning. Comput Electron Agric. 2019;164:104906.
Fang T, Chen P, Zhang J, Wang B. Crop leaf disease grade identification based on an improved convolutional neural network. J Electron Imaging. 2020;29(1):1.
Nagasubramanian K, Jones S, Singh AK, Sarkar S, Singh A, Ganapathysubramanian B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods. 2019;15(1):1–10.
Picon A, Seitz M, Alvarez-Gila A, Mohnke P, Echazarra J. Crop conditional convolutional neural networks for massive multi-crop plant disease classification over cell phone acquired images taken on real field conditions. Comput Electron Agric. 2019;167:105093.
Tianjiao C, Wei D, Juan Z, Chengjun X, Rujing W, Wancai L, et al. Intelligent identification system of disease and insect pests based on deep learning. China Plant Prot Guide. 2019;039(004):26–34.
Dechant C, Wiesner-Hanks T, Chen S, Stewart EL, Yosinski J, Gore MA, et al. Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology. 2017;107:1426–32.
Wiesner-Hanks T, Wu H, Stewart E, Dechant C, Nelson RJ. Millimeter-level plant disease detection from aerial photographs via deep learning and crowdsourced data. Front Plant Sci. 2019;10:1550.
Shougang R, Fuwei J, Xingjian G, Peishen Y, Wei X, Huanliang X. Deconvolution-guided tomato leaf disease identification and lesion segmentation model. J Agric Eng. 2020;36(12):186–95.
Fujita E, Kawasaki Y, Uga H, Kagiwada S, Iyatomi H. Basic investigation on a robust and practical plant diagnostic system. In: IEEE international conference on machine learning & applications. New York: IEEE; 2016.
Mohanty SP, Hughes DP, Salathé M. Using deep learning for image-based plant disease detection. Front Plant Sci. 2016;7:1419. https://doi.org/10.3389/fpls.2016.01419 .
Brahimi M, Arsenovic M, Laraba S, Sladojevic S, Boukhalfa K, Moussaoui A. Deep learning for plant diseases: detection and saliency map visualisation. In: Zhou J, Chen F, editors. Human and machine learning. Cham: Springer International Publishing; 2018. p. 93–117.
Chapter Google Scholar
Barbedo JG. Plant disease identification from individual lesions and spots using deep learning. Biosyst Eng. 2019;180:96–107.
Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39(6):1137–49.
Liu W, Anguelov D, Erhan D, Szegedy C, Berg AC. SSD: Single shot MultiBox detector. In: European conference on computer vision. Cham: Springer International Publishing; 2016.
Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: unified, real-time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015.
Redmon J, Farhadi A. Yolo9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. p. 6517–25.
Redmon J, Farhadi A. Yolov3: an incremental improvement. arXiv preprint. arXiv:1804.02767 . 2018.
Fuentes A, Yoon S, Kim SC, Park DS. A robust deep-learning-based detector for real-time tomato plant diseases and pests detection. Sensors. 2017;17(9):2022.
Ozguven MM, Adem K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Phys A Statal Mech Appl. 2019;535(2019):122537.
Zhou G, Zhang W, Chen A, He M, Ma X. Rapid detection of rice disease based on FCM-KM and faster R-CNN fusion. IEEE Access. 2019;7:143190–206. https://doi.org/10.1109/ACCESS.2019.2943454 .
Xie X, Ma Y, Liu B, He J, Wang H. A deep-learning-based real-time detector for grape leaf diseases using improved convolutional neural networks. Front Plant Sci. 2020;11:751.
Singh D, Jain N, Jain P, Kayal P, Kumawat S, Batra N. Plantdoc: a dataset for visual plant disease detection. In: Proceedings of the 7th ACM IKDD CoDS and 25th COMAD. 2019.
Sun J, Yang Y, He X, Wu X. Northern maize leaf blight detection under complex field environment based on deep learning. IEEE Access. 2020;8:33679–88. https://doi.org/10.1109/ACCESS.2020.2973658 .
Bhatt PV, Sarangi S, Pappula S. Detection of diseases and pests on images captured in uncontrolled conditions from tea plantations. In: Proc. SPIE 11008, autonomous air and ground sensing systems for agricultural optimization and phenotyping IV; 2019. p. 1100808. https://doi.org/10.1117/12.2518868 .
Zhang B, Zhang M, Chen Y. Crop pest identification based on spatial pyramid pooling and deep convolution neural network. Trans Chin Soc Agric Eng. 2019;35(19):209–15.
Ramcharan A, McCloskey P, Baranowski K, Mbilinyi N, Mrisho L, Ndalahwa M, Legg J, Hughes D. A mobile-based deep learning model for cassava disease diagnosis. Front Plant Sci. 2019;10:272. https://doi.org/10.3389/fpls.2019.00272 .
Selvaraj G, Vergara A, Ruiz H, Safari N, Elayabalan S, Ocimati W, Blomme G. AI-powered banana diseases and pest detection. Plant Methods. 2019. https://doi.org/10.1186/s13007-019-0475-z .
Tian Y, Yang G, Wang Z, Li E, Liang Z. Detection of apple lesions in orchards based on deep learning methods of CycleGAN and YOLOV3-dense. J Sens. 2019. https://doi.org/10.1155/2019/7630926 .
Zheng Y, Kong J, Jin X, Wang X, Zuo M. CropDeep: the crop vision dataset for deep-learning-based classification and detection in precision agriculture. Sensors. 2019;19:1058. https://doi.org/10.3390/s19051058 .
Arsenovic M, Karanovic M, Sladojevic S, Anderla A, Stefanović D. Solving current limitations of deep learning based approaches for plant disease detection. Symmetry. 2019;11:21. https://doi.org/10.3390/sym11070939 .
Fuentes AF, Yoon S, Lee J, Park DS. High-performance deep neural network-based tomato plant diseases and pests diagnosis system with refinement filter bank. Front Plant Sci. 2018;9:1162. https://doi.org/10.3389/fpls.2018.01162 .
Jiang P, Chen Y, Liu B, He D, Liang C. Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access. 2019. https://doi.org/10.1109/ACCESS.2019.2914929 .
Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2015;39(4):640–51.
He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. In: 2017 IEEE international conference on computer vision (ICCV). New York: IEEE; 2017.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Berlin: Springer; 2015. p. 234–41. https://doi.org/10.1007/978-3-319-24574-4_28 .
Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2019;39(12):2481–95.
Wang Z, Zhang S. Segmentation of corn leaf disease based on fully convolution neural network. Acad J Comput Inf Sci. 2018;1:9–18.
Wang X, Wang Z, Zhang S. Segmenting crop disease leaf image by modified fully-convolutional networks. In: Huang DS, Bevilacqua V, Premaratne P, editors. Intelligent computing theories and application. ICIC 2019, vol. 11643. Lecture Notes in Computer Science. Cham: Springer; 2019. https://doi.org/10.1007/978-3-030-26763-6_62 .
Lin K, Gong L, Huang Y, Liu C, Pan J. Deep learning-based segmentation and quantification of cucumber powdery mildew using convolutional neural network. Front Plant Sci. 2019;10:155.
Kerkech M, Hafiane A, Canals R. Vine disease detection in UAV multispectral images using optimized image registration and deep learning segmentation approach. Comput Electron Agric. 2020;174:105446.
Stewart EL, Wiesner-Hanks T, Kaczmar N, Dechant C, Gore MA. Quantitative phenotyping of northern leaf blight in UAV images using deep learning. Remote Sens. 2019;11(19):2209.
Wang Q, Qi F, Sun M, Qu J, Xue J. Identification of tomato disease types and detection of infected areas based on deep convolutional neural networks and object detection techniques. Comput Intell Neurosci. 2019. https://doi.org/10.1155/2019/9142753 .
Hughes DP, Salathe M. An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing. Comput Sci. 2015.
Shah JP, Prajapati HB, Dabhi VK. A survey on detection and classification of rice plant diseases. In: IEEE international conference on current trends in advanced computing. New York: IEEE; 2016.
Prajapati HB, Shah JP, Dabhi VK. Detection and classification of rice plant diseases. Intell Decis Technol. 2017;11(3):1–17.
Barbedo JGA, Koenigkan LV, Halfeld-Vieira BA, Costa RV, Nechet KL, Godoy CV, Junior ML, Patricio FR, Talamini V, Chitarra LG, Oliveira SAS. Annotated plant pathology databases for image-based detection and recognition of diseases. IEEE Latin Am Trans. 2018;16(6):1749–57.
Brahimi M, Arsenovic M, Laraba S, Sladojevic S, Boukhalfa K, Moussaoui A. Deep learning for plant diseases: detection and saliency map visualisation. In: Zhou J, Chen F, editors. Human and machine learning. Human–computer interaction series. Cham: Springer; 2018. https://doi.org/10.1007/978-3-319-90403-0_6 .
Tyr WH, Stewart EL, Nicholas K, Chad DC, Harvey W, Nelson RJ, et al. Image set for deep learning: field images of maize annotated with disease symptoms. BMC Res Notes. 2018;11(1):440.
Thapa R, Snavely N, Belongie S, Khan A. The plant pathology 2020 challenge dataset to classify foliar disease of apples. arXiv preprint. arXiv:2004.11958 . 2020.
Wu X, Zhan C, Lai YK, Cheng MM, Yang J. IP102: a large-scale benchmark dataset for insect pest recognition. In: 2019 IEEE/CVF conference on computer vision and pattern recognition (CVPR). New York: IEEE; 2019.
Huang M-L, Chuang TC. A database of eight common tomato pest images. Mendeley Data. 2020. https://doi.org/10.17632/s62zm6djd2.1 .
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. In: Proceedings of the 2014 conference on advances in neural information processing systems 27. Montreal: Curran Associates, Inc.; 2014. p. 2672–80.
Pu Y, Gan Z, Henao R, et al. Variational autoencoder for deep learning of images, labels and captions [EB/OL]. 2016–09–28. arxiv:1609.08976 .
Oppenheim D, Shani G, Erlich O, Tsror L. Using deep learning for image-based potato tuber disease detection. Phytopathology. 2018;109(6):1083–7.
Too EC, Yujian L, Njuki S, Yingchun L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput Electron Agric. 2018;161:272–9.
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran YA. Using deep transfer learning for image-based plant disease identification. Comput Electron Agric. 2020;173:105393.
Zhang S, Huang W, Zhang C. Three-channel convolutional neural networks for vegetable leaf disease recognition. Cogn Syst Res. 2018;53:31–41. https://doi.org/10.1016/j.cogsys.2018.04.006 .
Liu B, Ding Z, Tian L, He D, Li S, Wang H. Grape leaf disease identification using improved deep convolutional neural networks. Front Plant Sci. 2020;11:1082. https://doi.org/10.3389/fpls.2020.01082 .
Karthik R, Hariharan M, Anand S, et al. Attention embedded residual CNN for disease detection in tomato leaves. Appl Soft Comput J. 2020;86:105933.
Guan W, Yu S, Jianxin W. Automatic image-based plant disease severity estimation using deep learning. Comput Intell Neurosci. 2017;2017:2917536.
Barbedo JGA. Factors influencing the use of deep learning for plant disease recognition. Biosyst Eng. 2018;172:84–91.
Barbedo JGA. Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput Electron Agric. 2018;153:46–53.
Nawaz MA, Khan T, Mudassar R, Kausar M, Ahmad J. Plant disease detection using internet of thing (IOT). Int J Adv Comput Sci Appl. 2020. https://doi.org/10.14569/IJACSA.2020.0110162 .
Martinelli F, Scalenghe R, Davino S, Panno S, Scuderi G, Ruisi P, et al. Advanced methods of plant disease detection. A review. Agron Sustain Dev. 2015;35(1):1–25.
Liu J, Wang X. Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant Methods. 2020;16:83.
Article CAS PubMed PubMed Central Google Scholar
Liu J, Wang X. Tomato diseases and pests detection based on improved Yolo V3 convolutional neural network. Front Plant Sci. 2020;11:898.
Kamal KC, Yin Z, Wu M, Wu Z. Depthwise separable convolution architectures for plant disease classification. Comput Electron Agric. 2019;165:104948.
Download references
Acknowledgements
Appreciations are given to the editors and reviewer of the Journal Plant Method.
This study was supported by the Facility Horticulture Laboratory of Universities in Shandong with Project Numbers 2019YY003, 2018YY016, 2018YY043 and 2018YY044; school level High-level Talents Project 2018RC002; Youth Fund Project of Philosophy and Social Sciences of Weifang College of Science and Technology with project numbers 2018WKRQZ008 and 2018WKRQZ008-3; Key research and development plan of Shandong Province with Project Number 2019RKA07012, 2019GNC106034 and 2020RKA07036; Research and Development Plan of Applied Technology in Shouguang with Project Number 2018JH12; 2018 innovation fund of Science and Technology Development centre of the China Ministry of Education with Project Number 2018A02013; 2019 basic capacity construction project of private colleges and universities in Shandong Province; and Weifang Science and Technology Development Programme with project numbers 2019GX081 and 2019GX082, Special project of Ideological and political education of Weifang University of science and technology (W19SZ70Z01).
Author information
Authors and affiliations.
Shandong Provincial University Laboratory for Protected Horticulture, Blockchain Laboratory of Agricultural Vegetables, Weifang University of Science and Technology, Weifang, 262700, Shandong, China
Jun Liu & Xuewei Wang
You can also search for this author in PubMed Google Scholar
Contributions
JL designed the research. JL and XW conducted the experiments and data analysis and wrote the manuscript. XW revised the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Correspondence to Xuewei Wang .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.

Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Liu, J., Wang, X. Plant diseases and pests detection based on deep learning: a review. Plant Methods 17 , 22 (2021). https://doi.org/10.1186/s13007-021-00722-9
Download citation
Received : 20 September 2020
Accepted : 13 February 2021
Published : 24 February 2021
DOI : https://doi.org/10.1186/s13007-021-00722-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Deep learning
- Plant diseases and pests
- Classification
- Object detection
- Segmentation
Plant Methods
ISSN: 1746-4811
- Submission enquiries: [email protected]
- Open access
- Published: 07 December 2013
Digital image processing techniques for detecting, quantifying and classifying plant diseases
- Jayme Garcia Arnal Barbedo 1
SpringerPlus volume 2 , Article number: 660 ( 2013 ) Cite this article
214 Citations
12 Altmetric
Metrics details
This paper presents a survey on methods that use digital image processing techniques to detect, quantify and classify plant diseases from digital images in the visible spectrum. Although disease symptoms can manifest in any part of the plant, only methods that explore visible symptoms in leaves and stems were considered. This was done for two main reasons: to limit the length of the paper and because methods dealing with roots, seeds and fruits have some peculiarities that would warrant a specific survey. The selected proposals are divided into three classes according to their objective: detection, severity quantification, and classification. Each of those classes, in turn, are subdivided according to the main technical solution used in the algorithm. This paper is expected to be useful to researchers working both on vegetable pathology and pattern recognition, providing a comprehensive and accessible overview of this important field of research.
Introduction
Agriculture has become much more than simply a means to feed ever growing populations. Plants have become an important source of energy, and are a fundamental piece in the puzzle to solve the problem of global warming. There are several diseases that affect plants with the potential to cause devastating economical, social and ecological losses. In this context, diagnosing diseases in an accurate and timely way is of the utmost importance.
There are several ways to detect plant pathologies. Some diseases do not have any visible symptoms associated, or those appear only when it is too late to act. In those cases, normally some kind of sophisticated analysis, usually by means of powerful microscopes, is necessary. In other cases, the signs can only be detected in parts of the electromagnetic spectrum that are not visible to humans. A common approach in this case is the use of remote sensing techniques that explore multi and hyperspectral image captures. The methods that adopt this approach often employ digital image processing tools to achieve their goals. However, due to their many peculiarities and to the extent of the literature on the subject, they will not be treated in this paper. A large amount of information on the subject can be found in the papers by Bock et al. ( 2010 ), Mahlein et al. ( 2012 ) and Sankaran et al. ( 2010 ).
Most diseases, however, generate some kind of manifestation in the visible spectrum. In the vast majority of the cases, the diagnosis, or at least a first guess about the disease, is performed visually by humans. Trained raters may be efficient in recognizing and quantifying diseases, however they have associated some disadvantages that may harm the efforts in many cases. Bock et al. ( 2010 ) list some of those disadvantages:
Raters may tire and lose concentration, thus decreasing their accuracy.
There can be substantial inter- and intra-rater variability (subjectivity).
There is a need to develop standard area diagrams to aide assessment.
Training may need to be repeated to maintain quality. Raters are expensive.
Visual rating can be destructive if samples are collected in the field for assessment later in the laboratory.
Raters are prone to various illusions (for example, lesion number/size and area infected).
Besides those disadvantages, it is important to consider that some crops may extend for extremely large areas, making monitoring a challenging task.
Depending on the application, many of those problems may be solved, or at least reduced, by the use of digital images combined with some kind of image processing and, in some cases, pattern recognition and automatic classification tools. Many systems have been proposed in the last three decades, and this paper tries to organize and present those in a meaningful and useful way, as will be seen in the next section. Some critical remarks about the directions taken by the researches on this subject are presented in the concluding section.
Literature review
Vegetable pathologies may manifest in different parts of the plant. There are methods exploring visual cues present in almost all of those parts, like roots (Smith and Dickson 1991 ), kernels (Ahmad et al. 1999 ), fruits (Aleixos et al. 2002 ; Corkidi et al. 2005 ; López-García et al. 2010 ), stems and leaves. As commented before, this work concentrates in the latter two, particularly leaves.
This section is divided into three subsections according to the main purpose of the proposed methods. The subsections, in turn, are divided according to the main technical solution employed in the algorithm. A summarizing table containing information about the cultures considered and technical solutions adopted by each work is presented in the concluding section.
Some characteristics are shared by most methods presented in this section: the images are captured using consumer-level cameras in a controlled laboratory environment, and the format used for the images is RGB quantized with 8 bits. Therefore, unless stated otherwise, those are the conditions under which the described methods operate. Also, virtually all methods cited in this paper apply some kind of preprocessing to clean up the images, thus this information will be omitted from now on, unless some peculiarity warrants more detailing.
Because the information gathered by applying image processing techniques often allows not only detecting the disease, but also estimating its severity, there are not many methods focused only in the detection problem. There are two main situations in which simple detection applies:
Partial classification: when a disease has to be identified amidst several possible pathologies, it may be convenient to perform a partial classification, in which candidate regions are classified as being the result of the disease of interest or not, instead of applying a complete classification into any of the possible diseases. This is the case of the method by Abdullah et al. ( 2007 ), which is described in Section ‘Neural networks’.
Real-time monitoring: in this case, the system continuously monitor the crops, and issues an alarm as soon as the disease of interest is detected in any of the plants. The papers by Sena Jr et al. ( 2003 ) and Story et al. ( 2010 ) fit into this context. Both proposals are also described in the following.
Neural networks
The method proposed by Abdullah et al. ( 2007 ) tries to discriminate a given disease ( corynespora ) from other pathologies that affect rubber tree leaves. The algorithm does not employ any kind of segmentation. Instead, Principal Component Analysis is applied directly to the RGB values of the pixels of a low resolution (15×15 pixels) image of the leaves. The first two principal components are then fed to a Multilayer Perceptron (MLP) Neural Network with one hidden layer, whose output reveals if the sample is infected by the disease of interest or not.
Thresholding
The method proposed by Sena Jr et al. ( 2003 ) aims to discriminate between maize plants affected by fall armyworm from healthy ones using digital images. They divided their algorithm into two main stages: image processing and image analysis. In the image processing stage, the image is transformed to a grey scale, thresholded and filtered to remove spurious artifacts. In the image analysis stage, the whole image is divided into 12 blocks. Blocks whose leaf area is less than 5% of the total area are discarded. For each remaining block, the number of connected objects, representing the diseased regions, is counted. The plant is considered diseased if this number is above a threshold, which, after empirical evaluation, was set to ten.
Dual-segmented regression analysis
Story et al. ( 2010 ) proposed a method for monitoring and early detection of calcium deficiency in lettuce. The first step of the algorithm is the plant segmentation by thresholding, so the canopy region is isolated. The outlines of the region of interest are applied back to the original image, in such a way only the area of interest is considered. From that, a number of color features (RGB and HSL) and texture features (from the gray-level co-occurrence matrix) are extracted. After that, the separation point identifying the onset of stress due to the calcium deficiency is calculated by identifying the mean difference between the treatment and control containers at each measured time for all features. Dual-segmented regression analysis is performed to identify where in time a change point was present between the nutrient-deficit group of plants and the healthy group of plants. The authors concluded arguing that their system can be used to monitor plants in greenhouses during the night, but more research is needed for its use during the day, when lighting conditions vary more intensely.
Quantification
The methods presented in this section aim to quantify the severity of a given disease. Such a severity may be inferred either by the area of the leaves that are affected by the disease, or by how deeply rooted is the affection, which can be estimated by means of color and texture features. Most quantification algorithms include a segmentation step to isolate the symptoms, from which features can be extracted and properly processed in order to provide an estimate for the severity of the disease.
It is worth noting that the problem of determining the severity of a disease by analyzing and measuring its symptoms is difficult even if performed manually by one or more specialists, which have to pair the diagnosis guidelines with the symptoms as accurately as possible. As a result, the manual measurements will always contain some degree of subjectivity, which in turn means that references used to validate the automatic methods are not exactly “ground truth”. It is important to take this into consideration when assessing the performance of those methods.
The methods presented in the following are grouped according to the main strategies they employ to estimate the severity of the diseases.
One of the first methods to use digital image processing was proposed by Lindow and Webb ( 1983 ). The images were captured using an analog video camera, under a red light illumination to highlight the necrotic areas. Those images were later digitized and stored in a computer. The tests were performed using leaves from tomatoes, bracken fern, sycamore and California buckeye. The identification of the necrotic regions is done by a simple thresholding. The algorithm then apply a correction factor to compensate for pixel variations in the healthy parts of the leaves, so at least some of the pixels from healthy regions that were misclassified as part of the diseased areas can be reassigned to the correct set.
Price et al. ( 1993 ) compared visual and digital image-processing methods in quantifying the severity of coffee leaf rust. They tested two different imaging systems. In the first one, the images were captured by a black and white charge coupled device (CCD) camera, and in the second one, the images were captured with a color CCD camera. In both cases, the segmentation was performed by a simple thresholding. According to the authors, the image processing-based systems had better performance than visual evaluations, especially for cases with more severe symptoms. They also observed that the color imaging had greater potential in discriminating between rusted and non-rusted foliage.
The method proposed by Tucker and Chakraborty ( 1997 ) aims to quantify and identify diseases in sunflower and oat leaves. The first step of the algorithm is a segmentation whose threshold varies according to the disease being considered (blight or rust). The resulting pixels are connected into clusters representing the diseased regions. Depending on the characteristics of the lesions, they are classified into the appropriate category (type a or b in case of blight and by size in case of rust). The authors reported good results, but observed some errors due to inappropriate illumination during the capture of the images.
Martin and Rybicki ( 1998 ) proposed a method to quantify the symptoms caused by the maize streak virus. The thresholding scheme adopted by the authors was based on the strategy described by Lindow and Webb ( 1983 ) and briefly explained in the previous paragraph. The authors compared the results obtained by visual assessment, by using a commercial software package and by employing a custom system implemented by themselves. They concluded that the commercial and custom software packages had approximately the same performance, and that both computer-based methods achieved better accuracy and precision than the visual approach.
The method proposed by Skaloudova et al. ( 2006 ) measures the damage caused in leaves by spider mites. The algorithm is based on a two-stage thresholding. The first stage discriminates the leaf from the background, and the second stage separates damaged regions from healthy surface. The final estimate is given by the ratio between the number of pixels in damage regions divided by the total number of pixels of the leaf. The authors compared the results with two other methods based on the leaf damage index and chlorophyll fluorescence. They concluded that their method and the leaf damage index provided superior results when compared with the chlorophyll fluorescence.
In their work, Weizheng et al. ( 2008 ) presented a strategy to quantify lesions in soybean leaves. The algorithm is basically composed by a two-step thresholding. The first threshold aims to separate leaf from background. After that, the image containing only the leaf is converted to the HSI color space, and the Sobel operator is applied to identify the lesion edges. A second threshold is applied to the resulting Sobel gradient image. Finally, small objects in the binary image are discarded and holes enclosed by white pixels are filled. The resulting objects reveal the diseased regions.
Camargo and Smith ( 2009a ) proposed a method to identify regions of leaves containing lesions caused by diseases. The tests were performed using leaves from a variety of plants, like bananas, maize, alfalfa, cotton and soybean. Their algorithm is based on two main operations. First, a color transformation to the HSV and I1I2I3 spaces is performed, from which only H and two modified versions of I3 are used in the subsequent steps. After that, a thresholding based on the histogram of intensities technique (Prewitt 1970 ) is applied in order to separate healthy and diseased regions. According to the authors, their approach was able to properly discriminate between diseased and healthy areas for a wide variety of conditions and species of plants.
The method proposed by Macedo-Cruz et al. ( 2011 ) aimed to quantify the damage caused by frost in oat crops. The images used by the authors were captured directly in the crop fields. The first step of the algorithm is the conversion from RGB to the L*a*b* representation. The authors employed three different thresholding strategies: Otsu’s method, Isodata algorithm, and fuzzy thresholding. Each strategy generates a threshold value for each color channel, which are combined by a simple average so a single threshold value is assigned to each channel. If necessary, the resulting partitions may be thresholded again, and so on, until some stopping criteria are met. The final resulting partitions give rise to a number of classes that, after properly labeled, reveal the extent of the damage suffered by the crops.
Lloret et al. ( 2011 ) proposed a system to monitor the health of vineyards. The images were captured by means of webcams scattered throughout the field. The main objective was to detected and quantify diseased leaves. Their system has five stages: 1) leaf size estimation, which is necessary due to the variation of the distance between the cameras and the plants; 2) thresholding, which separates diseased leaves and ground from healthy leaves using both the RGB and HSV color representations of the image; 3) a set of morphological operations, aiming to reduce noise without eliminating useful features; 4) a detection step, which aims to discriminate between ground and actual diseased leaves; 5) calculation of the ratio of diseased leaves. Depending on the value of this ratio, the system emits a warning that the plant requires some attention.
Patil and Bodhe ( 2011 ) proposed a method for assessing the severity of fungi-related disease in sugar cane leaves. The method performs two segmentations. The first one aims to separate the leaves from the rest of the scene, and is performed by means of a simple thresholding. In the second segmentation, the image is converted from the RGB to the HSI color space, and a binarization is applied in order to separate the diseased regions. The threshold for the binarization is calculated by the so-called triangle thresholding method, which is based on the gray-scale histogram of the image. The binary image is finally used to determine the ratio of the infection with respect to the entire leaf.
Color analysis
Boese et al. ( 2008 ) proposed a method to estimate the severity of eelgrass leaf injury, which can be caused by desiccation, wasting disease, and micro herbivory feeding. The first step of the algorithm is the unsupervised segmentation of the leaves into a number of classes (six to ten). In the following, an expert labels the classes into one of five possibilities (the three types of injuries, plus healthy tissue and background). After that, the quantification is just a matter of measuring the areas occupied by each of the injuries. According to the authors, their approach still have a number of problems that limit its utility, but it is an improvement over other approaches to quantify complex leaf injuries from multiple stressors.
The method proposed by Pagola et al. ( 2009 ) deals with the problem of quantifying nitrogen deficiency in barley leaves. They use some color channel manipulations in the RGB space and apply Principal Component Analysis (PCA) to obtain a measure for the “greenness” of the pixels. In order to aggregate the results of all pixels into a single estimate, the authors tested four strategies, whose main goal was to emphasize relevant regions and reduce the influence of the regions that are not photosinthetically active, like veins and leaf spots. The authors concluded that their method had high correlation with the largely adopted approach based on non-destructive hand-held chlorophyll meters.
Contreras-Medina et al. ( 2012 ) proposed a system to quantify five different types of symptoms in plant leaves. Their system is actually composed of five independent modules: 1) chlorosis algorithm, which combines the red and green components of the image in order to determine the yellowness of the leaf, which indicates the severity of the chlorosis; 2) necrosis algorithm, which uses the blue component to discriminate leaves from background, and the green component to identify and quantify the necrotic regions; 3) leaf deformation algorithm, which uses the blue component to segment the leaf and calculates the sphericity of the leaf as a measure for its deformation; 4) white spots algorithm, which applies a thresholding to the blue component of the image to estimate the area occupied by those spots; 5) mosaic algorithm, which uses the blue channel, a number of morphological operations and the Canny edge detector to identify and quantify the venations present in the leaf.
Fuzzy logic
In their paper, Sannakki et al. ( 2011 ) presented a method to quantify disease symptoms based on Fuzzy logic. The tests were performed using pomegranate leaves. The algorithm begins converting the images to the L*a*b* color space. The pixels are grouped into a number of classes through K-means clustering. According to the authors, one of the groups will correspond to the diseased areas, however the paper does not provide any information on how the correct group is identified. In the following, the program calculates the percentage of the leaf that is infected. Finally, a Fuzzy Inference System is employed for the final estimation of the disease rating. The details on how such a system is applied are also absent.
Sekulska-Nalewajko and Goclawski ( 2011 ) method aims to detect and quantify disease symptoms in pumpkin and cucumber leaves. The images used in the tests were captured using a flatbed scanner. The leaves were detached from the plants, treated and stained prior to the imaging. The authors used functions present in the Matlab toolboxes to implement their ideas. The first step of the algorithm is the isolation of the leaf by thresholding. In the following, the image is transformed from RGB to HSV color space. The brightness component (V) is discarded. Then, a Fuzzy c-means algorithm is applied in order to group the pixels into two main clusters, representing healthy and diseased regions. The authors argued that their approach is a better solution than using third-party packages which, according to them, require too many operations to achieve the desired results.
Zhou et al. ( 2011 ) proposed a method to evaluate the degree of hopper infestation in rice crops. The presence of rice plant-hoppers manifests more intensely in the stem, so that was the part of the plant focused by the authors. In the algorithm, after the regions of interest are extracted, fractal-dimension value features are extracted using the box-counting dimension method. These features are used to derive a regression model. Finally, a fuzzy C-means algorithm is used to classify the regions into one of four classes: no infestation, mild infestation, moderate infestation and severe infestation.
Knowledge-based system
The aim of the work by Boissard et al. ( 2008 ) was a little different from the others presented in this paper, as their method tries to quantify the number of whiteflies in rose leaves as part of an early pest detection system. The method employs two knowledge-based systems (KBS) to estimate the number of insects. The first system, the so-called classification KBS, takes the numerical results from some image processing operations, and interprets them into higher level concepts which, in turn, are explored to assist the algorithm to choose and retain only the regions containing insects. The second system, the so-called supervision KBS, selects the image processing tools to be applied, as well as the parameters to be used, in order to collect and feed the most meaningful information to the first system. According to the authors, their proposal had some problems, but it was a good addition to the efforts towards the automation of greenhouse operations.
Region growing
Pang et al. ( 2011 ) proposed a method to segment lesions caused by six types of diseases that affect maize crops. The algorithm begins by identifying all pixels for which the level of the red channel (R) is higher than the level of the green channel (G). According to the authors, those pixels are part of a diseased region in 98% of the cases. The connected regions are then identified and labeled. The second part of the algorithm tries to identify the pixels for which R < G that are actually part of the lesions. To do that, the algorithm takes the connected regions as seeds and applies a region growing technique to more accurately define the diseased regions. The termination condition for the growing procedure is given by the threshold values obtained by applying Otsu’s method to each connected region.
Third party image processing packages
Olmstead et al. ( 2001 ) compared two different methods (one visual and one computational) for quantifying powdery mildew infection in sweet cherry leaves. The images were captured using a flatbed scanner. The image analysis, which is basically the application of thresholding, was performed using the SigmaScan Pro (v. 4.0) software package. In order to generate a standard for comparison of the two methods, the fungi colonies were manually painted white and submitted to the image analysis, providing the reference values. According to the authors, the visual assessment provided far superior estimates in comparison with the computational one.
The method proposed by Berner and Paxson ( 2003 ) aimed at quantifying the symptoms in infected yellow starthistle. The images were captured using a flatbed scanner, and the images were analyzed by the SigmaScan Pro (v.5.0) software package. The operations applied to the image are simple: brightness and contrast adjustments, transformation to gray scale, and application of color overlays. Those overlays emphasize both diseased regions (pustules) and dark areas along venations, so a shape-based selection is carried out in order to keep only the diseased regions. Finally, the pustules are counted.
Moya et al. ( 2005 ) compared the results obtained by visual and image processing-based assessment of squash leaves infected with powdery mildew. They used a commercial software package, the ArcView GIS 3.2, to segment the leaf images into either five or ten classes. The assigned classes were then manually compared to the original images, and the regions corresponding to disease were properly labeled and measured. Finally, the severity of the disease was given by the number of selected pixels divided by the total number of pixels in the leaf. The authors compared these results to those obtained entirely manually. They also compared the results according to the type of device used for capturing the images (digital camera or scanner).
In their proposals, Bock et al. ( 2008 2009 ) aimed at quantifying the severity of the Foliar Citrus Canker in Grapefruit leaves. To perform the image analysis, the authors employed a package called Assess V1.0: Image Analysis Software for plant disease quantification (Lamari 2002 ). In their approach, the images are first converted to the HSI format, and then thresholded to separate the diseased parts from the rest of the scene. The value of the threshold was initially tuned manually by visually comparing the resulting segmentation with the actual image. After the ideal segmentation is achieved, estimating the severity is just a matter of calculating the healthy and diseased areas and finding their ratio. The authors later tried to automate the thresholding process, achieving some mixed results due to tone and lighting variations that prevent fixed thresholds to be valid in all cases.
Goodwin and Hsiang ( 2010 ) and Wijekoon et al. ( 2008 ) used a freely available software called Scion Image to quantify fungal infection in leaves of lilies-of-the-valley, apple trees, phlox and golden rod. The images were captured both in laboratory and in situ , using flatbed scanners for detached leaves and consumer level digital cameras for attached leaves. The use of the Scion software was almost entirely based upon the method proposed by Murakami ( 2005 ), in which the color of a targeted area is manually adjusted in order to maximize the discrimination between healthy and diseased surfaces. The symptoms of several fungal diseases were tested, like powdery mildew, rust, anthracnose and scab.
The Assess software (v. 2.0) was used by Coninck et al. ( 2012 ) to determine the severity of Cercospora leaf spot (CLS) disease in sugar beet breeding. Their approach was related to that used by Bock et al. ( 2009 ), with the images being converted to the HSI representation and with a properly threshold being determined by means of practical experiments. The main purpose of the authors was not to develop a novel method for disease symptom quantification, but to compare the accuracy of three very different ways of estimating the disease severity: visual assessment, real-time Polymerase Chain Reaction (PCR) and image processing. The authors concluded stating that the use of both image analysis and real-time PCR had the potential to increase accuracy and sensitivity of assessments of CLS in sugar beet, while reducing bias in the evaluations.
The software package ImageJ was used by Peressotti et al. ( 2011 ) to quantify grapevine downy mildew sporulation. The authors wrote a macro for ImageJ, which properly adjusts color balance and contrast prior to presenting the image to the user. After that, the user can test several different values of threshold to segment the image, until a satisfactory result is achieved. The authors reported good correlation between the results obtained by their method and by visual assessment.
Classification
The classification methods can be seen as extensions of the detection methods, but instead of trying to detect only one specific disease amidst different conditions and symptoms, these ones try to identify and label whichever pathology that is affecting the plant. As in the case of quantification, classification methods almost always include a segmentation step, which is normally followed by the extraction of a number of features that will feed some kind of classifier. The methods presented in the following are grouped according to the kind of classification strategy employed.
A very early attempt to monitor plant health was carried out by Hetzroni et al. ( 1994 ). Their system tried to identify iron, zinc and nitrogen deficiencies by monitoring lettuce leaves. The capture of the images was done by an analog video camera, and only afterwards the images would be digitized. The first step of the proposed algorithm is the segmentation of the images into leaf and background. In the following a number of size and color features are extracted from both the RGB and HSI representations of the image. Those parameters are finally fed to neural networks and statistical classifiers, which are used to determine the plant condition.
Pydipati et al. ( 2005 ) compared two different approaches to detect and classify three types of citrus diseases. The authors collected 39 texture features, and created four different subsets of those features to be used in two different classification approaches. The first approach was based on a Mahalanobis minimum distance classifier, using the nearest neighbor principle. The second approach used radial basis functions (RBF) neural network classifiers trained with the backpropagation algorithm. According to the authors, both classification approaches performed equally well when using the best of the four subsets, which contained ten hue and saturation texture features.
Huang ( 2007 ) proposed a method to detect and classify three different types of diseases that affect Phalaenopsis orchid seedlings. The segmentation procedure adopted by the author is significantly more sophisticated than those found in other papers, and is composed by four steps: removal of the plant vessel using a Bayes classifier, equalization of the image using an exponential transform, a rough estimation for the location of the diseased region, and equalization of the sub-image centered at that rough location. A number of color and texture features are then extracted from the gray level co-occurrence matrix (Haralick et al. 1973 ). Finally, those features are submitted to an MLP artificial neural network with one hidden layer, which performs the final classification.
Sanyal et al. ( 2007 ) tackled the problem of detecting and classifying six types of mineral deficiencies in rice crops. First, the algorithm extracts a number of texture and color features. Each kind of feature (texture and color) is submitted to its own specific MLP neural network. Both networks have one hidden layer, but the number of neurons in the hidden layer is different (40 for texture and 70 for color). The results returned by both networks are then combined, yielding the final classification. A very similar approach is used by the same authors in another paper (Sanyal and Patel 2008 ), but in this case the objective is to identify two kinds of diseases (blast and brown spots) that affect rice crops.
The method proposed by Al Bashish et al. ( 2010 ) tries to identify five different plant diseases. The authors did not specify the species of plants used in the tests, and the images were captured in situ . After a preprocessing stage to clean up the image, a K-means clustering algorithm is applied in order to divide the image into four clusters. According to the authors, at least one of the clusters must correspond to one of the diseases. After that, for each cluster a number of color and texture features are extracted by means of the so-called Color Co-Occurrence Method, which operates with images in the HSI format. Those features are fed to a MLP Neural Network with ten hidden layers, which performs the final classification.
Kai et al. ( 2011 ) proposed a method to identify three types of diseases in maize leaves. First, the images are converted to the YCbCr color representation. Apparently, some rules are applied during the thresholding in order to properly segment the diseased regions. However, due to a lack of clarity, it is not possible to infer exactly how this is done. The authors then extract a number of texture features from the gray level co-occurrence matrix. Finally, the features are submitted to an MLP neural network with one hidden layer.
Wang et al. ( 2012 ) proposed a method to discriminate between pairs of diseases in wheat and grapevines. The images are segmented by a K-means algorithm, and then 50 color, shape and texture features are extracted. For the purpose of classification, the authors tested four different kinds of neural networks: Multilayer Perceptron, Radial Basis Function, Generalized Regression, and Probabilistic. The authors reported good results for all kinds of neural networks.
Suport vector machines
Meunkaewjinda et al. ( 2008 ) proposed a method to identify and classify diseases that affect grapevines. The method uses several color representations (HSI, L*a*b*, UVL and YCbCr) throughout its execution. The separation between leaves and background is performed by an MLP neural network, which is coupled with a color library built a priori by means of an unsupervised self organizing map (SOM). The colors present on the leaves are then clustered by means of an unsupervised and untrained self-organizing map. A genetic algorithm determines the number of clusters to be adopted in each case. Diseased and healthy regions are then separated by a Support Vector Machine (SVM). After some additional manipulations, the segmented image is submitted to a multiclass SVM, which performs the classification into either scab, rust, or no disease.
Youwen et al. ( 2008 ) proposed a method to identify two diseases that can manifest in cucumber leaves. The segmentation into healthy and diseased regions is achieved using a statistic pattern recognition approach. In the following, some color, shape and texture features are extracted. Those features feed an SVM, which performs the final classification. The authors stated that the results provided by the SVM are far better than those achieved using neural networks.
The system proposed by Yao et al. ( 2009 ) aimed to identify and classify three types of diseases that affect rice crops. The algorithm first applies a particular color transformation to the original RGB image, resulting in two channels ( y 1 and y 2). Then, the image is segmented by Otsu’s method, after which the diseased regions are isolated. Color, shape and texture features are extracted, the latter one from the HSV color space. Finally, the features are submitted to a Support Vector Machine, which performs the final classification.
The method proposed by Camargo and Smith ( 2009b ) tries to identify three different kinds of diseases that affect cotton plants. The authors used images not only of leaves, but also of fruits and stems. The segmentation of the image is performed using a technique developed by the authors (Camargo and Smith 2009a ), which was described earlier in this paper (Section ‘Thresholding’). After that, a number of features is extracted from the diseased regions. Those features are then used to feed an SVM. The one-against-one method (Hsu and Lin 2002 ) was used to allow the SVM to deal with multiple classes. The authors concluded that the texture features have the best discrimination potential.
Jian and Wei ( 2010 ) proposed a method to recognize three kinds of cucumber leaf diseases. As in most approaches, the separation between healthy and diseased regions is made by a simple thresholding procedure. In the following, a variety of color, shape and texture features are extracted. Those features are submitted to an SVM with Radial Basis Function (RBF) as kernel, which performs the final classification.
Fuzzy classifier
The method proposed by Hairuddin et al. ( 2011 ) tries to identify four different nutritional deficiencies in oil palm plants. The image is segmented according to color similarities, but the authors did not provide any detail on how this is done. After the segmentation, a number of color and texture features are extracted and submitted to a fuzzy classifier which, instead of outputting the deficiencies themselves, reveals the amounts of fertilizers that should be used to correct those deficiencies. Unfortunately, the technical details provided in this paper are superficial, making it difficult to reach a clear understanding about the approach adopted by the authors.
Xu et al. ( 2011 ) proposed a method to detect nitrogen and potassium deficiencies in tomato plants. The algorithm begins extracting a number of features from the color image. The color features are all based on the b* component of the L*a*b* color space. The texture features are extracted using three different methods: difference operators, Fourier transform and Wavelet packet decomposition. The selection and combination of the features was carried out by means of a genetic algorithm. Finally, the optimized combination of features is used as the input of a fuzzy K-nearest neighbor classifier, which is responsible for the final identification.
Feature-based rules
In their two papers, Kurniawati et al. ( 2009a 2009b ) proposed a method to identify and label three different kinds of diseases that affect paddy crops. As in many other methods, the segmentation of healthy and diseased regions is performed by means of thresholding. The authors tested two kinds of thresholding, Otsu’s and local entropy, with the best results being achieved by the latter one. Afterwards, a number of shape and color features are extracted. Those features are the basis for a set of rules that determine the disease that best fits the characteristics of the selected region.
Zhang ( 2010 ) proposed a method for identifying and classifying lesions in citrus leaves. The method is mostly based on two sets of features. The first set was selected having as main goal to separate lesions from the rest of the scene, which is achieved by setting thresholds to each feature and applying a weighted voting scheme. The second set aims to provide as much information as possible about the lesions, so a discrimination between diseases becomes possible. The final classification is, again, achieved by means of feature thresholds and a weighted voting system. A more detailed version of (Zhang 2010 ) can be found in (Zhang and Meng 2011 ).
The method proposed by Wiwart et al. ( 2009 ) aims to detect and discriminate among four types of mineral deficiencies (nitrogen, phosphorus, potassium and magnesium). The tests were performed using faba bean, pea and yellow lupine leaves. Prior to the color analysis, the images are converted to the HSI and L*a*b* color spaces. The presence or absence of the deficiencies is then determined by the color differences between healthy leaves and the leaves under test. Those differences are quantified by Euclidean distances calculated in both color spaces.
Pugoy and Mariano ( 2011 ) proposed a system to identify two different types of diseases that attack rice leaves. The algorithm first converts the image from RGB to HSI color space. The K-means technique is applied to cluster the pixels into a number of groups. Those groups are then compared to a library that relates colors to the respective diseases. This comparison results in values that indicate the likelihood of each region being affected by each of the diseases.
Self organizing maps
The method proposed by Phadikar and Sil ( 2008 ) detects and differentiates two diseases that affect rice crops, blast and brown spots. First, the image is converted to the HSI color space. Then, a entropy-based thresholding is used to segment the image. An edge detector is applied to the segmented image, and the intensity of the green components is used to detect the spots. Each region containing each detected spot is then resized by interpolation, so all regions have a size of 80×100 pixels. The pixel values (gray scale) are finally fed to a self organizing map (SOM), which performs the final classification.
Discriminant analysis
Pydipati et al. ( 2006 ) method aims to detect and classify three different types of citrus diseases. The method relies heavily on the color co-occurrence method (CCM), which, in turn, was developed through the use of spatial gray-level dependence matrices (SGDM’s) (Shearer and Holmes 1990 ). The resulting CCM matrices, which are generated from the HSI color representation of the images, are used to extract 39 texture features. The number of features was then reduced by means of a redundancy reduction procedure. The authors observed that the elimination of intensity features improved the results, as hue and saturation features are more robust to ambient light variations than the former ones. The final classification was performed using discriminant analysis.
Membership function
Anthonys and Wickramarachchi ( 2009 ) proposed a method to discriminate among three different diseases that attack paddy plants. The image is segmented by a thresholding procedure – the grayscale version of the image used in such a procedure is obtained after assigning different weights to each component of its RGB representation. The resulting images, containing only the regions supposedly containing the symptoms of the diseases, are then converted to the L*a*b* format, and a number of color and shape features are extracted. The values of those features are compared to some reference value intervals stored in a lookup table by means of the so-called Membership Function, which outputs a single similarity score for each possible disease. The highest score determines the disease affecting the plant.
Table 1 shows an overview of all methods presented in this paper, together with the type of plant considered in each research and the main technical solution used in the algorithm.
Despite the importance of the subject of identifying plant diseases using digital image processing, and although this has been studied for at least 30 years, the advances achieved seem to be a little timid. Some facts lead to this conclusion:
Methods are too specific. The ideal method would be able to identify any disease in any kind of plant. Evidently, this is unfeasible given the current technological level. However, many of the methods that are being proposed not only are able to deal with only one species of plant, but those plants need to be at a certain growth stage in order to the algorithm to be effective. That is acceptable if the disease only attacks the plant in that specific stage, but it is very limiting otherwise. Many of the papers do not state this kind of information explicitly, but if their training and test sets include only images of a certain growth stage, which is often the case, the validity of the results cannot be extended to other stages.
Operation conditions are too strict. Many images used to develop new methods are collected under very strict conditions of lighting, angle of capture, distance between object and capture device, among others. This is a common practice and is perfectly acceptable in the early stages of research. However, in most real world applications, those conditions are almost impossible to be enforced, especially if the analysis is expected to be carried out in a non-destructive way. Thus, it is a problem that many studies never get to the point of testing and upgrading the method to deal with more realistic conditions, because this limits their scope greatly.
Lack of technical knowledge about more sophisticated technical tools. The simplest solution for a problem is usually the preferable one. In the case of image processing, some problems can be solved by using only morphological mathematical operations, which are easy to implement and understand. However, more complex problems often demand more sophisticated approaches. Techniques like neural networks, genetic algorithms and support vector machines can be very powerful if properly applied. Unfortunately, that is often not the case. In many cases, it seems that the use of those techniques is founded more in the hype they generate in the scientific community than in their technical appropriateness with respect to the problem at hand. As a result, problems like overfitting, overtraining, undersized sample sets, sample sets with low representativeness, bias, among others, seem to be a widespread plague. Those problems, although easily identifiable by a knowledgeable individual on the topic, seem to go widely overlooked by the authors, probably due to the lack of knowledge about the tools they are employing. The result is a whole group of technically flawed solutions.
Evidently, there are some high quality manuscripts in which the authors rigorously take into account most factors that could harm the validity of their results, but unfortunately those still seem to be the exception, not the rule. As a result, the technology evolves slower than it could. The underlying conclusion is that the authors should spend a little more time learning about the tools they intend to use. A better understand about the concepts behind those tools can potentially lead to more solid results and to less time wasted, improving the overall quality of the literature of the area.
The wide-ranging variety of applications on the subject of counting objects in digital images makes it difficult for someone to prospect all possible useful ideas present in the literature, which can cause potential solutions for problematic issues to be missed. In this context, this paper tried to present a comprehensive survey on the subject, aiming at being a starting point for those conducting research on the issue. Due to the large number of references, the descriptions are short, providing a quick overview of the ideas underlying each of the solutions. It is important to highlight that the work on the subject is not limited to what was shown here. Many papers on the subject could not be included in order to keep the paper length under control – the papers were selected as to consider the largest number of different problems as possible. Thus, if the reader wishes to attain a more complete understanding on a given application or problem, he/she can refer to the bibliographies of the respective articles.
Abdullah NE, Rahim AA, Hashim H, Kamal MM: Classification of rubber tree leaf diseases using multilayer perceptron neural network. In 2007 5th student conference on research and development . Selangor: IEEE; 2007:1-6.
Google Scholar
Ahmad IS, Reid JF, Paulsen MR, Sinclair JB: Color classifier for symptomatic soybean seeds using image processing. Plant Dis 1999, 83(4):320-327. 10.1094/PDIS.1999.83.4.320
Article Google Scholar
Al Bashish D, Braik M, Bani-Ahmad S: A framework for detection and classification of plant leaf and stem diseases. In 2010 international conference on signal and image processing . Chennai: IEEE; 2010:113-118.
Chapter Google Scholar
Aleixos N, Blasco J, Navarron F, Molto E: Multispectral inspection of citrus in real-time using machine vision and digital signal processors. Comput Electron Agric 2002, 33(2):121-137. 10.1016/S0168-1699(02)00002-9
Anthonys G, Wickramarachchi N: An image recognition system for crop disease identification of paddy fields in Sri Lanka. In 2009 International Conference on Industrial and Information Systems (ICIIS) . Sri Lanka: IEEE; 2009:403-407.
Berner DK, Paxson LK: Use of digital images to differentiate reactions of collections of yellow starthistle (Centaurea solstitialis) to infection by Puccinia jaceae. Biol Control 2003, 28(2):171-179. 10.1016/S1049-9644(03)00096-3
Bock CH, Parker PE, Cook AZ, Gottwald TR: Visual rating and the use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Dis 2008, 92(4):530-541. 10.1094/PDIS-92-4-0530
Bock CH, Cook AZ, Parker PE, Gottwald TR: Automated image analysis of the severity of foliar citrus canker symptoms. Plant Dis 2009, 93(6):660-665. 10.1094/PDIS-93-6-0660
Bock CH, Poole GH, Parker PE, Gottwald TR: Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Critical Rev Plant Sci 2010, 29(2):59-107. 10.1080/07352681003617285
Boese BL, Clinton PJ, Dennis D, Golden RC, Kim B: Digital image analysis of Zostera marina leaf injury. Aquat Bot 2008, 88: 87-90. 10.1016/j.aquabot.2007.08.016
Boissard P, Martin V, Moisan S: A cognitive vision approach to early pest detection in greenhouse crops. Comput Electron Agric 2008, 62(2):81-93. 10.1016/j.compag.2007.11.009
Camargo A, Smith JS: An image-processing based algorithm to automatically identify plant disease visual symptoms. Biosyst Eng 2009a, 102: 9-21. 10.1016/j.biosystemseng.2008.09.030
Camargo A, Smith JS: Image pattern classification for the identification of disease causing agents in plants. Comput Electron Agric 2009b, 66(2):121-125. 10.1016/j.compag.2009.01.003
Coninck BMA, Amand O, Delauré SL, Lucas S, Hias N, Weyens G, Mathys J, De Bruyne E, Cammue BPA: The use of digital image analysis and real-time PCR fine-tunes bioassays for quantification of Cercospora leaf spot disease in sugar beet breeding. Plant Pathol 2012, 61: 76-84. 10.1111/j.1365-3059.2011.02497.x
Contreras-Medina LM, Osornio-Rios RA, Torres-Pacheco I, Romero-Troncoso RJ, Guevara-González RG, Millan-Almaraz JR: Smart sensor for real-time quantification of common symptoms present in unhealthy plants. Sensors (Basel, Switzerland) 2012, 12: 784-805. 10.3390/s120100784
Corkidi G, Balderas-Ruíz KA, Taboada B, Serrano-Carreón L, Galindo E: Assessing mango anthracnose using a new three-dimensional image-analysis technique to quantify lesions on fruit. Plant Pathol 2005, 55(2):250-257.
Goodwin PH, Hsiang T: Quantification of fungal infection of leaves with digital images and Scion Image software. Methods Mol Biol 2010, 638: 125-135. 10.1007/978-1-60761-611-5_9
Hairuddin MA, Tahir NM, Baki SRS: Overview of image processing approach for nutrient deficiencies detection in Elaeis Guineensis. In 2011 IEEE international conference on system engineering and technology . Shah Alam: IEEE; 2011:116-120.
Haralick RM, Shanmugam K, Dinstein I: Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3 1973, 3: 610-621.
Hetzroni A, Miles GE, Engel BA, Hammer PA, Latin RX: Machine vision monitoring of plant health. Adv Space Res 1994, 14(11):203-212. 10.1016/0273-1177(94)90298-4
Hsu CW, Lin CJ: A comparison of methods for multi-class support vector machines. IEEE Trans Neural Netw 2002, 13: 415-425. 10.1109/72.991427
Huang KY: Application of artificial neural network for detecting Phalaenopsis seedling diseases using color and texture features. Comput Electron Agric 2007, 57: 3-11. 10.1016/j.compag.2007.01.015
Jian Z, Wei Z: Support vector machine for recognition of cucumber leaf diseases. In 2010 2nd international conference on advanced computer control . Shenyang: IEEE; 2010:264-266.
Kai S, Zhikun L, Hang S, Chunhong G: A research of maize disease image recognition of corn based on BP networks. In 2011 third international conference on measuring technology and mechatronics automation . Shangshai: IEEE; 2011:246-249.
Kurniawati NN, Abdullah SNHS, Abdullah S, Abdullah S: Investigation on image processing techniques for diagnosing paddy diseases. In 2009 international conference of soft computing and pattern recognition . Malacca: IEEE; 2009a:272-277.
Kurniawati NN, Abdullah SNHS, Abdullah S, Abdullah S: Texture analysis for diagnosing paddy disease. In 2009 International conference on electrical engineering and informatics . Selangor: IEEE; 2009b:23-27.
Lamari L: Assess: image analysis software for plant disease quantification . St. Paul: APS Press; 2002.
Lindow SE, Webb RR: Quantification of foliar plant disease symptoms by microcomputer-digitized video image analysis. Phytopathology 1983, 73(4):520-524. 10.1094/Phyto-73-520
Lloret J, Bosch I, Sendra S, Serrano A: A wireless sensor network for vineyard monitoring that uses image processing. Sensors 2011, 11(6):6165-6196.
López-García F, Andreu-García G, Blasco J, Aleixos N, Valiente JM: Automatic detection of skin defects in citrus fruits using a multivariate image analysis approach. Comput Electron Agric 2010, 71(2):189-197. 10.1016/j.compag.2010.02.001
Macedo-Cruz A, Pajares G, Santos M, Villegas-Romero I: Digital image sensor-based assessment of the status of oat (Avena sativa L.) crops after frost damage. Sensors 2011, 11(6):6015-6036.
Mahlein AK, Oerke EC, Steiner U, Dehne HW: Recent advances in sensing plant diseases for precision crop protection. Eur J Plant Pathol 2012, 133: 197-209. 10.1007/s10658-011-9878-z
Martin DP, Rybicki EP: Microcomputer-based quantification of maize streak virus symptoms in zea mays. Phytopathology 1998, 88(5):422-427. 10.1094/PHYTO.1998.88.5.422
Meunkaewjinda A, Kumsawat P, Attakitmongcol K, Srikaew A: Grape leaf disease detection from color imagery using hybrid intelligent system. In 2008 5th international conference on electrical engineering/electronics, computer, telecommunications and information technology . Krabi: IEEE; 2008:513-516.
Moya EA, Barrales LR, Apablaza GE: Assessment of the disease severity of squash powdery mildew through visual analysis, digital image analysis and validation of these methodologies. Crop Protect 2005, 24(9):785-789. 10.1016/j.cropro.2005.01.003
Murakami PF: An instructional guide for leaf color analysis using digital imaging software. 2005.
Olmstead JW, Lang GA, Grove GG: Assessment of severity of powdery mildew infection of sweet cherry leaves by digital image analysis. Hortscience 2001, 36: 107-111.
Pagola M, Ortiz R, Irigoyen I, Bustince H, Barrenechea E, Aparicio-Tejo P, Lamsfus C, Lasa B: New method to assess barley nitrogen nutrition status based on image colour analysis. Comput Electron Agric 2009, 65(2):213-218. 10.1016/j.compag.2008.10.003
Pang J, Bai Zy, Lai Jc, Li Sk: Automatic segmentation of crop leaf spot disease images by integrating local threshold and seeded region growing. In 2011 international conference on image analysis and signal processing . Hubei: IEEE; 2011:590-594.
Patil SB, Bodhe SK: Leaf disease severity measyrement using image processing. Int J Eng Technol 2011, 3(5):297-301.
Peressotti E, Duchêne E, Merdinoglu D, Mestre P: A semi-automatic non-destructive method to quantify grapevine downy mildew sporulation. J Microbiol Methods 2011, 84(2):265-271. 10.1016/j.mimet.2010.12.009
Phadikar S, Sil J: Rice disease identification using pattern recognition techniques . Khulna: IEEE; 2008. pp 420–423
Book Google Scholar
Prewitt J: Object enhancement and extraction. In Picture processing and psychopictorics . Orlando: Academic Press; 1970.
Price TV, Gross R, Wey JH, Osborne CF: A comparison of visual and digital image-processing methods in quantifying the severity of coffee leaf rust (Hemileia vastatrix). Aust J Exp Agric 1993, 33: 97-101. 10.1071/EA9930097
Pugoy RADL, Mariano VY: Automated rice leaf disease detection using color image analysis. In 3rd international conference on digital image processing, volume 8009 . Chengdu: SPIE; 2011:F1-F7.
Pydipati R, Burks TF, Lee WS: Statistical and neural network classifiers for citrus disease detection using machine vision. Trans ASAE 2005, 48(5):2007-2014.
Pydipati R, Burks TF, Lee WS: Identification of citrus disease using color texture features and discriminant analysis. Comput Electron Agric 2006, 52(1–2):49-59.
Sankaran S, Mishra A, Ehsani R, Davis C: A review of advanced techniques for detecting plant diseases. Comput Electron Agric 2010, 72: 1-13. 10.1016/j.compag.2010.02.007
Sannakki SS, Rajpurohit VS, Nargund VB, Kumar A: Leaf disease grading by machine vision and fuzzy logic. Int J 2011, 2(5):1709-1716.
Sanyal P, Patel SC: Pattern recognition method to detect two diseases in rice plants. Imaging Sci J 2008, 56(6):7.
Sanyal P, Bhattacharya U, Parui SK, Bandyopadhyay SK, Patel S: Color texture analysis of rice leaves diagnosing deficiency in the balance of mineral levels towards improvement of crop productivity. In 10th International Conference on Information Technology (ICIT 2007) . Orissa: IEEE; 2007:85-90.
Sekulska-Nalewajko J, Goclawski J: A semi-automatic method for the discrimination of diseased regions in detached leaf images using fuzzy c-means clustering. In VII international conference on perspective technologies and methods in MEMS design . Polyana-Svalyava: IEEE; 2011:172-175.
Sena DG Jr, Pinto FAC, Queiroz DM, Viana PA: Fall armyworm damaged maize plant identification using digital images. Biosyst Eng 2003, 85(4):449-454. 10.1016/S1537-5110(03)00098-9
Shearer SA, Holmes RG: Plant identification using color co-occurrence matrices. Trans ASAE 1990, 33(6):2037-2044.
Skaloudova B, Krvan V, Zemek R: Computer-assisted estimation of leaf damage caused by spider mites. Comput Electron Agric 2006, 53(2):81-91. 10.1016/j.compag.2006.04.002
Smith SE, Dickson S: Quantification of active vesicular-arbuscular mycorrhizal infection using image analysis and other techniques. Aust J Plant Physiol 1991, 18(6):637-648. 10.1071/PP9910637
Story D, Kacira M, Kubota C, Akoglu A, An L: Lettuce calcium deficiency detection with machine vision computed plant features in controlled environments. Comput Electron Agric 2010, 74(2):238-243. 10.1016/j.compag.2010.08.010
Tucker CC, Chakraborty S: Quantitative assessment of lesion characteristics and disease severity using digital image processing. J Phytopathol 1997, 145(7):273-278. 10.1111/j.1439-0434.1997.tb00400.x
Wang H, Li G, Ma Z, Li X: Application of neural networks to image recognition of plant diseases. In Proceedings of the 2012 International Conference on Systems and Informatics (ICSAI) . Yantai: IEEE; 2012:2159-2164.
Weizheng S, Yachun W, Zhanliang C, Hongda W: Grading method of leaf spot disease based on image processing. In 2008 international conference on computer science and software engineering . Wuhan: IEEE; 2008:491-494.
Wijekoon CP, Goodwin PH, Hsiang T: Quantifying fungal infection of plant leaves by digital image analysis using scion image software. J Microbiol Methods 2008, 74(2–3):94-101.
Wiwart M, Fordonski G, Zuk-Golaszewska K, Suchowilska E: Early diagnostics of macronutrient deficiencies in three legume species by color image analysis. Comput Electron Agric 2009, 65: 125-132. 10.1016/j.compag.2008.08.003
Xu G, Zhang F, Shah SG, Ye Y, Mao H: Use of leaf color images to identify nitrogen and potassium deficient tomatoes. Pattern Recognit Lett 2011, 32(11):1584-1590. 10.1016/j.patrec.2011.04.020
Yao Q, Guan Z, Zhou Y, Tang J, Hu Y, Yang B: Application of support vector machine for detecting rice diseases using shape and color texture features. In 2009 international conference on engineering computation . Hong Kong: IEEE; 2009:79-83.
Youwen T, Tianlai L, Yan N: The recognition of cucumber disease based on image processing and support vector machine. In 2008 congress on image and signal processing . Sanya: IEEE; 2008:262-267.
Zhang M: Citrus canker detection based on leaf images analysis. In The 2nd international conference on information science and engineering . Hangzhou: IEEE; 2010:3584-3587.
Zhang M, Meng Q: Automatic citrus canker detection from leaf images captured in field. Pattern Recognit Lett 2011, 32(15):2036-2046. 10.1016/j.patrec.2011.08.003
Zhou Z, Zang Y, Li Y, Zhang Y, Wang P, Luo X: Rice plant-hopper infestation detection and classification algorithms based on fractal dimension values and fuzzy C-means. Math Comput Model 2011, 58: 701-709.
Download references
Author information
Authors and affiliations.
Embrapa Agricultural Informatics, Campinas, SP, Brazil
Jayme Garcia Arnal Barbedo
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Jayme Garcia Arnal Barbedo .
Additional information
Competing interests.
The author declares that he has no competing interests.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Reprints and permissions
About this article
Cite this article.
Arnal Barbedo, J.G. Digital image processing techniques for detecting, quantifying and classifying plant diseases. SpringerPlus 2 , 660 (2013). https://doi.org/10.1186/2193-1801-2-660
Download citation
Received : 14 June 2013
Accepted : 26 September 2013
Published : 07 December 2013
DOI : https://doi.org/10.1186/2193-1801-2-660
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Radial Basis Function
- Powdery Mildew
- Texture Feature
- Diseased Region
Plant Leaf Disease Detection using Image Processing
Ieee account.
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
Plant Disease Detection Using Image Classification
- Conference paper
- First Online: 23 December 2020
- Cite this conference paper

- Ankit Chhillar 14 &
- Sanjeev Thakur 14
Part of the book series: Lecture Notes in Networks and Systems ((LNNS,volume 150))
392 Accesses
3 Citations
Indian agriculture yields have been facing major problems that lead to the fall in overall production. The major factor can be listed as proper irrigation, availability of water, fertile land, and plant disease. Farmers invest a huge sum of money on disease management, regularly without effective technical support, causing poor disease control, contamination, and harmful outcomes. Early detection of disease not only helps in imposing quick protective major but as well as increase overall farm yield. In earlier times, the process of disease detection was done manually with the help of experts, costing a fortune for the local farmers, and sometimes, unavailability of any expertise help leads to a huge loss in farmlands. Digital image processing works as a time and cost-efficient alternative to the manual process. This process is proven to be much faster and gives better results than the manual method. Deep learning and CNN are widely used for the purpose of image classification (Eldem et al. in A model of deep neural network for iris classification with different activation functions, 2018; Abu et al. in Int J Eng Res Technol 12(4):563–569, 2019) [ 1 , 2 ]. This paper focuses on developing an artificial intelligence (M. Approach in A modern approach) [ 3 ]-based plant disease detection technique based upon computer vision (Sladojevic et al. in Comput Intell Neurosci 2016, 2016) [ 4 ]. The dataset for the study was downloaded from the kaggle which contains 14 different categories of plants. For this paper, we mainly focused on “4” category of plants (corn, pepper, potato, and tomato). “50” images from each category were taken, and the maximum accuracy which was achieved through the algorithm is “96.54%”. This algorithm is suitable getting maximum accuracy from a small dataset.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Similar content being viewed by others

Plant Disease Detection and Classification Using Artificial Intelligence Approach

Plant Disease Prediction Using Image Processing and Deep Convolutional Neural Network

Deep Learning Model for Automated Image Based Plant Disease Classification
Eldem, A., Eldem, H., & Üstün, D. (2018). A model of deep neural network for iris classification with different activation functions.
Google Scholar
Abu, M. A., Indra, N. H., Halim, A., Rahman, A., Sapiee, N. A., & Ahmad, I. (2019). A study on image classification based on deep learning and tensorflow. International Journal of Engineering Research and Technology, 12 (4), 563–569.
Stuart, R. (2003) Artificial Intelligence A Modern Approach . vol. 3 , Prentice Hall.
Sladojevic, S., Arsenovic, M., Anderla, A., Culibrk, D., & Stefanovic, D. (2016). Deep neural networks based recognition of plant diseases by leaf image classification. Computational Intelligence and Neuroscience, 2016 .
Dhillon, A., & Verma, G. K. (2019). Convolutional neural network: A review of models, methodologies and applications to object detection. Progress in Artificial Intelligence , no. 0123456789.
Mohanty, S. P., Hughes, D. P., Salathé, M. (2016). Using deep learning for image-based plant disease detection. Frontiers in Plant Science, 7 , 1–10.
Wahab, A. H. B. A., Zahari, R., & Lim, T. H. (2019). Detecting diseases in chilli plants using K-means segmented support vector machine. In 2019 3rd International Conference on Imaging, Signal Processing and Communications (pp. 57–61).
Khirade, S. D. (2015). Plant disease detection using image processing (pp. 1–4).
Ramcharan, A., Baranowski, K., Mccloskey, P., Ahmed, B., Legg, J., & Hughes, D. P. (2017). Deep learning for image-based cassava disease detection. Frontiers in Plant Science, 8 , 1–7.
Ferentinos, K. P. (2018). Deep learning models for plant disease detection and diagnosis. Computers and Electronics in Agriculture, 145 , 311–318.
Article Google Scholar
Sheikh, H., Mim, T. T., Reza, S., Shahariar, A. K. M., Rabby, A., & Hossain, S. A. (2019). Detection of maize and peach leaf diseases using image processing. In 2019 10th International Conference on Communication and Network Technology (pp. 1–7).
Valdoria, J. C., Caballeo, A. R., Condino, J. M. M., & Fernandez, B. I. D. (2019). iDahon: An android based terrestrial plant disease detection mobile application through digital image processing using deep learning neural network algorithm. In 2019 4th Interantional Conference on Information Technology (pp. 94–98).
Agostinelli, F., Hoffman, M., Sadowski, P., & Baldi, P. (2015). Learning activation functions, (2013), 1–9.
Luo, H., & Hanagud, S. (1997). Dynamic learning rate neural network training and composite structural damage detection. AIAA Journal, 35 (9).
Recognition, I., & Postalcioglu, S. (2019). Accepted manuscript to appear in IJPRAI International Journal of Pattern Recognition and Artificial Intelligence .
Schaul, T. (2011). No more Pesky learning rates.
Hawkins, D. M. (2004). The problem of overfitting. Journal of Chemical Information and Modeling, 44 , 1–12.
Allamy, H. K. (2016). Methods to avoid over-fitting and under-fitting in supervised machine learning (comparative study).
Parker, J. R. (2001). Rank and response combination from confusion matrix data. Information Fusion, 2 .
https://www.kaggle.com/vipoooool/new-plant-diseases-dataset
Download references
Author information
Authors and affiliations.
Amity School of Engineering and Technology, Amity University, Noida, Uttar Pradesh, India
Ankit Chhillar & Sanjeev Thakur
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Sanjeev Thakur .
Editor information
Editors and affiliations.
Department of Computer Science and Engineering, ABES Engineering College, Ghaziabad, Uttar Pradesh, India
Shailesh Tiwari
Department of Information Systems, Institut Teknologi Sepuluh Nopember, Surabaya, Indonesia
Erma Suryani
Singapore Institute of Technology, Singapore, Singapore
Andrew Keong Ng
Department of Computer Science and Engineering, Motilal Nehru National Institute of Technology Allahabad, Prayagraj, Uttar Pradesh, India
K. K. Mishra
Department of Electrical Engineering, Motilal Nehru National Institute of Technology Allahabad, Prayagraj, Uttar Pradesh, India
Nitin Singh
Rights and permissions
Reprints and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper.
Chhillar, A., Thakur, S. (2021). Plant Disease Detection Using Image Classification. In: Tiwari, S., Suryani, E., Ng, A.K., Mishra, K.K., Singh, N. (eds) Proceedings of International Conference on Big Data, Machine Learning and their Applications. Lecture Notes in Networks and Systems, vol 150. Springer, Singapore. https://doi.org/10.1007/978-981-15-8377-3_23
Download citation
DOI : https://doi.org/10.1007/978-981-15-8377-3_23
Published : 23 December 2020
Publisher Name : Springer, Singapore
Print ISBN : 978-981-15-8376-6
Online ISBN : 978-981-15-8377-3
eBook Packages : Intelligent Technologies and Robotics Intelligent Technologies and Robotics (R0)
Share this paper
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research

IMAGES
VIDEO
COMMENTS
The detection of the disease includes methods including. image segregation, pre-processing data, fragmentation of the image, detection, and recognition of characteristics. This paper also examines ...
Disease detection involves the steps like image acquisition, image pre-processing, image segmentation, feature extraction and classification. This paper discussed the methods used for the detection of plant diseases using their leaves images. This paper also discussed some segmentation and feature extraction algorithm used in the plant disease ...
Here, we demonstrate the technical feasibility using a deep learning approach utilizing 54,306 images of 14 crop species with 26 diseases (or healthy) made openly available through the project PlantVillage ( Hughes and Salathé, 2015 ). An example of each crop—disease pair can be seen in Figure 1. Figure 1.
This paper proposes a smart and efficient technique for detection of crop disease which uses computer vision and machine learning techniques. The proposed system is able to detect 20 different diseases of 5 common plants with 93% accuracy. Keywords: Digital image processing, Foreground detection, Machine learning, Plant disease detection.
The process of disease detection system primarily involves four phases as shown in Fig. 3.The primary part involves acquisition of pictures either through smart devices [11, 16, 21] such as camera and mobile or from internet.The second part segments the image [5, 15] into varied numbers of clusters that completely different techniques will be applied.
Plant diseases and pests are important factors determining the yield and quality of plants. Plant diseases and pests identification can be carried out by means of digital image processing. In recent years, deep learning has made breakthroughs in the field of digital image processing, far superior to traditional methods. How to use deep learning technology to study plant diseases and pests ...
This paper is focused on summarizing the articles that utilized the features of soft computing along with image processing. Therefore, the task of plant disease detection using image processing techniques along with soft computing is categorized in the following modules: image acquisition, image preprocessing, image segmentation and ...
Extract colour, shape, texture, colour texture, and random rework. SVM Associate in Nursing closest neigh-bour classifiers achieved 83.6% accuracy. A processed image of a cold plant leaf shows its health. Their strategy is to limit Chemicals to the morbid cold plant. MATLAB extracts features and recognises images.
Disease detection involves the steps like image acquisition, image pre-processing, image segmentation, feature extraction and classification. This paper discussed the methods used for the detection of plant diseases using their leaves images. This paper also discussed some segmentation and feature extraction algorithm used in the plant disease ...
In the field of agriculture, image processing is a constantly evolving field of research and progress. Currently, several plant disease identification studies are underway. Identifying plant diseases can not only help farmers increase yields, but also promote a variety of agricultural practices. This paper proposes an algorithmic program for the diseases detection and categorization with the ...
This paper presents a survey on methods that use digital image processing techniques to detect, quantify and classify plant diseases from digital images in the visible spectrum. Although disease symptoms can manifest in any part of the plant, only methods that explore visible symptoms in leaves and stems were considered. This was done for two main reasons: to limit the length of the paper and ...
[2] Prof. Sanjay B. et al., Agricultural plant leaf disease detection using image processing (2013) Vision-based detection algorithm with masking the green-pixels and color co-occurrence method: NN's can be used to increase the recognition rate of classification process [3] Mrunalini R. et al., An application of K-means clustering and ...
In this paper, we provide a review on methods developed by various researchers for detection of diseases in plants, in the field of image processing. It includes research in disease detection of plants such as apple, grapes, pepper, pomegranate, tomato etc.
By identifying the color features of the leaves image processing helps in the detection of the diseases and also provides prevention to the particular diseases.[4]At the very first stage the image is segmented by taking the picture of the plant in the RGB form and later the green pixel is detached. Texture statistics is an arrangement of ...
The paper is divided into four major parts. The first part gives the introduction and literature overview. The second part discusses the method of implementation in plant leaf disease detection using image processing and AI-based technique and further using this information, the hardware implementation to spray the pesticide.
The use of technologies like Computer vision and Machine Learning (ML) helps to fight against diseases [6], [7], [8]. In this paper, we are using ML to give a solution to Plant Diseases. In this method, we have divided the process into three stages Identity, Analyse and Verify with the Available database [9].
Plant Leaf Disease Detection using Image Processing ... So to answer that I am writing this research paper because all the foods and grains that we eat is controlled by pesticides and insecticides that harms our body and not good for our health. In this paper I have defined the techniques to detect the diseases in the leaf of the plant by image ...
This paper presents an image-based plant disease detection technique. This study mainly focuses on vegetable plant for the purpose of disease detection. The plants are listed as corn, pepper, potato, and tomato. Both the healthy and infected plant leaf images were used for training the classification model: