March 28, 2024

Kombucha Tea-associated microbes remodel host metabolic pathways to suppress lipid accumulation
Image credit: pgen.1011003
Research Article
Genomic analyses of Symbiomonas scintillans show no evidence for endosymbiotic bacteria but does reveal the presence of giant viruses
A multi-gene tree showed the three SsV genome types branched within highly supported clades with each of BpV2, OlVs, and MpVs, respectively.
Image credit: pgen.1011218
Recently Published Articles
- Drosophila melanogaster ">The impact of developmental stage, tissue type, and sex on DNA double-strand break repair in Drosophila melanogaster
- Transcriptional control of visual neural circuit development by GS homeobox 1
- Unraveling the genetics of arsenic toxicity with cellular morphology QTL
Current Issue March 2024
A natural bacterial pathogen of C . elegans uses a small RNA to induce transgenerational inheritance of learned avoidance
A mechanism of learning and remembering pathogen avoidance likely happens in the wild.
Image credit: pgen.1011178
Spoink , a LTR retrotransposon, invaded D. melanogaster populations in the 1990s
Evidence of Spoink retrotransposon's horizontal transfer into D. melanogaster populations post-1993, suggesting its origin from D.willistoni .
Image credit: pgen.1011201
Comparison of clinical geneticist and computer visual attention in assessing genetic conditions
Understanding AI, specifically Deep Learning, in facial diagnostics for genetic conditions can enhance the design and utilization of AI tools, facilitating more meaningful interactions between clinicians and AI technologies.
Image credit: pgen.1011168
Maintenance of proteostasis by Drosophila Rer1 is essential for competitive cell survival and Myc-driven overgrowth
Loss of Rer1 induces proteotoxic stress, leading to cell competition and elimination, while increased Rer1 levels provide cytoprotection and support Myc-driven overgrowth.
Image credit: pgen.1011171
Anthracyclines induce cardiotoxicity through a shared gene expression response signature
TOP2i induce thousands of shared gene expression changes in cardiomyocytes.
Image credit: pgen.1011164
CryptoCEN: A Co-Expression Network for Cryptococcus neoformans reveals novel proteins involved in DNA damage repair
Co-expression analysis of CryptoCEN network identifys 13 new DNA damage response genes.
Image credit: pgen.1011158
TRPS1 modulates chromatin accessibility to regulate estrogen receptor alpha (ER) binding and ER target gene expression in luminal breast cancer cells
TRPS1 orchestrates gene expression, estrogen signaling, and chromatin dynamics …
Image credit: pgen.1011159
IntroUNET: Identifying introgressed alleles via semantic segmentation
Deep learning algorithm accurately identifies introgressed alleles at the individual level, unveiling insights into the extent and fitness effects of introgression.
Image credit: pgen.1010657
New PLOS journals accepting submissions
Five new journals unified in addressing global health and environmental challenges are now ready to receive submissions: PLOS Climate , PLOS Sustainability and Transformation , PLOS Water , PLOS Digital Health , and PLOS Global Public Health
COVID-19 Collection
The COVID-19 Collection highlights all content published across the PLOS journals relating to the COVID-19 pandemic.
Submit your Lab and Study Protocols to PLOS ONE !
PLOS ONE is now accepting submissions of Lab Protocols, a peer-reviewed article collaboration with protocols.io, and Study Protocols, an article that credits the work done prior to producing and publishing results.
PLOS Reviewer Center
A collection of free training and resources for peer reviewers of PLOS journals—and for the peer review community more broadly—drawn from research and interviews with staff editors, editorial board members, and experienced reviewers.
Ten Simple Rules
PLOS Computational Biology 's "Ten Simple Rules" articles provide quick, concentrated guides for mastering some of the professional challenges research scientists face in their careers.
Welcome New Associate Editors!
PLOS Genetics welcomes several new Associate Editors to our board: Nicolas Bierne, Julie Simpson, Yun Li, Hongbin Ji, Hongbing Zhang, Bertrand Servin, & Benjamin Schwessinger
Expanding human variation at PLOS Genetics
The former Natural Variation section at PLOS Genetics relaunches as Human Genetic Variation and Disease. Read the editors' reasoning behind this change.
PLOS Genetics welcomes new Section Editors
Quanjiang Ji (ShanghaiTech University) joined the editorial board and Xiaofeng Zhu (Case Western Reserve University) was promoted as new Section Editors for the PLOS Genetics Methods section.
PLOS Genetics editors elected to National Academy of Sciences
Congratulations to Associate Editor Michael Lichten and Consulting Editor Nicole King, who are newly elected members of the National Academy of Sciences.
Harmit Malik receives Novitski Prize
Congratulations to Associate Editor Harmit Malik, who was awarded the Edward Novitski Prize by the Genetics Society of America for his work on genetic conflict. Harmit has also been elected as a new member of the American Academy of Arts & Sciences.
Publish with PLOS
- Submission Instructions
- Submit Your Manuscript
Connect with Us
- PLOS Genetics on Twitter
- PLOS on Facebook
Get new content from PLOS Genetics in your inbox
Thank you you have successfully subscribed to the plos genetics newsletter., sorry, an error occurred while sending your subscription. please try again later..
- Search Menu
- Advance Articles
- Perspectives
- Knowledgebase and Database Resources
- Nobel Laureates Collection
China Virtual Outreach Webinar
Neurogenetics, fungal genetics and genomics.
- Multiparental Populations
- Genomic Prediction
- Plant Genetics and Genomics
Genetic Models of Rare Diseases
- Genomic Data Analyses In Biobanks
- Why Publish
- Author Guidelines
- Submission Site
- Open Access Options
- Full Data Policy
- Self-Archiving Policy
- About Genetics
- About Genetics Society of America
- Editorial Board
- Early Career Reviewers
- Guidelines for Reviewers
- Advertising & Corporate Services
- Journals on Oxford Academic
- Books on Oxford Academic

Editor-in-Chief
Howard Lipshitz
Executive Editor
Tracey DePellegrin
Managing Editor
Ruth Isaacson
Scientific Editor and Program Manager
Opportunities and challenges for genomic data analyses in biobanks: a call for papers.
The GSA Journals are calling for submissions of papers on biobank-scale genomic data analyses. The closing date for submissions is May 31 2024.
Why publish with GENETICS?
Why publish in genetics.
Learn more about why GENETICS is the perfect home for your research, and submit today to join our celebrated author community.
Why publish?
Series and Collections accepting papers
Submit your work to one of GSA’s ongoing series and collections.
Currently accepting submissions
Meet the Editorial Board
See who handles papers for GENETICS by topic.
Editorial board
Re-watch the recent China Virtual Outreach Webinar where you will learn more about publishing your work in the journal.
Watch the webinar
Latest articles
Series & collections.

Genes and variants of interest in rare diseases often benefit from modelling in cellular assays or genetic models to aid in understanding molecular and cellular mechanisms of disfunction. Model organisms are useful for the discovery of new genetic diseases and key to understanding variant effects, and modelling a disease gene in a genetic model means that researchers can perform an in-depth exploration of gene or variant function. The GSA Journals are pleased to publish a series highlighting ongoing advances in rare disease discovery and mechanisms by presenting key research findings and new discoveries.

Plant Genetics and Genomics
Plant science has generated many discoveries and advances in genetics and genomics research. These contributions reflect the ingenuity and rigor of the plant science community, as well as the rich diversity of plants and their biology. To showcase this critical work, GENETICS and G3: Genes|Genomes|Genetics has launched the Plant Genetics and Genomics series with a collection of fourteen research articles and an accompanying editorial.

Neurogenetics lies at the intersection of Neuroscience and Genetics, where genetic approaches are applied to the study of nervous system development, function, and plasticity. Overseen by Series Editors Oliver Hobert, Cecilia Moens, and Kate O’Connor Giles, this new series aims to make the GSA Journals a home for cutting-edge, robust research in neurogenetics.

The fungal kingdom is remarkable in its breadth and depth of impact on global health, agriculture, biodiversity, ecology, manufacturing, and biomedical research. Overseen by editors Leah Cowen and Joseph Heitman, this series aims to report and thereby further stimulate advances in genetics and genomics across a diversity of fungal species.

FlyBook from GENETICS is a comprehensive compendium of review articles presenting the current state of knowledge in Drosophila research.
Browse FlyBook

WormBook from GENETICS features a comprehensive compendium of review articles presenting the current state of knowledge in C. elegans research. WormBook articles will span the breadth of the biology, genetics, genomics, and evolutionary biology of C. elegans .
Browse WormBook

The YeastBook series from GENETICS features a comprehensive compendium of reviews that presents the current state of knowledge of the molecular biology, cellular biology, and genetics of the yeast Saccharomyces cerevisiae .
Browse YeastBook
More from GSA

G3: Genes|Genomes|Genetics
G3, a Genetics Society of America journal, provides a forum for the publication of high-quality foundational research-particularly research that generates useful genetic and genomic information, as well as genome reports, mutant screens, and advances in methods and technology.
Find out more

GSA members of all career stages receive member benefits including access to professional development programs, discounted meeting registration, and eligibility for travel awards. Members also receive a personal subscription to GENETICS, as well as discounted publication fees in both GSA journals.

Conferences
GSA conferences have long served as community hubs for researchers focused on particular organisms or topics. GSA also hosts The Allied Genetics Conference (TAGC) , a unique meeting that brings together multiple research communities for collaboration and synthesis.
Attend a conference

Career Development
GSA professional development programs provide rich opportunities for scientists to gain skills, experience, mentors, and networks. Our initiatives and resources range from peer review training to inclusive public engagement, newsletters, webinars, a job board, leadership programs, and much more.
Browse Opportunities

Email alerts
Register to receive email alerts as soon as new content from GENETICS is published online.

Recommend to your library
Fill out our simple online form to recommend GENETICS to your library. Recommend now

Author resources
Learn about how to submit your article, our publishing process, and tips on how to promote your article.
Related Titles
- Recommend to Your Librarian
- Advertising and Corporate Services
- Journals Career Network
Affiliations
- Online ISSN 1943-2631
- Copyright © 2024 Genetics Society of America
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
- Open access
- Published: 29 May 2019
Genomics and data science: an application within an umbrella
- Fábio C. P. Navarro 1 , 2 ,
- Hussein Mohsen 1 , 2 ,
- Chengfei Yan 1 , 2 ,
- Shantao Li 5 , 6 ,
- Mengting Gu 1 , 2 ,
- William Meyerson 1 , 2 &
- Mark Gerstein ORCID: orcid.org/0000-0002-9746-3719 1 , 2 , 3 , 4
Genome Biology volume 20 , Article number: 109 ( 2019 ) Cite this article
35k Accesses
40 Citations
49 Altmetric
Metrics details
Data science allows the extraction of practical insights from large-scale data. Here, we contextualize it as an umbrella term, encompassing several disparate subdomains. We focus on how genomics fits as a specific application subdomain, in terms of well-known 3 V data and 4 M process frameworks (volume-velocity-variety and measurement-mining-modeling-manipulation, respectively). We further analyze the technical and cultural “exports” and “imports” between genomics and other data-science subdomains (e.g., astronomy). Finally, we discuss how data value, privacy, and ownership are pressing issues for data science applications, in general, and are especially relevant to genomics, due to the persistent nature of DNA.
Introduction
Data science as a formal discipline is currently popular because of its tremendous commercial utility. Large companies have used several well-established computational and statistical techniques to mine high volumes of commercial and social data [ 1 ]. The broad interest across many applications stirred the birth of data science as a field that acts as an umbrella, uniting a number of disparate disciplines using a common set of computational approaches and techniques [ 2 ]. In some cases, these techniques were created, developed, or established in other data-driven fields (e.g., astronomy and earth science). In fact, some of these disciplines significantly predate the formal foundation of data science and have contributed to several techniques to cope with knowledge extraction from large amounts of data.
Many scholars have probed the origins of data science. For example, in 1960 Tukey described a new discipline called data analysis, which some consider being a forerunner of data science. He defined data analysis as the interplay between statistics, computer science, and mathematics [ 3 ]. Jim Gray also introduced the concept of data-intensive science in his book The Fourth Paradigm [ 4 ], and discussed how the developments in computer science would shape and transform segments of science to a data-driven exercise. More practically, the maturation of modern data science from an amorphous discipline can be tracked to the expansion of the technology industry and its adoption of several concepts at the confluence of statistics and algorithmic computer science, such as machine learning [ 5 ]. Somewhat less explored is the fact that several applied disciplines have contributed to a collection of techniques and cultural practices that today comprise data science.
Contextualizing natural science within the data science umbrella
Long before the development of formal data science, and even computer science or statistics, traditional fields of natural sciences established an extensive culture around data management and analytics. For instance, physics has a long history of contributions of several concepts that are now at the foundation of data science. In particular, physicists such as Laplace, Gauss, Poisson, and Dirichlet have led the way for the development of hypothesis testing, least squares fits, and Gaussian, Poisson, and Dirichlet distributions [ 6 ].
More recently, physics also has contributed new data techniques and data infrastructure. For example, Ulam originally invented the Monte Carlo sampling method while he was working on the hydrogen bomb [ 7 ] and Berners-Lee, from the CERN (European Organization for Nuclear Research), developed the World Wide Web [ 8 ] to enable distributed collaboration in particle physics. While most disciplines are now experiencing issues with rapid data growth [ 9 , 10 ], we find it interesting that physics had issues with data management long before most disciplines. As early as the 1970s, for example, Jashcek introduced the term “information explosion” to describe the rapid data growth in astrophysics [ 11 ].
Fundamental contributions to data management and analytics have not been exclusive to physics. The biological sciences, perhaps most prominently genetics, also have significantly influenced data science. For instance, many of the founders of modern statistics, including Galton, Pearson, and Fisher, pioneered principal component analysis, linear regression, and linear discriminant analysis while they were also preoccupied with analyzing large amounts of biological data [ 6 ]. More recently, methods such as logistic regression [ 12 ], clustering [ 13 ], decision trees [ 14 ], and neural networks [ 15 ] were either conceptualized or developed by researchers focused on biological questions. Even Shannon, a central figure in information theory, completed a short PhD in population genetics [ 16 ].
Genomics and data science
More recent biological disciplines such as macromolecular structure and genomics have inherited many of these data analytics features from genetics and other natural sciences. Genomics, for example, emerged in the 1980s at the confluence of genetics, statistics, and large-scale datasets [ 17 ]. The tremendous advancements in nucleic acid sequencing allowed the discipline to swiftly assume one of the most prominent positions in terms of raw data scale across all the sciences [ 18 ]. This pre-eminent role of genomics also inspired the emergence of many “-omics” terms inside and outside academia [ 19 , 20 ]. Although today genomics is pre-eminent in terms of data scale, this may change over time due to technological developments in other areas, such as cryo-electron microscopy [ 21 ] and personal wearable devices [ 22 ]. Moreover, it is important to realize that many other existing data-rich areas in the biological sciences are also rapidly expanding, including image processing (including neuroimaging), macromolecular structure, health records analysis, proteomics, and the inter-relation of these large data sets, in turn, is giving rise to a new subfield termed biomedical data science (Fig. 1 a).
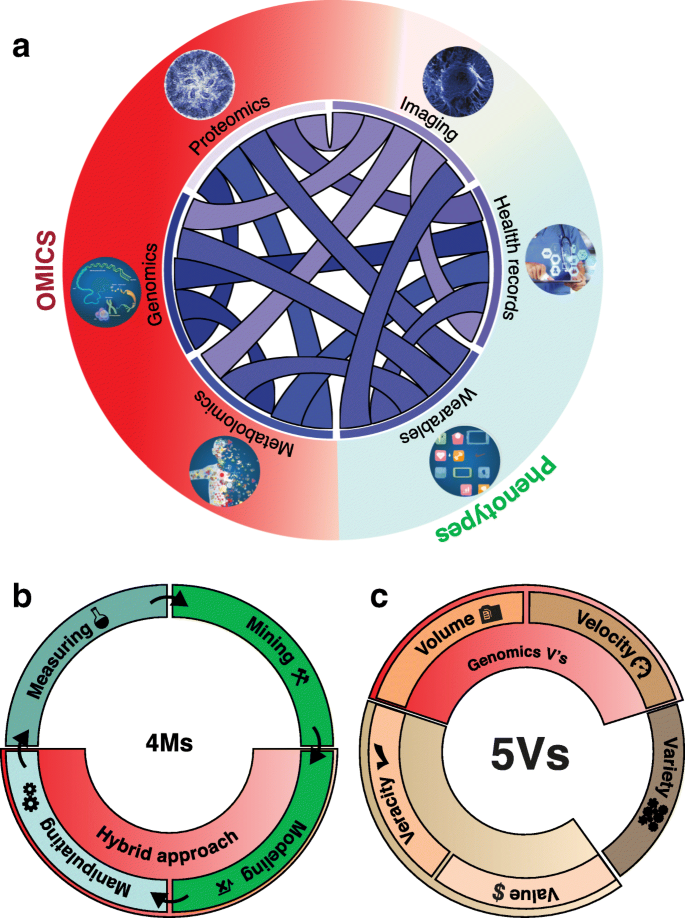
A holistic view of biomedical data science. a Biomedical data science emerged at the confluence of large-scale datasets connecting genomics, metabolomics, wearable devices, proteomics, health records, and imaging to statistics and computer science. b The 4 M processes framework. c The 5 V data framework
Here, we explore how genomics has been, and probably will continue to be, a pre-eminent data science subdiscipline in terms of data growth and availability. We first explore how genomics data can be framed in terms of the 3Vs (data volume, velocity, and variety) to contextualize the discipline in the “big-data world”. We also explore how genomics processes can be framed in terms of the 4Ms (measurement, mining, modeling, and manipulating) to discuss how physical and biological modeling can be leveraged to generate better predictive models. Genomics researchers have been exchanging ideas with those from other data science subfields; we review some of these “imports” and “exports” in a third section. Finally, we explore issues related to data availability in relation to data ownership and privacy. Altogether, this perspective discusses the past, present, and future of genomics as a subfield of data science.
Genomics versus other data science applications in terms of the V framework
One way of categorizing the data in data science disciplines is in terms of its volume, velocity, and variety. Within data science, this is broadly referred to as the V framework [ 23 ]. Over the years, the V framework has been expanded from its original 3Vs [ 24 ] (volume, velocity, and variety) to the most recent versions with four and five Vs (3 V + value and veracity; Fig. 1 c) [ 25 ]. In general, the distinct V frameworks use certain data-related parameters to recognize issues and bottlenecks that might require a new set of tools and techniques to cope with unstructured and high-volume data. Here, we explore how we can use the original 3 V framework to evaluate the current state of data in genomics in relation to other applications in data sciences.
One of the key aspects of genomics as a data science is the sheer amount of data being generated by sequencers. As shown in Fig. 2 , we tried to put this data volume into context by comparing genomics datasets with other data-intensive disciplines. Figure 2 a shows that the total volume of data in genomics is considerably smaller than the data generated by earth science [ 26 ], but orders of magnitude larger than the social sciences. The data growth trend in genomics, however, is greater than in other disciplines. In fact, some researchers have suggested that if the genomics data generation growth trend remains constant, genomics will soon generate more data than applications such as social media, earth sciences, and astronomy [ 27 ].

Data volume growth in genomics versus other disciplines. a Data volume growth in genomics in the context of other domains and data infrastructure (computing power and network throughput). Continuous lines indicate the amount of data archived in public repositories in genomics (SRA), astronomy (Earth Data, NASA), and sociology (Harvard dataverse). Data infrastructure such as computing power ( TOP500 SuperComputers ) and network throughput ( IPTraffic ) are also included. Dashed lines indicate projections of future growth in data volume and infrastructure capacity for the next decade. b Cumulative number of datasets being generated for whole genome sequencing ( WGS ) and whole exome sequencing ( WES ) in comparison with molecular structure datasets such as X-ray and electron microscopy ( EM ). PDB Protein Data Base, SRA Sequence Read Archive
Many strategies have been used to address the increase in data volume in genomics. For example, researchers are now tending to discard primary data (e.g., FASTQ) and prioritizing the storage of secondary data such as compressed mapped reads (BAMs), variant calls (VCFs), or even only quantifications such as gene expression [ 28 ].
In Fig. 2 b, we compare genomics to other data-driven disciplines in the biological sciences. This analysis clearly shows that the large amount of early biological data was not in genomics, but rather in macromolecular structure. Only in 2001, for example, did the number of datasets in genomics finally surpass protein-structure data. More recently, new trends have emerged with the rapidly increasing amount of electron microscopy data, due to the advent of cryo-electron microscopy, and of mass-spectrometry-based proteomics data. Perhaps these trends will shift the balance of biomedical data science in the future.
There are two widely accepted interpretations of data velocity: (i) the speed of data generation (Fig. 2 ) and (ii) the speed at which data are processed and made available [ 29 ].
We explored the growth of data generation in the previous section in relation to genomics. The sequencing of a human genome could soon take less than 24 h, down from 2 to 8 weeks by currently popular technologies and 13 years of uninterrupted sequencing work by the Human Genome Project (HGP) [ 30 ]. Other technologies, such as diagnostic imaging and microarrays, have also experienced remarkable drops in cost and complexity and, therefore, resulting data are much quicker to generate.
The second definition of data velocity speaks to the speed at which data are processed. A remarkable example is the speed of fraud detection during a credit card transaction or some types of high-frequency trading in finance [ 31 ]. In contrast, genomics data and data processing have been traditionally static, relying on fixed snapshots of genomes or transcriptomes. However, new fields leveraging rapid sequencing technologies, such as rapid diagnosis, epidemiology, and microbiome research, are beginning to use nucleic acid sequences for fast, dynamic tracking of diseases [ 32 ] and pathogens [ 33 ]. For these and other near-future technologies, we envision that fast, real-time processing might be necessary.
The description of the volume and velocity of genomics data has great implications for what types of computations are possible. For instance, when looking at the increase of genomics and other types of data relative to network traffic and bandwidth, one must decide whether to store, compute, or transfer datasets. This decision-making process can also be informed by the 3 V framework. In Fig. 2 , we show that the computing power deployed for research and development (using the top 500 supercomputers as a proxy) is growing at a slower pace than genomic data growth. Additionally, while the global web traffic throughput has no foreseeable bottlenecks (Fig. 2 a) [ 34 ], for researchers the costs of transferring such large-scale datasets might hinder data sharing and processing of large-scale genomics projects. Cloud computing is one way of addressing this bottleneck. Large consortia already tend to process and store most of their datasets on the cloud [ 35 , 36 , 37 ]. We believe genomics should consider the viability of public repositories that leverage cloud computing more broadly. At the current rate, the field will soon reach a critical point at which cloud solutions might be indispensable for large-scale analysis.
Genomics data have a two-sided aspect. On one side is the monolithic sequencing data, ordered lists of nucleotides. In human genomics, traditionally these are mapped to the genome and are used to generate coverage or variation data. The monolithic nature of sequencing output, however, hides a much more varied set of assays that are used to measure many aspects of genomes. In Fig. 3 we illustrate this issue by showing the growth in the diversity of sequencing assays over time and displaying a few examples. We also display how different sequencing methods are connected to different omes [ 19 ]. The other side of genomics data is the complex phenotypic data with which the nucleotides are being correlated. Phenotypic data can consist of such diverse entities as simple and unstructured text descriptions from electronic health records, quantitative measurements from laboratories, sensors, and electronic trackers, and imaging data. The varied nature of the phenotypic data is more complicated; as the scale and diversity of sequencing data grow larger, more attention is being paid to the importance of standardizing and scaling the phenotypic data in a complementary fashion. For example, mobile devices can be used to harness large-scale consistent digital phenotypes [ 38 ].
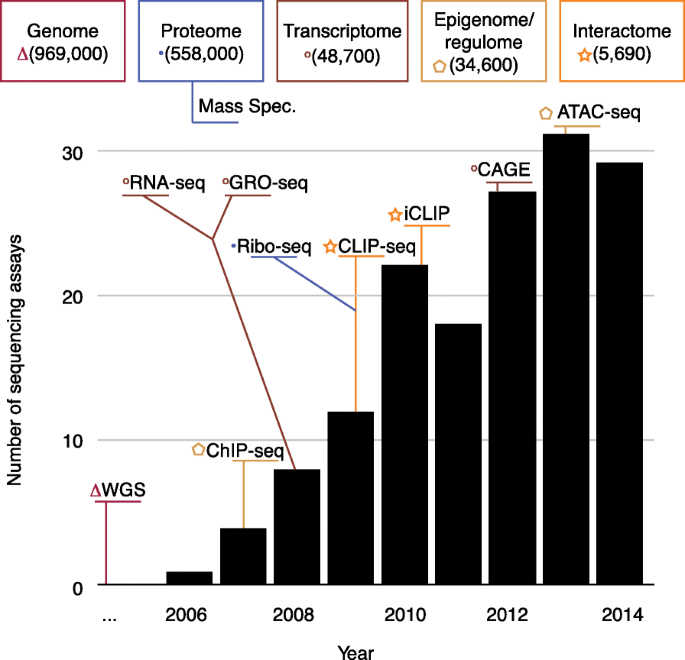
Variety of sequencing assays. Number of new sequencing protocols published per year. Popular protocols are highlighted in their year of publication and their connection to omes
Genomics and the 4 M framework
Two aspects distinguish data science in the natural sciences from social science context. First, in the natural sciences much of the data are quantitative and structured; they often derive from sensor readings from experimental systems and observations under well-controlled conditions. In contrast, data in the social sciences are more frequently unstructured and derived from more subjective observations (e.g., interviews and surveys). Second, the natural sciences also have underlying chemical, physical, and biological models that are often highly mathematized and predictive.
Consequently, data science mining in the natural sciences is intimately associated with mathematical modeling. One succinct way of understanding this relationship is the 4 M framework, developed by Lauffenburger [ 39 ]. This concept describes the overall process in systems biology, closely related to genomics, in terms of (i) Measuring the quantity, (ii) large-scale Mining, which is what we often think of as data science, (3) Modeling the mined observations, and finally (4) Manipulating or testing this model to ensure it is accurate.
The hybrid approach of combining data mining and biophysical modeling is a reasonable way forward for genomics (Fig. 1 b). Integrating physical–chemical mechanisms into machine learning provides valuable interpretability, boosts the data-efficiency in learning (e.g., through training-set augmentation and informative priors) and allows data extrapolation when observations are expensive or impossible [ 40 ]. On the other hand, data mining is able to accurately estimate model parameters, replace some complex parts of the models where theories are weak, and emulate some physical models for computational efficiency [ 41 ].
Short-term weather forecasting as an exemplar of this hybrid approach is perhaps what genomics is striving for. For this discipline, predictions are based on sensor data from around the globe and then fused with physical models. Weather forecasting was, in fact, one of the first applications of large-scale computing in the 1950s [ 42 , 43 ]. However, it was an abject flop, trying to predict the weather solely based on physical models. Predictions were quickly found to only be correct for a short time, mostly because of the importance of the initial conditions. That imperfect attempt contributed to the development of the fields of nonlinear dynamics and chaos, and to the coining of the term “butterfly effect” [ 43 ]. However, subsequent years dramatically transformed weather prediction into a great success story, thanks to integrating physically based models with large datasets measured by satellites, weather balloons, and other sensors [ 43 ]. Moreover, the public’s appreciation for the probabilistic aspects of a weather forecast (i.e., people readily dress appropriately based on a chance of rain) foreshadows how it might respond to probabilistic “health forecasts” based on genomics.
Imports and exports
Thus far, we have analyzed how genomics sits with other data-rich subfields in terms of data (volume, velocity, and variety) and processes. We argue that another aspect of genomics as an applied data science subfield is the frequent exchange of techniques and cultural practices. Over the years, genomics has imported and exported several concepts, practices, and techniques from other applied data science fields. While listing all of the movements is impossible in this piece, we will highlight a few key examples.
Technical imports
A central aspect of genomics—the process of mapping reads to the human reference genome—relies on a foundational technique within data science: fast and memory-efficient string-processing algorithms. Protein pairwise alignment predates DNA sequence alignment. One of the first successful implementations of sequence alignment was based on Smith–Waterman [ 44 ] and dynamic programming [ 45 , 46 ]. These methods were highly reliant on computing power and required substantial memory. With advances in other string-alignment techniques and the explosion of sequencing throughput, the field of genomics saw a surge in the performance of sequence alignment. As most sequencing technologies produce short reads, researchers generated several new methods using index techniques, starting around 2010. Several methods are now based on the Burrows–Wheeler transformation (BWA, bowtie) [ 47 , 48 ], De Bruijn graphs (Kallisto, Salmon) [ 49 , 50 ], and the Maximal Mappable Prefix (STAR) [ 51 ].
Hidden Markov models (HMMs) are well-known algorithms used for modeling the sequential or time-series correlations between symbols or events. HMMs have been widely adopted in fields such as speech recognition and digital communication [ 52 ]. Data scientists also have long used HMMs to smooth a series of events in a varied number of datasets, such as the stock market, text suggestions, and in silico diagnosis [ 53 ]. The field of genomics has applied HMMs to predict chromatin states, annotate genomes, and study ancestry/population genetics [ 54 ]. Figure 4 a displays the adoption of HMMs in genomics compared with other disciplines. It shows that the fraction of HMM papers related to genomics has been growing over time and today it corresponds to more than a quarter of the scientific publications related to the topic.
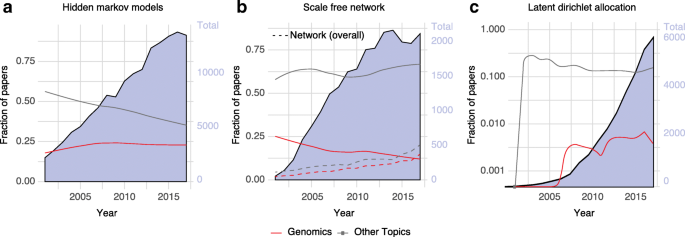
Technical exchanges between genomics and other data science subdisciplines. The background area displays the total number of publications per year for the terms. a Hidden Markov model, b Scale-free network, c latent Dirichlet allocation. Continuous lines indicate the fraction of papers related to topics in genomics and in other disciplines
Another major import into genomics has been network science and, more broadly, graphs. Other subfields have been using networks for many tasks, including algorithm development [ 55 ], social network research [ 56 ], and modeling transportation systems [ 57 ]. Many subfields of genomics rely heavily on networks to model different aspects of the genome and subsequently generate new insights [ 58 ]. One of the first applications of networks within genomics and proteomics was protein–protein interaction networks [ 59 ]. These networks are used to describe the interaction between several protein(s) and protein domains within a genome to ultimately infer functional pathways [ 60 ]. After the development of large-scale transcriptome quantification and chromatin immunoprecipitation sequencing (ChIP-Seq), researchers built regulatory networks to describe co-regulated genes and learn more about pathways and hub genes [ 61 ]. Figure 4 b shows the usage of “scale-free networks” and “networks” as a whole. While the overall use of networks has continued to grow in popularity in genomics after their introduction, the specific usage of scale-free has been falling, reflecting the brief moment of popularity of this concept.
Given the abundance of protein structures and DNA sequences, there has been an influx of deep-learning solutions imported from machine learning [ 62 ]. Many neural network architectures can be transferred to biological research. For example, the convolutional neural network (CNN) is widely applied in computer vision to detect objects in a positional invariant fashion. Similarly, convolution kernels in CNN are able to scan biological sequences and detect motifs, resembling position weight matrices (PWMs). Researchers are developing intriguing implementations of deep-learning networks to integrate large datasets, for instance, to detect gene homology [ 63 ], annotate and predict regulatory regions in the genome [ 64 ], predict polymer folding [ 65 ], predict protein binding [ 66 ], and predict the probability of a patient developing certain diseases from genetic variants [ 67 ]. While neural networks offer a highly flexible and powerful tool for data mining and machine learning, they are usually “black box” models and often very difficult to interpret.
Cultural imports
The exchanges between genomics and other disciplines are not limited to methods and techniques, but also include cultural practices. As a discipline, protein-structure prediction pioneered concepts such as the Critical Assessment of protein Structure Prediction (CASP) competition format. CASP is a community-wide effort to evaluate predictions. Every 2 years since 1994, a committee of researchers has selected a group of proteins for which hundreds of research groups around the world will (i) experimentally describe and (ii) predict in silico its structure. CASP aims to determine the state of the art in modeling protein structure from amino acid sequences [ 68 ]. After research groups submit their predictions, independent assessors compare the models with the experiments and rank methods. In the most recent instantiation of CASP, over 100 groups submitted over 50,000 models for 82 targets. The success of the CASP competition has inspired more competitions in the biological community, including genomics. DREAM Challenges, for example, have played a leading role in organizing and catalyzing data-driven competitions to evaluate the performance of predictive models in genomics. Challenge themes have included “Genome-Scale Network Inference”, “Gene Expression Prediction”, “Alternative Splicing”, and “in vivo Transcription Factor Binding Site Prediction” [ 69 ]. DREAM Challenges was initiated in 2006, shortly before the well-known Netflix Challenge and the Kaggle platform, which were instrumental in advancing machine-learning research [ 70 ].
Technical exports
A few methods exported from genomics to other fields were initially developed to address specific biological problems. However, these methods were later generalized for a broader set of applications. A notable example of such an export is the latent Dirichlet allocation (LDA) model. Pritchard et al. [ 71 ] initially proposed this unsupervised generative model to find a group of latent processes that, in combination, can be used to infer and predict individuals’ population ancestry based on single nucleotide variants. Blei, Ng, and Jordan [ 72 ] independently proposed the same model to learn the latent topics in natural language processing (NLP). Today, LDA and its countless variants have been widely adapted in, for example, text mining and political science. In fact, when we compare genomics with other topics such as text mining we observe that genomics currently accounts for a very small percentage of work related to LDA (Fig. 4 c).
Genomics has also contributed to new methods of data visualization. One of the best examples is the Circos plot [ 73 ], which is related to the import above of network science. Circos was initially conceptualized as a circular representation of linear genomes. In its conception, this method displayed chromosomal translocations or large syntenic regions. As this visualization tool evolved to display more generic networks , it was also used to display highly connected datasets. In particular, the media has used Circos to display and track customer behavior, political citations, and migration patterns [ 73 ]. In genomics, networks and graphs are also being used in order to represent the human genome. For instance, researchers are attempting to represent the reference genome and its variants as a graph [ 74 ].
Another prominent idea exported from genomics is the notion of family classification based on large-scale datasets. This derives from the biological taxonomies dating back to Linnaeus, but also impacts the generation of protein and gene family databases [ 75 , 76 ]. Other disciplines, for example, linguistics and neuroimaging, have also addressed similar issues by constructing semantic and brain region taxonomies [ 77 , 78 ]. This concept has even made its way into pop culture; for example, Pandora initially described itself as the music genome project [ 79 ]. Another example is the art genome project [ 80 ], which maps characteristics (referred to as “genes”) that connect artists, artworks, architecture, and design objects across history.
Cultural exports
Genomics has also tested and exported several cultural practices that can serve as a model for other data-rich disciplines [ 81 ]. On a fundamental level, these practices promote data openness and re-use, which are central issues to data science disciplines.
Most genomics datasets, and most prominently datasets derived from sequencing, are frequently openly accessible to the public. This practice is evidenced by the fact that most genomics journals require a public accession identifier for any dataset associated with a publication. This broad adoption of data openness is perhaps a reflection of how genomics evolved as a discipline. Genomics mainly emerged after the conclusion of HGP—a public initiative that, at its core, was dedicated to release a draft of the human genome that was not owned or patented by a company. It is also notable that the public effort was in direct competition with a private effort by Celera Genomics, which aimed to privatize and patent sections of the genome. Thus, during the development of the HGP, researchers elaborated the Bermuda principles, a set of rules that called for public releases of all data produced by HGP within 24 h of generation [ 82 ]. The adoption of the Bermuda principles had two main benefits for genomics. First, it facilitated the exchange of data between many of the dispersed researchers involved in the HGP. Second, perhaps due to the central role of the HGP, it spurred the adoption of open-data frameworks more broadly. In fact, today most large projects in genomics adopt Bermuda-like standards. For example, the 1000 Genomes [ 83 ] and the ENCODE [ 35 ] projects release their datasets openly before publication to allow other researchers to use their datasets [ 84 ]. Other subfields such as neuroscience (e.g., the human connectome) were also inspired by the openness and setup of the genomics community [ 81 ].
In order to attain a broad distribution of open datasets, genomics has also adopted the usage of central, large-scale public dataset repositories. Unlike several other applied fields, genomics data are frequently hosted on free and public platforms. The early adoption of these central dataset resources, such as the Sequence Read Archive (SRA), European Nucleotide Archive (ENA), GenBank, and Protein Data Base (PDB), to host large amounts of all sorts of genetics data, including microarray and sequencing data, has allowed researchers to easily query and promote re-use datasets produced by others [ 85 ].
The second effect of these large-scale central dataset repositories, such as the National Center for Biotechnology Information (NCBI) and ENA, is the incentive for early adoption of a small set of standard data formats. This uniformity of file formats encouraged standardized and facilitated access to genomics datasets. Most computations in genomics data are hosted as FASTA/FASTQ, BED, BAM, VCF, or bigwig files, which respectively represent sequences, coordinates, alignments, variants, and coverage of DNA or amino acid sequences. Furthermore, as previously discussed, the monolithic nature of genomic sequences also contributes to the standardization of pipelines and allows researchers to quickly test, adapt, and switch to other methods using the same input format [ 86 ].
The open-data nature of many large-scale genomics projects may also have spurred the adoption of open-source software within genomics. For example, most genomics journals require public links to source codes to publish in silico results or computational methods. To evaluate the adoption of open source in genomics, we used the growth of GitHub repositories and activity (commits) over time (Fig. 5 ). Compared with many fields of similar scale (e.g., astronomy and ecology) genomics has a particularly large representation on GitHub and this is growing rapidly.
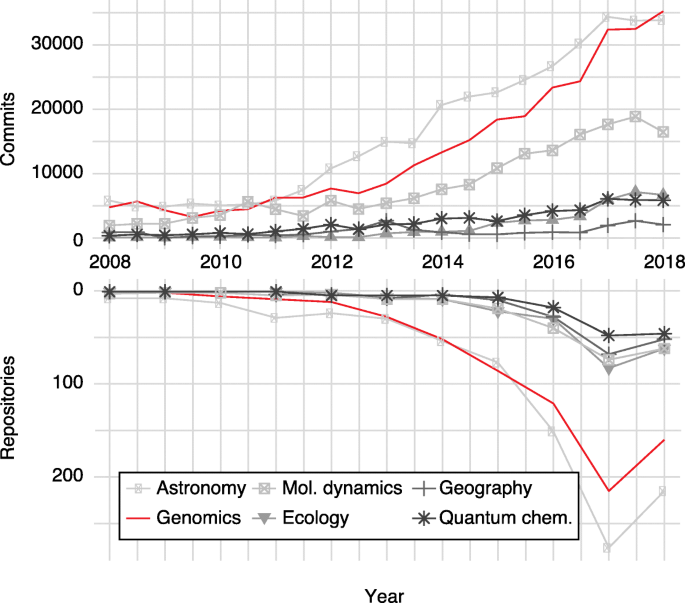
Open source adoption in genomics and other data science subdisciplines. The number of GitHub commits ( upper panel ) and new GitHub repositories ( lower panel ) per year for a variety of subfields. Subfield repositories were selected by GitHub topics such as genomics, astronomy, geography, molecular dynamics ( Mol. Dynamics ), quantum chemistry ( Quantum Chem. ), and ecology
Data science issues with which genomics is grappling
In closing, we consider the issues that genomics and, more broadly, data science face both now and in the future. One of the major issues related to data science is privacy. Indeed, the current privacy concerns related to email, financial transactions, and surveillance cameras are critically important to the public [ 87 ]. The potential to cross-reference large datasets (e.g., via quasi-identifiers) can make privacy leaks non-intuitive [ 70 ]. Although genomics-related privacy overlaps with data science-related privacy, the former has some unique aspects given that the genome is passed down through generations and is fundamentally important to the public [ 88 ]. Leaking genomic information might be considered more damaging than leaking other types of information. Although we may not know everything about the genome today, we will know much more in 50 years. At that time, a person would not be able to take their or their children’s variants back after they have been released or leaked [ 88 ]. Finally, genomic data are considerably larger in scale than many other bits of individual information; that is, the genome carries much more individual data than a credit card or social security number. Taken together, these issues make genomic privacy particularly problematic.
However, in order to carry out several types of genomic calculations, particularly for phenotypic associations like genome-wide association studies, researchers can get better power and a stronger signal by using larger numbers of data points (i.e., genomes). Therefore, sharing and aggregating large amounts of information can result in net benefits to the group even if the individual’s privacy is slightly compromised. The Global Alliance for Genomics and Health (GA4GH) has made strides in developing technical ways to balance the concerns of individual privacy and social benefits of data sharing [ 89 ]. This group has discussed the notion of standardized consents associated with different datasets. The fields of security and privacy are undertaking projects like homomorphic encryption, where one can make certain calculations on an encrypted dataset without accessing its underlying contents [ 90 ].
Data ownership
Privacy is an aspect of a larger issue of data ownership and control. Although the individual or patient typically is thought to own their personal data, a countervailing trend in biomedical research is the idea that the researcher who generates a dataset owns it. There is a longstanding tradition among researchers who have generated large datasets to progressively analyze their data over the course of several papers, even a career, to extract interesting stories and discoveries [ 91 ]. There is also the notion that human data, particularly health data, have obvious medical and commercial value, and thus companies and nations often seek ownership and control over large datasets.
From the data miner’s perspective, all information should be free and open, since such a practice would lead to the easy aggregation of a large amount of information, the best statistical power, and optimally mined results. Intuitively, aggregating larger datasets will, most frequently, give progressively better genotypes being associated to phenotypes.
Furthermore, even in an ideal scenario in which individuals consent to free access and the resulting dataset is completely open and freely shared by users, we imagine complications will arise from collection and sharing biases such as particular cohort ethnicity, diseases, and phenotypes being more open to share their genetic data. Socioeconomic status, education, and access to healthcare can all possibly cause skew in datasets, which would further bias mining efforts such as machine learning algorithms and knowledge extraction. For example, ImageNet, a heavily used dataset in image classification, has nearly half of the images coming from the USA. Similarly, about 80% of genome-wide association study catalog participants are of European descent, a group which only makes up 16% of the world population [ 92 ].
For this reason, completely open data sharing will probably not be reasonable for the best future genomic association studies. One possible technical solution for sharing genomics data might be the creation of a massive private enclave. This is very different from the World Wide Web, which is fundamentally a public entity. A massive private enclave would be licensed only to certified biomedical researchers to enable data sharing and provide a way to centralize the storage and computation of large datasets for maximum efficiency. We believe this is the most practical viewpoint going forward.
On the other hand, the positive externality of data sharing behaviors will become more significant as genomic science develops and becomes more powerful in aggregating and analyzing data. We believe that, in the future, introducing data property rights, Pigouvian subsidies, and regulation may be necessary to encourage a fair and efficient data trading and use environment. Furthermore, we imagine a future where people will grapple with complex data science issues such as sharing limited forms of data within certain contexts and pricing of data accordingly.
Lastly, data ownership is also associated with extracting profit and credit from the data. Companies and the public are realizing that the value of data does not only come from generating it per se, but also from analyzing the data in meaningful and innovative new ways. We need to recognize the appropriate approaches to not only recognize the generation of the data but also to value the analysis of large amounts of data and appropriately reward analysts as well as data generators.
In this piece, we have described how genomics fits into the emergence of modern data science. We have characterized data science as an umbrella term that is increasingly connecting disparate application subdisciplines. We argue that several applied subdisciplines considerably predate formal data science and, in fact, were doing large-scale data analysis before it was “cool”. We explore how genomics is perhaps the most prominent biological science discipline to connect to data science. We investigate how genomics fits in with many of the other areas of data science, in terms of its data volume, velocity, and variety. Furthermore, we discuss how genomics may be able to leverage modeling (both physical and biological) to enhance predictive power, similar in a sense to what has been achieved in weather forecasting. Finally, we discuss how many data science ideas have been both imported to and exported from genomics. In particular, we explore how the HGP might have inspired many cultural practices that led to large-scale adoption of open-data standards.
We conclude by exploring some of the more urgent issues related to data, and how they are impacting data in genomics and other disciplines. Several of these issues do not relate to data analytics per se but are associated with the flow of data. In particular, we discuss how individual privacy concerns, more specifically data ownership, are central issues in many data-rich fields, and especially in genomics. We think grappling with several of these issues of data ownership and privacy will be central to scaling genomics to an even greater size in the future.
Abbreviations
Critical Assessment of Protein Structure Prediction
Convolutional Neural Network
European Nucleotide Archive
Human Genome Project
Hidden Markov model
Latent Dirichlet allocation
Davenport TH, Patil DJ. Data scientist: the sexiest job of the 21st century. Harv Bus Rev. 2012;90:70–6.
PubMed Google Scholar
Provost F, Fawcett T. Data science and its relationship to big data and data-driven decision making. Big Data. 2013;1:51–9.
Article PubMed Google Scholar
Tukey JW. The future of data analysis. Ann Math Stat. 1962;33:1–67.
Article Google Scholar
Tansley S, Tolle KM. The fourth paradigm: Microsoft Press; 2009.
Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015;349:255–60.
Article CAS PubMed Google Scholar
Fienberg SE. A brief history of statistics in three and one-half chapters: a review essay. Stat Sci. 1992;7:208–25.
Robert C, Casella G. A short history of Markov chain Monte Carlo: subjective recollections from incomplete data. Stat Sci. 2011;26:102–15.
Lee TB, Cailliau R, Groff JF, Pollermann B. World-wide web: the information universe. Internet Res. 2013;2:52–8.
Google Scholar
Kodama Y, Shumway M, Leinonen R. International nucleotide sequence database collaboration. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–6.
Hey T, Trefethen A. The data deluge: an e-science perspective. In: Berman F, Fox G, Hey T, editors. Grid computing: making the global infrastructure a reality. Chichester: Wiley-Blackwell; 2003. p. 809–24.
Chapter Google Scholar
Jaschek C. Data in astronomy. Cambridge: Cambridge University Press; 1989.
Cox DR. Analysis of binary data. New York: Routledge; 1970.
Blashfield RK, Aldenderfer MS. The methods and problems of cluster analysis. In: Nesselroade JR, Cattell RB, editors. Handbook of multivariate experimental psychology. Boston: Springer; 1988. p. 447–73.
Belson WA. Matching and prediction on the principle of biological classification. App Stat. 1959;8:65.
McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biol. 1943:99–115 discussion 73–97.
Shannon CE. An algebra for theoretical genetics. PhD thesis. Cambridge: Massachusetts Institute of Technology; 1940.
Kuska B. Beer, Bethesda, and biology: how “genomics” came into being. J Natl Cancer Inst. 1998;90:93.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51.
Greenbaum D, Luscombe NM, Jansen R, Qian J, Gerstein M. Interrelating different types of genomic data, from proteome to secretome: ‘oming in on function. Genome Res. 2001;11:1463–8.
Eisen JA. Badomics words and the power and peril of the ome-meme. Gigascience. 2012;1:6.
Article PubMed PubMed Central Google Scholar
Cheng Y. Single-particle cryo-EM – how did it get here and where will it go. Science. 2018;361:876–80.
Article CAS PubMed PubMed Central Google Scholar
Althoff T, Sosič R, Hicks JL, King AC, Delp SL, Leskovec J. Large-scale physical activity data reveal worldwide activity inequality. Nature. 2017;547:336–9.
Wamba SF, Akter S, Edwards A, Chopin G, Gnanzou D. How “big data” can make big impact: findings from a systematic review and a longitudinal case study. Int J Prod Econ. 2015;165:234–46.
McAfee A, Brynjolfsson E. Big data: the management revolution. Harv Bus Rev. 2012;90:61–7.
White M. Digital workplaces: vision and reality. Bus Inf Rev. 2012;29:205–14.
NASA. https://earthdata.nasa.gov . Accessed 10 May 2019.
Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al. Big Data: astronomical or genomical? PLoS Biol. 2015;13:e1002195.
Article PubMed PubMed Central CAS Google Scholar
Marx V. Biology: The big challenges of big data. Nature. 2013;498:255–60.
Zikopoulos P, Eaton C. IBM. Understanding big data: analytics for enterprise class hadoop and streaming data. India: McGraw-Hill; 2011.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
Gandomi A, Haider M. 2015. Beyond the hype: big data concepts, methods, and analytics. Int J Inf. 2015;35:137–44.
Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135.
Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–32.
Cisco Visual Networking Index: forecast and trends, 2017–2022 White Paper. 2018. https://www.cisco.com/c/en/us/solutions/collateral/service-provider/visual-networking-index-vni/white-paper-c11-741490.html . Accessed 10 May 2019.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Article CAS Google Scholar
Campbell PJ, Getz G, Stuart JM, Korbel JO, Stein LD. ICGC/TCGA Pan-Cancer analysis of whole genomes net. Pan-cancer analysis of whole genomes. BioRxiv. 2018:1–29.
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73.
Onnela J-P, Rauch SL. Harnessing smartphone-based digital phenotyping to enhance behavioral and mental health. Neuropsychopharmacology. 2016;41:1691–6.
Ideker T, Winslow LR, Lauffenburger DA. Bioengineering and systems biology. Ann Biomed Eng. 2006;34:1226–33.
Reichstein M, Camps-Valls G, Stevens B, Jung M, Denzler J, Carvalhais N, et al. Deep learning and process understanding for data-driven earth system science. Nature. 2019;566:195–204.
Artificial intelligence alone won't solve the complexity of Earth sciences [Comment]. Nature. 2019;566:153.
Murphy AH. The early history of probability forecasts: some extensions and clarifications. Wea Forecasting. 1998;13:5–15.
Bauer P, Thorpe A, Brunet G. The quiet revolution of numerical weather prediction. Nature. 2015;525:47–55.
Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–7.
Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–41.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Li H, Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009;25:1754–60.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nature. 2012;9:357–9.
CAS Google Scholar
Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Gales M, Young S. The application of hidden Markov models in speech recognition. FNT in Signal Processing. 2007;1:195–304.
Gagniuc PA. Markov chains. Hoboken: John Wiley; 2017.
Book Google Scholar
Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–63.
Mealy GH. A method for synthesizing sequential circuits. Bell Syst Tech J. 1955;34:1045–79.
Ediger D, Jiang K, Riedy J, Bader DA, Corley C. Massive social network analysis: mining twitter for social good. 2010. 39th International Conference on Parallel Processing (ICPP) IEEE; p 583–593.
Guimera R, Mossa S, Turtschi A, Amaral LA. The worldwide air transportation network: anomalous centrality, community structure, and cities’ global roles. Proc Natl Acad Sci U S A. 2005;102:7794–9.
McGillivray P, Clarke D, Meyerson W, Zhang J, Lee D, Gu M, et al. Network analysis as a grand unifier in biomedical data science. Annu Rev Biomed Data Sci. 2018;1:153–80.
Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52.
Marbach D, Costello JC, Küffner R, Vega NM, Prill RJ, Camacho DM, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804.
Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–55.
Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. A primer on deep learning in genomics. Nature. 2018;12:878.
Hochreiter S, Heusel M, Obermayer K. Fast model-based protein homology detection without alignment. Bioinformatics. 2007;23:1728–36.
Jia C, He W. EnhancerPred: a predictor for discovering enhancers based on the combination and selection of multiple features. Sci Rep. 2016;6:38741.
Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, Wang J, et al. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Sci Rep. 2015;5:11476.
Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–8.
Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362:eaat8464.
Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23:ii–v.
Prill RJ, Marbach D, Saez-Rodriguez J, Sorger PK, Alexopoulos LG, Xue X, et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS One. 2010;5:e9202.
Narayanan A, Shi E, Rubinstein BIP. Link prediction by de-anonymization: how we won the Kaggle Social Network Challenge. 2011 International Joint Conference on Neural Networks (IJCNN 2011, San Jose). IEEE; p. 1825–34.
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59.
CAS PubMed PubMed Central Google Scholar
Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45.
Paten B, Novak AM, Eizenga JM, Garrison E. Genome graphs and the evolution of genome inference. Genome Res. 2017;27:665–76.
Schreiber F, Patricio M, Muffato M, Pignatelli M, Bateman A. TreeFam v9: a new website, more species and orthology-on-the-fly. Nucleic Acids Res. 2014;42:D922–5.
Lam HYK, Khurana E, Fang G, Cayting P, Carriero N, Cheung K-H, et al. Pseudofam: the pseudogene families database. Nucleic Acids Res. 2009;37:D738–43.
Panagiotaki E, Schneider T, Siow B, Hall MG, Lythgoe MF, Alexander DC. Compartment models of the diffusion MR signal in brain white matter: a taxonomy and comparison. Neuroimage. 2012;59:2241–54.
Ponzetto SP, Strube M. Deriving a large-scale taxonomy from Wikipedia. Proceedings of the National Conference on Artificial Intelligence, 2007. Palo Alto: Association for the Advancement of Artificial Intelligence; 2007. p. 440–5.
Prockup M, Ehmann AF, Gouyon F, Schmidt EM, Kim YE. Modeling musical rhythmatscale with the music genome project. 2015 IEEE workshop on applications of signal processing to audio and acoustics (WASPAA). Piscataway: IEEE; 2015. p. 1–5.
Artsy. www.artsy.net . Accessed 10 May 2019.
Choudhury S, Fishman JR, McGowan ML, Juengst ET. Big data, open science and the brain: lessons learned from genomics. Front Hum Neurosci. 2014;8:239.
Cook-Deegan R, Ankeny RA, Maxson Jones K. Sharing data to build a medical information commons: from Bermuda to the global alliance. Annu Rev Genomics Hum Genet. 2017;18:389–415.
1000 Genomes Project Consortium, Auton A, Brooks LD, Garrison EP, Kang HM, Marchini JL, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Wang D, Yan K-K, Rozowsky J, Pan E, Gerstein M. Temporal dynamics of collaborative networks in large scientific consortia. Trends Genet. 2016;32:251–3.
Article PubMed CAS Google Scholar
Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14:89–99.
Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–8.
Acquisti A, Gross R. Imagined communities: awareness, information sharing, and privacy on the Facebook. In: Danezis G, Golle P, editors. Privacy enhancing technologies. PET 2006. Lecture notes in computer science, vol 4258. Berlin: Springer; 2006. p. 36–58.
Greenbaum D, Sboner A, Mu XJ, Gerstein M. Genomics and privacy: implications of the new reality of closed data for the field. PLoS Comput Biol. 2011;7:e1002278.
Knoppers BM. International ethics harmonization and the global alliance for genomics and health. Genome Med. 2014;6:13.
Erlich Y, Narayanan A. Routes for breaching and protecting genetic privacy. Nat Rev Genet. 2014;15:409–21.
Longo DL, Drazen JM. Data sharing. N Engl J Med. 2016;374:276–7.
Zou J, Schiebinger L. AI can be sexist and racist – it's time to make it fair. Nature. 2018;559:324–6.
Download references
The authors acknowledge the generous funding from the US National Science Foundation DBI 1660648 for MBG.
Author information
Authors and affiliations.
Program in Computational Biology and Bioinformatics, Yale University, Bass 432, 266 Whitney Avenue, New Haven, CT, 06520, USA
Fábio C. P. Navarro, Hussein Mohsen, Chengfei Yan, Mengting Gu, William Meyerson & Mark Gerstein
Department of Molecular Biophysics and Biochemistry, Yale University, Bass 432, 266 Whitney Avenue, New Haven, CT, 06520, USA
Department of Computer Science, Yale University, Bass 432, 266 Whitney Avenue, New Haven, CT, 06520, USA
Mark Gerstein
Department of Statistics and Data Science, Yale University, Bass 432, 266 Whitney Avenue, New Haven, CT, 06520, USA
Department of Computer Science, Stanford University, Stanford, CA, 94305, USA
Department of Biomedical Data Sciences, Stanford University, Stanford, CA, 94305, USA
You can also search for this author in PubMed Google Scholar
Contributions
FCPN and MBG conceived and planned the study, prepared the figures, and wrote the manuscript. HM prepared the figures and wrote the manuscript. CY, SL, MG, and WM collected data and wrote the manuscript. All authors discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Mark Gerstein .
Ethics declarations
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License ( http://creativecommons.org/licenses/by/4.0/ ), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
Reprints and permissions
About this article
Cite this article.
Navarro, F.C.P., Mohsen, H., Yan, C. et al. Genomics and data science: an application within an umbrella. Genome Biol 20 , 109 (2019). https://doi.org/10.1186/s13059-019-1724-1
Download citation
Published : 29 May 2019
DOI : https://doi.org/10.1186/s13059-019-1724-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
EDITORIAL article
Editorial: the genetics and epigenetics of mental health.

- 1 Instituto de Pesquisa Pelé Pequeno Príncipe, Curitiba, Brazil
- 2 Faculdades Pequeno Príncipe, Curitiba, Brazil
- 3 Department of Genetics, Federal University of Parana, Post-graduation Program in Genetics, Curitiba, Brazil
- 4 Translational Research in Respiratory Medicine, Hospital Universitari Arnau de Vilanova-Santa Maria, Biomedical Research Institute of Lleida (IRBLleida), Lleida, Spain
- 5 CIBER of Respiratory Diseases (CIBERES), Institute of Health Carlos III, Madrid, Spain
Editorial on the Research Topic The genetics and epigenetics of mental health
Mental health conditions cover a broad spectrum of disturbances, including neurological and substance use disorders, suicide risk, and associated psychosocial, cognitive, and intellectual disabilities (WHO, 2022). Despite a substantial amount of evidence, the interaction of genetic variants, epigenetic mechanisms, and environmental risk factors involved in mental health is poorly understood. Through distinct perspectives and different experimental approaches, the genetics and epigenetics of mental health were addressed in seven relevant articles included in this Research Topic, briefly summarized below.
Stress has severe consequences on the epigenome, but the timing of its occurrence, as well as the intensity and number of events, are critical for the severity of mental health symptoms. In particular, Serpeloni et al. demonstrated that stress generated in the form of intimate partner violence (IPV) during and/or after pregnancy impacts the offspring’s epigenome, shaping its resilience. They observed that individuals exposed to maternal IPV after birth presented psychiatric issues similar to their mothers, with different outcomes if the exposure to maternal IPV occurred both prenatally and postnatally. Prenatal IPV was associated with differential methylation in CpG sites in the genes encoding the glucocorticoid receptor ( NR3C1 ) and its repressor FKBP51 ( FKBP5 ), associated with the ability to terminate hormonal stress responses. Also considering early-life experiences and data from 2008 to 2016 of the Health and Retirement Study, Shin et al. concluded that early life experiences and relationships have a significant influence, attenuating or exacerbating the risk of suffering from mental health problems among individuals with a higher polygenic risk score predisposing to autism.
Environmental and developmental factors are also strongly linked to obsessive-compulsive disorder (OCD). They may explain the apparent discrepancy between the relatively high heritability scores and the inconsistent results found in genetic association studies, owing to their impact on gene expression and regulation. Based on this, Deng et al. stratified OCD patients by the age of disease onset. The findings revealed associations between the early onset and variants of genes whose products play a role in neural development, corroborating the age-associated genetic heterogeneity of OCD.
Further exploring environmental and genetic etiological clues, Li et al. used genome-wide association study (GWAS) data to calculate polygenic risk scores for salivary and tongue dorsum microbiomes associated with anxiety and depression. Additionally, causal relationships between the oral microbiome, anxiety, and depression were detected through Mendelian randomization, unraveling potential pathogenic mechanisms and interventional targets. Constructing a similar line of evidence, Becerra et al. found associations between the epigenetic regulation of inflammatory processes, the composition of gut microbiome, and modified Rosenberg self-esteem scores in samples from the Native Hawaiian and other Pacific Islander (NHPI) populations, which present a high prevalence and mortality from chronic and immunometabolic diseases, as well as mental health problems. This warrants further investigation into the relationship of microbiota to brain activity and mental health.
There is a lot of debate regarding suicidal behavior and its relationship with psychiatric disorders, but the extent to which they share the same genetic architecture is unknown. This Research Topic was investigated by Kootbodien et al. through the use of genomic structural equation modeling and Mendelian randomization with a large genomic dataset. The authors observed a strong genetic correlation between suicidal ideation, attempts, and self-harm, as well as a moderate to strong genetic correlation between suicidal behavioral traits and a range of psychiatric disorders, most notably major depressive disorder, involving pathways related to developmental biology, signal transduction, and RNA degradation. In conclusion, the study provided evidence of a shared etiology between suicidal behavior and psychiatric disorders, with overlapping pathophysiological pathways.
Malekpour et al. , in their investigation of psychogenic non-epileptic seizures (PNES), also uncovered shared pathways with psychiatric conditions. PNES, the most prevalent non-epileptic disorder among patients referring to epilepsy centers, carries a mortality rate akin to drug-resistant epilepsy. Employing a systems biology approach, the authors pinpointed several key components influencing the disease pathogenesis network. These include brain-derived neurotrophic factor (BDNF), cortisol, norepinephrine, proopiomelanocortin (POMC), neuropeptide Y (NPY), the growth hormone receptor signaling pathway, phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling, and the neurotrophin signaling pathway.
In general, these studies have some limitations: small sample sizes, leading to low statistical power in some cases, environmental confounding factors (such as diet and physical activity), which were not considered in the microbiome studies, incomplete phenotype descriptions, and partial coverages of human genetic diversity. Childhood adversities and adult comorbidities are among the variables that were not controlled for as possible causes of the investigated psychiatric and neurological disorders, and some results still claim for functional studies to be validated. Thus, the findings brought more elaborated questions, each of which shed some light on knowledge gaps that remain very difficult to fill. How do early-life epigenetic processes regulate our mental health resilience and disease resistance? What is the role of the microbiome in this process and how do genetic variants influence its composition? How does the impact of all these elements shape the resistance of human populations to psychiatric and neurological diseases and, most importantly, translate into public health measures in the future? We hope to engage more researchers in the pursuit of these answers.
Author contributions
GCK: Conceptualization, Data curation, Writing–original draft, Writing–review and editing. ABWB: Writing–original draft, Writing–review and editing. ADST: Conceptualization, Data curation, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Empresa Brasileira de Serviços Hospitalares (Ebserh) grant numbers 423317/2021-0 and 313741/2021-2 (8520137521584230), Research for the United Health SUS System (PPSUS-MS), CNPq, Fundação Araucária and SESA-PR, Protocol N°: SUS2020131000106. ABWB receives CNPq research productivity scholarships (protocols 313741/2021). ADST receives financial support from Instituto de Salud Carlos III (Miguel Servet, 2023: CP23/00095), co-funded by Fondo Social Europeo Plus (FSE+).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: methylation, GWAS-genome-wide association study, microbiome & dysbiosis, poligenic risk score, neurological conditions, epigenome, genome
Citation: Kretzschmar GC, Boldt ABW and Targa ADS (2024) Editorial: The genetics and epigenetics of mental health. Front. Genet. 15:1402495. doi: 10.3389/fgene.2024.1402495
Received: 17 March 2024; Accepted: 26 March 2024; Published: 09 April 2024.
Edited and reviewed by:
Copyright © 2024 Kretzschmar, Boldt and Targa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriela Canalli Kretzschmar, [email protected] ; Angelica Beate Winter Boldt, [email protected] ; Adriano D. S. Targa, [email protected]
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Advertisement
A review on genetic algorithm: past, present, and future
- Published: 31 October 2020
- Volume 80 , pages 8091–8126, ( 2021 )
Cite this article

- Sourabh Katoch 1 ,
- Sumit Singh Chauhan 1 &
- Vijay Kumar ORCID: orcid.org/0000-0002-3460-6989 1
152k Accesses
1758 Citations
14 Altmetric
Explore all metrics
In this paper, the analysis of recent advances in genetic algorithms is discussed. The genetic algorithms of great interest in research community are selected for analysis. This review will help the new and demanding researchers to provide the wider vision of genetic algorithms. The well-known algorithms and their implementation are presented with their pros and cons. The genetic operators and their usages are discussed with the aim of facilitating new researchers. The different research domains involved in genetic algorithms are covered. The future research directions in the area of genetic operators, fitness function and hybrid algorithms are discussed. This structured review will be helpful for research and graduate teaching.
Similar content being viewed by others

Evolutionary algorithms and their applications to engineering problems

Genetic algorithms: theory, genetic operators, solutions, and applications

A tutorial on multiobjective optimization: fundamentals and evolutionary methods
Avoid common mistakes on your manuscript.
1 Introduction
In the recent years, metaheuristic algorithms are used to solve real-life complex problems arising from different fields such as economics, engineering, politics, management, and engineering [ 113 ]. Intensification and diversification are the key elements of metaheuristic algorithm. The proper balance between these elements are required to solve the real-life problem in an effective manner. Most of metaheuristic algorithms are inspired from biological evolution process, swarm behavior, and physics’ law [ 17 ]. These algorithms are broadly classified into two categories namely single solution and population based metaheuristic algorithm (Fig. 1 ). Single-solution based metaheuristic algorithms utilize single candidate solution and improve this solution by using local search. However, the solution obtained from single-solution based metaheuristics may stuck in local optima [ 112 ]. The well-known single-solution based metaheuristics are simulated annealing, tabu search (TS), microcanonical annealing (MA), and guided local search (GLS). Population-based metaheuristics utilizes multiple candidate solutions during the search process. These metaheuristics maintain the diversity in population and avoid the solutions are being stuck in local optima. Some of well-known population-based metaheuristic algorithms are genetic algorithm (GA) [ 135 ], particle swarm optimization (PSO) [ 101 ], ant colony optimization (ACO) [ 47 ], spotted hyena optimizer (SHO) [ 41 ], emperor penguin optimizer (EPO) [ 42 ], and seagull optimization (SOA) [ 43 ].
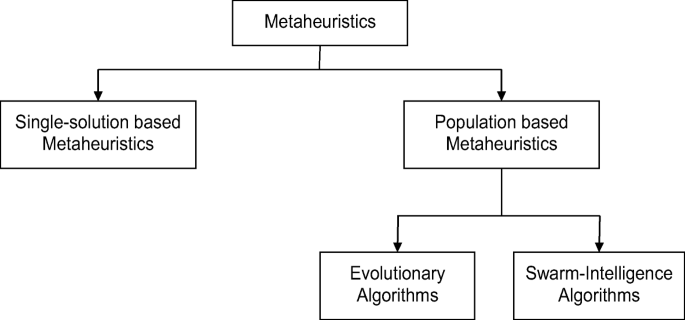
Classification of metaheuristic Algorithms
Among the metaheuristic algorithms, Genetic algorithm (GA) is a well-known algorithm, which is inspired from biological evolution process [ 136 ]. GA mimics the Darwinian theory of survival of fittest in nature. GA was proposed by J.H. Holland in 1992. The basic elements of GA are chromosome representation, fitness selection, and biological-inspired operators. Holland also introduced a novel element namely, Inversion that is generally used in implementations of GA [ 77 ]. Typically, the chromosomes take the binary string format. In chromosomes, each locus (specific position on chromosome) has two possible alleles (variant forms of genes) - 0 and 1. Chromosomes are considered as points in the solution space. These are processed using genetic operators by iteratively replacing its population. The fitness function is used to assign a value for all the chromosomes in the population [ 136 ]. The biological-inspired operators are selection, mutation, and crossover. In selection, the chromosomes are selected on the basis of its fitness value for further processing. In crossover operator, a random locus is chosen and it changes the subsequences between chromosomes to create off-springs. In mutation, some bits of the chromosomes will be randomly flipped on the basis of probability [ 77 , 135 , 136 ]. The further development of GA based on operators, representation, and fitness has diminished. Therefore, these elements of GA are focused in this paper.
The main contribution of this paper are as follows:
The general framework of GA and hybrid GA are elaborated with mathematical formulation.
The various types of genetic operators are discussed with their pros and cons.
The variants of GA with their pros and cons are discussed.
The applicability of GA in multimedia fields is discussed.
The main aim of this paper is two folds. First, it presents the variants of GA and their applicability in various fields. Second, it broadens the area of possible users in various fields. The various types of crossover, mutation, selection, and encoding techniques are discussed. The single-objective, multi-objective, parallel, and hybrid GAs are deliberated with their advantages and disadvantages. The multimedia applications of GAs are elaborated.
The remainder of this paper is organized as follows: Section 2 presents the methodology used to carry out the research. The classical genetic algorithm and genetic operators are discussed in Section 3 . The variants of genetic algorithm with pros and cons are presented in Section 4 . Section 5 describes the applications of genetic algorithm. Section 6 presents the challenges and future research directions. The concluding remarks are drawn in Section 7 .
2 Research methodology
PRISMA’s guidelines were used to conduct the review of GA [ 138 ]. A detailed search has been done on Google scholar and PubMed for identification of research papers related to GA. The important research works found during the manual search were also added in this paper. During search, some keywords such as “Genetic Algorithm” or “Application of GA” or “operators of GA” or “representation of GA” or “variants of GA” were used. The selection and rejection of explored research papers are based on the principles, which is mentioned in Table 1 .
Total 27,64,792 research papers were explored on Google Scholar, PubMed and manual search. The research work related to genetic algorithm for multimedia applications were also included. During the screening of research papers, all the duplicate papers and papers published before 2007 were discarded. 4340 research papers were selected based on 2007 and duplicate entries. Thereafter, 4050 research papers were eliminated based on titles. 220 research papers were eliminated after reading of abstract. 70 research papers were left after third round of screening. 40 more research papers were discarded after full paper reading and facts found in the papers. After the fourth round of screening, final 30 research papers are selected for review.
Based on the relevance and quality of research, 30 papers were selected for evaluation. The relevance of research is decided through some criteria, which is mentioned in Table 1 . The selected research papers comprise of genetic algorithm for multimedia applications, advancement of their genetic operators, and hybridization of genetic algorithm with other well-established metaheuristic algorithms. The pros and cons of genetic operators are shown in preceding section.
3 Background
In this section, the basic structure of GA and its genetic operators are discussed with pros and cons.
3.1 Classical GA
Genetic algorithm (GA) is an optimization algorithm that is inspired from the natural selection. It is a population based search algorithm, which utilizes the concept of survival of fittest [ 135 ]. The new populations are produced by iterative use of genetic operators on individuals present in the population. The chromosome representation, selection, crossover, mutation, and fitness function computation are the key elements of GA. The procedure of GA is as follows. A population ( Y ) of n chromosomes are initialized randomly. The fitness of each chromosome in Y is computed. Two chromosomes say C1 and C2 are selected from the population Y according to the fitness value. The single-point crossover operator with crossover probability (C p ) is applied on C1 and C2 to produce an offspring say O . Thereafter, uniform mutation operator is applied on produced offspring ( O ) with mutation probability (M p ) to generate O′ . The new offspring O′ is placed in new population. The selection, crossover, and mutation operations will be repeated on current population until the new population is complete. The mathematical analysis of GA is as follows [ 126 ]:
GA dynamically change the search process through the probabilities of crossover and mutation and reached to optimal solution. GA can modify the encoded genes. GA can evaluate multiple individuals and produce multiple optimal solutions. Hence, GA has better global search capability. The offspring produced from crossover of parent chromosomes is probable to abolish the admirable genetic schemas parent chromosomes and crossover formula is defined as [ 126 ]:
where g is the number of generations, and G is the total number of evolutionary generation set by population. It is observed from Eq.( 1 ) that R is dynamically changed and increase with increase in number of evolutionary generation. In initial stage of GA, the similarity between individuals is very low. The value of R should be low to ensure that the new population will not destroy the excellent genetic schema of individuals. At the end of evolution, the similarity between individuals is very high as well as the value of R should be high.
According to Schema theorem, the original schema has to be replaced with modified schema. To maintain the diversity in population, the new schema keep the initial population during the early stage of evolution. At the end of evolution, the appropriate schema will be produced to prevent any distortion of excellent genetic schema [ 65 , 75 ]. Algorithm 1 shows the pseudocode of classical genetic algorithm.
Algorithm 1: Classical Genetic Algorithm (GA)
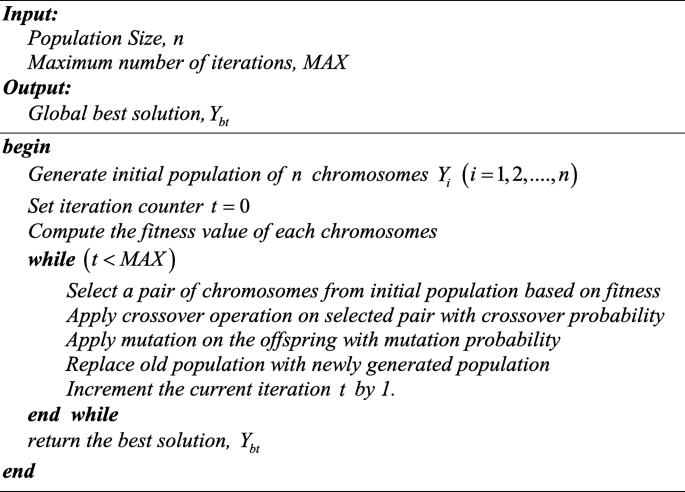
3.2 Genetic operators
GAs used a variety of operators during the search process. These operators are encoding schemes, crossover, mutation, and selection. Figure 2 depicts the operators used in GAs.
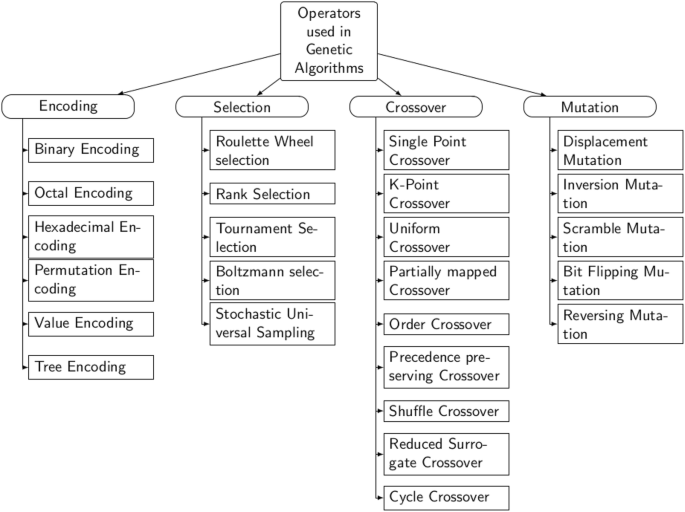
Operators used in GA
3.2.1 Encoding schemes
For most of the computational problems, the encoding scheme (i.e., to convert in particular form) plays an important role. The given information has to be encoded in a particular bit string [ 121 , 183 ]. The encoding schemes are differentiated according to the problem domain. The well-known encoding schemes are binary, octal, hexadecimal, permutation, value-based, and tree.
Binary encoding is the commonly used encoding scheme. Each gene or chromosome is represented as a string of 1 or 0 [ 187 ]. In binary encoding, each bit represents the characteristics of the solution. It provides faster implementation of crossover and mutation operators. However, it requires extra effort to convert into binary form and accuracy of algorithm depends upon the binary conversion. The bit stream is changed according the problem. Binary encoding scheme is not appropriate for some engineering design problems due to epistasis and natural representation.
In octal encoding scheme, the gene or chromosome is represented in the form of octal numbers (0–7). In hexadecimal encoding scheme, the gene or chromosome is represented in the form of hexadecimal numbers (0–9, A-F) [ 111 , 125 , 187 ]. The permutation encoding scheme is generally used in ordering problems. In this encoding scheme, the gene or chromosome is represented by the string of numbers that represents the position in a sequence. In value encoding scheme, the gene or chromosome is represented using string of some values. These values can be real, integer number, or character [ 57 ]. This encoding scheme can be helpful in solving the problems in which more complicated values are used. As binary encoding may fail in such problems. It is mainly used in neural networks for finding the optimal weights.
In tree encoding, the gene or chromosome is represented by a tree of functions or commands. These functions and commands can be related to any programming language. This is very much similar to the representation of repression in tree format [ 88 ]. This type of encoding is generally used in evolving programs or expressions. Table 2 shows the comparison of different encoding schemes of GA.
3.2.2 Selection techniques
Selection is an important step in genetic algorithms that determines whether the particular string will participate in the reproduction process or not. The selection step is sometimes also known as the reproduction operator [ 57 , 88 ]. The convergence rate of GA depends upon the selection pressure. The well-known selection techniques are roulette wheel, rank, tournament, boltzmann, and stochastic universal sampling.
Roulette wheel selection maps all the possible strings onto a wheel with a portion of the wheel allocated to them according to their fitness value. This wheel is then rotated randomly to select specific solutions that will participate in formation of the next generation [ 88 ]. However, it suffers from many problems such as errors introduced by its stochastic nature. De Jong and Brindle modified the roulette wheel selection method to remove errors by introducing the concept of determinism in selection procedure. Rank selection is the modified form of Roulette wheel selection. It utilizes the ranks instead of fitness value. Ranks are given to them according to their fitness value so that each individual gets a chance of getting selected according to their ranks. Rank selection method reduces the chances of prematurely converging the solution to a local minima [ 88 ].
Tournament selection technique was first proposed by Brindle in 1983. The individuals are selected according to their fitness values from a stochastic roulette wheel in pairs. After selection, the individuals with higher fitness value are added to the pool of next generation [ 88 ]. In this method of selection, each individual is compared with all n-1 other individuals if it reaches the final population of solutions [ 88 ]. Stochastic universal sampling (SUS) is an extension to the existing roulette wheel selection method. It uses a random starting point in the list of individuals from a generation and selects the new individual at evenly spaced intervals [ 3 ]. It gives equal chance to all the individuals in getting selected for participating in crossover for the next generation. Although in case of Travelling Salesman Problem, SUS performs well but as the problem size increases, the traditional Roulette wheel selection performs relatively well [ 180 ].
Boltzmann selection is based on entropy and sampling methods, which are used in Monte Carlo Simulation. It helps in solving the problem of premature convergence [ 118 ]. The probability is very high for selecting the best string, while it executes in very less time. However, there is a possibility of information loss. It can be managed through elitism [ 175 ]. Elitism selection was proposed by K. D. Jong (1975) for improving the performance of Roulette wheel selection. It ensures the elitist individual in a generation is always propagated to the next generation. If the individual having the highest fitness value is not present in the next generation after normal selection procedure, then the elitist one is also included in the next generation automatically [ 88 ]. The comparison of above-mentioned selection techniques are depicted in Table 3 .
3.2.3 Crossover operators
Crossover operators are used to generate the offspring by combining the genetic information of two or more parents. The well-known crossover operators are single-point, two-point, k-point, uniform, partially matched, order, precedence preserving crossover, shuffle, reduced surrogate and cycle.
In a single point crossover, a random crossover point is selected. The genetic information of two parents which is beyond that point will be swapped with each other [ 190 ]. Figure 3 shows the genetic information after swapping. It replaced the tail array bits of both the parents to get the new offspring.
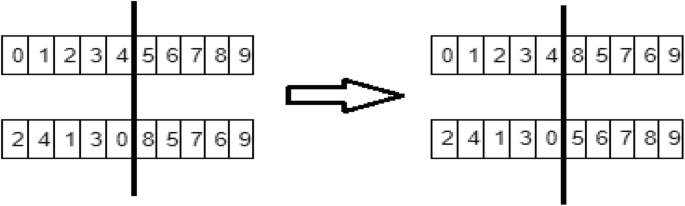
Swapping genetic information after a crossover point
In a two point and k-point crossover, two or more random crossover points are selected and the genetic information of parents will be swapped as per the segments that have been created [ 190 ]. Figure 4 shows the swapping of genetic information between crossover points. The middle segment of the parents is replaced to generate the new offspring.
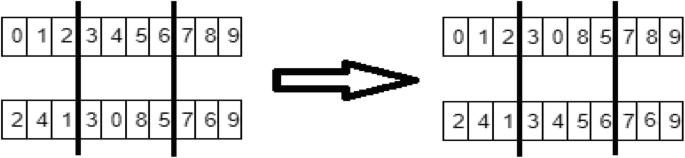
Swapping genetic information between crossover points
In a uniform crossover, parent cannot be decomposed into segments. The parent can be treated as each gene separately. We randomly decide whether we need to swap the gene with the same location of another chromosome [ 190 ]. Figure 5 depicts the swapping of individuals under uniform crossover operation.
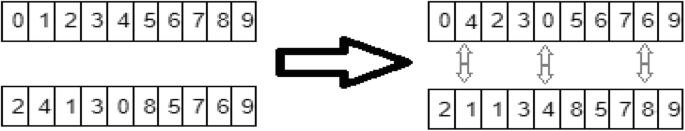
Swapping individual genes
Partially matched crossover (PMX) is the most frequently used crossover operator. It is an operator that performs better than most of the other crossover operators. The partially matched (mapped) crossover was proposed by D. Goldberg and R. Lingle [ 66 ]. Two parents are choose for mating. One parent donates some part of genetic material and the corresponding part of other parent participates in the child. Once this process is completed, the left out alleles are copied from the second parent [ 83 ]. Figure 6 depicts the example of PMX.
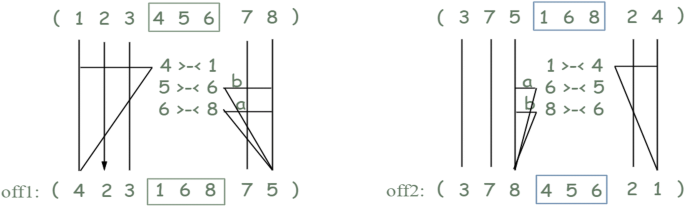
Partially matched crossover (PMX) [ 117 ]
Order crossover (OX) was proposed by Davis in 1985. OX copies one (or more) parts of parent to the offspring from the selected cut-points and fills the remaining space with values other than the ones included in the copied section. The variants of OX are proposed by different researchers for different type of problems. OX is useful for ordering problems [ 166 ]. However, it is found that OX is less efficient in case of Travelling Salesman Problem [ 140 ]. Precedence preserving crossover (PPX) preserves the ordering of individual solutions as present in the parent of offspring before the application of crossover. The offspring is initialized to a string of random 1’s and 0’s that decides whether the individuals from both parents are to be selected or not. In [ 169 ], authors proposed a modified version of PPX for multi-objective scheduling problems.
Shuffle crossover was proposed by Eshelman et al. [ 20 ] to reduce the bias introduced by other crossover techniques. It shuffles the values of an individual solution before the crossover and unshuffles them after crossover operation is performed so that the crossover point does not introduce any bias in crossover. However, the utilization of this crossover is very limited in the recent years. Reduced surrogate crossover (RCX) reduces the unnecessary crossovers if the parents have the same gene sequence for solution representations [ 20 , 139 ]. RCX is based on the assumption that GA produces better individuals if the parents are sufficiently diverse in their genetic composition. However, RCX cannot produce better individuals for those parents that have same composition. Cycle crossover was proposed by Oliver [ 140 ]. It attempts to generate an offspring using parents where each element occupies the position by referring to the position of their parents [ 140 ]. In the first cycle, it takes some elements from the first parent. In the second cycle, it takes the remaining elements from the second parent as shown in Fig. 7 .
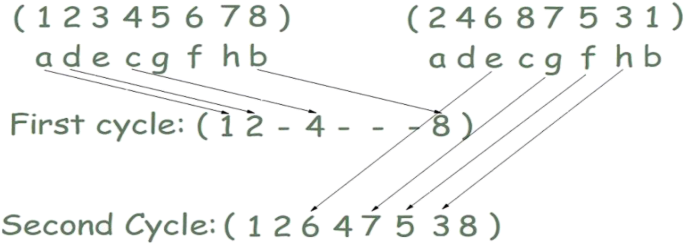
Cycle Crossover (CX) [ 140 ]
Table 4 shows the comparison of crossover techniques. It is observed from Table 4 that single and k-point crossover techniques are easy to implement. Uniform crossover is suitable for large subsets. Order and cycle crossovers provide better exploration than the other crossover techniques. Partially matched crossover provides better exploration. The performance of partially matched crossover is better than the other crossover techniques. Reduced surrogate and cycle crossovers suffer from premature convergence.
3.2.4 Mutation operators
Mutation is an operator that maintains the genetic diversity from one population to the next population. The well-known mutation operators are displacement, simple inversion, and scramble mutation. Displacement mutation (DM) operator displaces a substring of a given individual solution within itself. The place is randomly chosen from the given substring for displacement such that the resulting solution is valid as well as a random displacement mutation. There are variants of DM are exchange mutation and insertion mutation. In Exchange mutation and insertion mutation operators, a part of an individual solution is either exchanged with another part or inserted in another location, respectively [ 88 ].
The simple inversion mutation operator (SIM) reverses the substring between any two specified locations in an individual solution. SIM is an inversion operator that reverses the randomly selected string and places it at a random location [ 88 ]. The scramble mutation (SM) operator places the elements in a specified range of the individual solution in a random order and checks whether the fitness value of the recently generated solution is improved or not [ 88 ]. Table 5 shows the comparison of different mutation techniques.
Table 6 shows the best combination of encoding scheme, mutation, and crossover techniques. It is observed from Table 6 that uniform and single-point crossovers can be used with most of encoding and mutation operators. Partially matched crossover is used with inversion mutation and permutation encoding scheme provides the optimal solution.
4 Variants of GA
Various variants of GA’s have been proposed by researchers. The variants of GA are broadly classified into five main categories namely, real and binary coded, multiobjective, parallel, chaotic, and hybrid GAs. The pros and cons of these algorithms with their application has been discussed in the preceding subsections.
4.1 Real and binary coded GAs
Based on the representation of chromosomes, GAs are categorized in two classes, namely binary and real coded GAs.
4.1.1 Binary coded GAs
The binary representation was used to encode GA and known as binary GA. The genetic operators were also modified to carry out the search process. Payne and Glen [ 153 ] developed a binary GA to identify the similarity among molecules. They used binary representation for position of molecule and their conformations. However, this method has high computational complexity. Longyan et al. [ 203 ] investigated three different method for wind farm design using binary GA (BGA). Their method produced better fitness value and farm efficiency. Shukla et al. [ 185 ] utilized BGA for feature subset selection. They used mutual information maximization concept for selecting the significant features. BGAs suffer from Hamming cliffs, uneven schema, and difficulty in achieving precision [ 116 , 199 ].
4.1.2 Real-coded GAs
Real-coded GAs (RGAs) have been widely used in various real-life applications. The representation of chromosomes is closely associated with real-life problems. The main advantages of RGAs are robust, efficient, and accurate. However, RGAs suffer from premature convergence. Researchers are working on RGAs to improve their performance. Most of RGAs are developed by modifying the crossover, mutation and selection operators.
Crossover operators
The searching capability of crossover operators are not satisfactory for continuous search space. The developments in crossover operators have been done to enhance their performance in real environment. Wright [ 210 ] presented a heuristics crossover that was applied on parents to produce off-spring. Michalewicz [ 135 ] proposed arithmetical crossover operators for RGAs. Deb and Agrawal [ 34 ] developed a real-coded crossover operator, which is based on characteristics of single-point crossover in BGA. The developed crossover operator named as simulated binary crossover (SBX). SBX is able to overcome the Hamming cliff, precision, and fixed mapping problem. The performance of SBX is not satisfactory in two-variable blocked function. Eshelman et al. [ 53 ] utilized the schemata concept to design the blend crossover for RGAs. The unimodal normal distribution crossover operator (UNDX) was developed by Ono et al. [ 144 ]. They used ellipsoidal probability distribution to generate the offspring. Kita et al. [ 106 ] presented a multi-parent UNDX (MP-UNDX), which is the extension of [ 144 ]. However, the performance of RGA with MP-UNDX is much similar to UNDX. Deep and Thakur [ 39 ] presented a Laplace crossover for RGAs, which is based on Laplacian distribution. Chuang et al. [ 27 ] developed a direction based crossover to further explore the all possible search directions. However, the search directions are limited. The heuristic normal distribution crossover operator was developed by Wang et al. [ 207 ]. It generates the cross-generated offspring for better search operation. However, the better individuals are not considered in this approach. Subbaraj et al. [ 192 ] proposed Taguchi self-adaptive RCGA. They used Taguchi method and simulated binary crossover to exploit the capable offspring.
Mutation operators
Mutation operators generate diversity in the population. The two main challenges have to tackle during the application of mutation. First, the probability of mutation operator that was applied on population. Second, the outlier produced in chromosome after mutation process. Michalewicz [ 135 ] presented uniform and non-uniform mutation operators for RGAs. Michalewicz and Schoenauer [ 136 ] developed a special case of uniform mutation. They developed boundary mutation. Deep and Thakur [ 38 ] presented a novel mutation operator based on power law and named as power mutation. Das and Pratihar [ 30 ] presented direction-based exponential mutation operator. They used direction information of variables. Tang and Tseng [ 196 ] presented a novel mutation operator for enhancing the performance of RCGA. Their approach was fast and reliable. However, it stuck in local optima for some applications. Deb et al. [ 35 ] developed polynomial mutation that was used in RCGA. It provides better exploration. However, the convergence speed is slow and stuck in local optima. Lucasius et al. [ 129 ] proposed a real-coded genetic algorithm (RCGA). It is simple and easy to implement. However, it suffers from local optima problem. Wang et al. [ 205 ] developed multi-offspring GA and investigated their performance over single point crossover. Wang et al. [ 206 ] stated the theoretical basis of multi-offspring GA. The performance of this method is better than non-multi-offspring GA. Pattanaik et al. [ 152 ] presented an improvement in the RCGA. Their method has better convergence speed and quality of solution. Wang et al. [ 208 ] proposed multi-offspring RCGA with direction based crossover for solving constrained problems.
Table 7 shows the mathematical formulation of genetic operators in RGAs.
4.2 Multiobjective GAs
Multiobjective GA (MOGA) is the modified version of simple GA. MOGA differ from GA in terms of fitness function assignment. The remaining steps are similar to GA. The main motive of multiobjective GA is to generate the optimal Pareto Front in the objective space in such a way that no further enhancement in any fitness function without disturbing the other fitness functions [ 123 ]. Convergence, diversity, and coverage are main goal of multiobjective GAs. The multiobjective GAs are broadly categorized into two categories namely, Pareto-based, and decomposition-based multiobjective GAs [ 52 ]. These techniques are discussed in the preceding subsections.
4.2.1 Pareto-based multi-objective GA
The concept of Pareto dominance was introduced in multiobjective GAs. Fonseca and Fleming [ 56 ] developed first multiobjective GA (MOGA). The niche and decision maker concepts were proposed to tackle the multimodal problems. However, MOGA suffers from parameter tuning problem and degree of selection pressure. Horn et al. [ 80 ] proposed a niched Pareto genetic algorithm (NPGA) that utilized the concept of tournament selection and Pareto dominance. Srinivas and Deb [ 191 ] developed a non-dominated sorting genetic algorithm (NSGA). However, it suffers from lack of elitism, need of sharing parameter, and high computation complexity. To alleviate these problems, Deb et al. [ 36 ] developed a fast elitist non-dominated sorting genetic algorithm (NSGA-II). The performance of NSGA-II may be deteriorated for many objective problems. NSGA-II was unable to maintain the diversity in Pareto-front. To alleviate this problem, Luo et al. [ 130 ] introduced a dynamic crowding distance in NSGA-II. Coello and Pulido [ 28 ] developed a multiobjective micro GA. They used an archive for storing the non-dominated solutions. The performance of Pareto-based approaches may be deteriorated in many objective problems [ 52 ].
4.2.2 Decomposition-based multiobjective GA
Decomposition-based MOGAs decompose the given problem into multiple subproblems. These subproblems are solved simultaneously and exchange the solutions among neighboring subproblems [ 52 ]. Ishibuchi and Murata [ 84 ] developed a multiobjective genetic local search (MOGLS). In MOGLS, the random weights were used to select the parents and local search for their offspring. They used generation replacement and roulette wheel selection method. Jaszkiewicz [ 86 ] modified the MOGLS by utilizing different selection mechanisms for parents. Murata and Gen [ 141 ] proposed a cellular genetic algorithm for multiobjective optimization (C-MOGA) that was an extension of MOGA. They added cellular structure in MOGA. In C-MOGA, the selection operator was performed on the neighboring of each cell. C-MOGA was further extended by introducing an immigration procedure and known as CI-MOGA. Alves and Almeida [ 11 ] developed a multiobjective Tchebycheffs-based genetic algorithm (MOTGA) that ensures convergence and diversity. Tchebycheff scalar function was used to generate non-dominated solution set. Patel et al. [ 151 ] proposed a decomposition based MOGA (D-MOGA). They integrated opposition based learning in D-MOGA for weight vector generation. D-MOGA is able to maintain the balance between diversity of solutions and exploration of search space.
4.3 Parallel GAs
The motivation behind the parallel GAs is to improve the computational time and quality of solutions through distributed individuals. Parallel GAs are categorized into three broad categories such as master-slave parallel GAs, fine grained parallel GAs, and multi-population coarse grained parallel Gas [ 70 ]. In master-slave parallel GA, the computation of fitness functions is distributed over the several processors. In fine grained GA, parallel computers are used to solve the real-life problems. The genetic operators are bounded to their neighborhood. However, the interaction is allowed among the individuals. In coarse grained GA, the exchange of individuals among sub-populations is performed. The control parameters are also transferred during migration. The main challenges in parallel GAs are to maximize memory bandwidth and arrange threads for utilizing the power of GPUs [ 23 ]. Table 8 shows the comparative analysis of parallel GAs in terms of hardware and software. The well-known parallel GAs are studied in the preceding subsections.
4.3.1 Master slave parallel GA
The large number of processors are utilized in master-slave parallel GA (MS-PGA) as compared to other approaches. The computation of fitness functions may be increased by increasing the number of processors. Hong et al. [ 79 ] used MS-PGA for solving data mining problems. Fuzzy rules are used with parallel GA. The evaluation of fitness function was performed on slave machines. However, it suffers from high computational time. Sahingzo [ 174 ] implemented MS-PGA for UAV path finding problem. The genetic operators were executed on processors. They used multicore CPU with four cores. Selection and fitness evaluation was done on slave machines. MS-PGA was applied on traffic assignment problem in [ 127 ]. They used thirty processors to solve this problem at National University of Singapore. Yang et al. [ 213 ] developed a web-based parallel GA. They implemented the master slave version of NSGA-II in distributed environment. However, the system is complex in nature.
4.3.2 Fine grained parallel GA
In last few decades, researchers are working on migration policies of fine grained parallel GA (FG-PGA). Porta et al. [ 161 ] utilized clock-time for migration frequency, which is independent of generations. They used non-uniform structure and static configuration. The best solution was selected for migration and worst solution was replaced with migrant solution. Kurdi [ 115 ] used adaptive migration frequency. The migration procedure starts until there is no change in the obtained solutions after ten successive generations. The non-uniform and dynamic structure was used. In [ 209 ], local best solutions were synchronized and formed a global best solutions. The global best solutions were transferred to all processors for father execution. The migration frequency depends upon the number of generation. They used uniform structure with fixed configuration. Zhang et al. [ 220 ] used parallel GA to solve the set cover problem of wireless networks. They used divide-and-conquer strategy to decompose the population into sub-populations. Thereafter, the genetic operators were applied on local solutions and Kuhn-Munkres was used to merge the local solutions.
4.3.3 Coarse grained parallel GA
Pinel et al. [ 158 ] proposed a GraphCell. The population was initialized with random values and one solution was initialized with Min-min heuristic technique. 448 processors were used to implement the proposed approach. However, coarse grained parallel GAs are less used due to complex in nature. The hybrid parallel GAs are widely used in various applications. Shayeghi et al. [ 182 ] proposed a pool-based Birmingham cluster GA. Master node was responsible for managing global population. Slave node selected the solutions from global population and executed it. 240 processors are used for computation. Roberge et al. [ 170 ] used hybrid approach to optimize switching angle of inverters. They used four different strategies for fitness function computation. Nowadays, GPU, cloud, and grid are most popular hardware for parallel GAs [ 198 ].
4.4 Chaotic GAs
The main drawback of GAs is premature convergence. The chaotic systems are incorporated into GAs to alleviate this problem. The diversity of chaos genetic algorithm removes premature convergence. Crossover and mutation operators can be replaced with chaotic maps. Tiong et al. [ 197 ] integrated the chaotic maps into GA for further improvement in accuracy. They used six different chaotic maps. The performance of Logistic, Henon and Ikeda chaotic GA performed better than the classical GA. However, these techniques suffer from high computational complexity. Ebrahimzadeh and Jampour [ 48 ] used Lorenz chaotic for genetic operators of GA to eliminate the local optima problem. However, the proposed approach was unable to find relationship between entropy and chaotic map. Javidi and Hosseinpourfard [ 87 ] utilized two chaotic maps namely logistic map and tent map for generating chaotic values instead of random selection of initial population. The proposed chaotic GA performs better than the GA. However, this method suffers from high computational complexity. Fuertes et al. [ 60 ] integrated the entropy into chaotic GA. The control parameters are modified through chaotic maps. They investigated the relationship between entropy and performance optimization.
Chaotic systems have also used in multiobjective and hybrid GAs. Abo-Elnaga and Nasr [ 5 ] integrated chaotic system into modified GA for solving Bi-level programming problems. Chaotic helps the proposed algorithm to alleviate local optima and enhance the convergence. Tahir et al. [ 193 ] presented a binary chaotic GA for feature selection in healthcare. The chaotic maps were used to initialize the population and modified reproduction operators were applied on population. Xu et al. [ 115 ] proposed a chaotic hybrid immune GA for spectrum allocation. The proposed approach utilizes the advantages of both chaotic and immune operator. However, this method suffers from parameter initialization problem.
4.5 Hybrid GAs
Genetic Algorithms can be easily hybridized with other optimization methods for improving their performance such as image denoising methods, chemical reaction optimization, and many more. The main advantages of hybridized GA with other methods are better solution quality, better efficiency, guarantee of feasible solutions, and optimized control parameters [ 51 ]. It is observed from literature that the sampling capability of GAs is greatly affected from population size. To resolve this problem, local search algorithms such as memetic algorithm, Baldwinian, Lamarckian, and local search have been integrated with GAs. This integration provides proper balance between intensification and diversification. Another problem in GA is parameter setting. Finding appropriate control parameters is a tedious task. The other metaheuristic techniques can be used with GA to resolve this problem. Hybrid GAs have been used to solve the issues mentioned in the preceding subsections [ 29 , 137 , 186 ].
4.5.1 Enhance search capability
GAs have been integrated with local search algorithms to reduce the genetic drift. The explicit refinement operator was introduced in local search for producing better solutions. El-Mihoub et al. [ 54 ] established the effect of probability of local search on the population size of GA. Espinoza et al. [ 50 ] investigated the effect of local search for reducing the population size of GA. Different search algorithms have been integrated with GAs for solving real-life applications.
4.5.2 Generate feasible solutions
In complex and high-dimensional problems, the genetic operators of GA generate infeasible solutions. PMX crossover generates the infeasible solutions for order-based problems. The distance preserving crossover operator was developed to generate feasible solutions for travelling salesman problem [ 58 ]. The gene pooling operator instead of crossover was used to generate feasible solution for data clustering [ 19 ]. Konak and Smith [ 108 ] integrated a cut-saturation algorithm with GA for designing the communication networks. They used uniform crossover to produce feasible solutions.
4.5.3 Replacement of genetic operators
There is a possibility to replace the genetic operators which are mentioned in Section 3.2 with other search techniques. Leng [ 122 ] developed a guided GA that utilizes the penalties from guided local search. These penalties were used in fitness function to improve the performance of GA. Headar and Fukushima [ 74 ] used simplex crossover instead of standard crossover. The standard mutation operator was replaced with simulated annealing in [ 195 ]. The basic concepts of quantum computing are used to improve the performance of GAs. The heuristic crossover and hill-climbing operators can be integrated into GA for solving three-matching problem.
4.5.4 Optimize control parameters
The control parameters of GA play a crucial role in maintaining the balance between intensification and diversification. Fuzzy logic has an ability to estimate the appropriate control parameters of GA [ 167 ]. Beside this, GA can be used to optimize the control parameters of other techniques. GAs have been used to optimize the learning rate, weights, and topology of neutral networks [ 21 ]. GAs can be used to estimate the optimal value of fuzzy membership in controller. It was also used to optimize the control parameters of ACO, PSO, and other metaheuristic techniques [ 156 ]. The comparative analysis of well-known GAs are mentioned in Table 9 .
5 Applications
Genetic Algorithms have been applied in various NP-hard problems with high accuracy rates. There are a few application areas in which GAs have been successfully applied.
5.1 Operation management
GA is an efficient metaheuristic for solving operation management (OM) problems such as facility layout problem (FLP), supply network design, scheduling, forecasting, and inventory control.
5.1.1 Facility layout
Datta et al. [ 32 ] utilized GA for solving single row facility layout problem (SRFLP). For SRFLP, the modified crossover and mutation operators of GA produce valid solutions. They applied GA to large sized problems that consists of 60–80 instances. However, it suffers from parameter dependency problem. Sadrzadeh [ 173 ] proposed GA for multi-line FLP have multi products. The facilities were clustered using mutation and heuristic operators. The total cost obtained from the proposed GA was decreased by 7.2% as compared to the other algorithms. Wu et al. [ 211 ] implemented hierarchical GA to find out the layout of cellular manufacturing system. However, the performance of GA is greatly affected from the genetic operators. Aiello et al. [ 7 ] proposed MOGA for FLP. They used MOGA on the layout of twenty different departments. Palomo-Romero et al. [ 148 ] proposed an island model GA to solve the FLP. The proposed technique maintains the population diversity and generates better solutions than the existing techniques. However, this technique suffers from improper migration strategy that can be utilized for improving the population. GA and its variants has been successfully applied on FLP [ 103 , 119 , 133 , 201 ].
5.1.2 Scheduling
GA shows the superior performance for solving the scheduling problems such as job-shop scheduling (JSS), integrated process planning and scheduling (IPPS), etc. [ 119 ]. To improve the performance in the above-mentioned areas of scheduling, researchers developed various genetic representation [ 12 , 159 , 215 ], genetic operators, and hybridized GA with other methods [ 2 , 67 , 147 , 219 ].
5.1.3 Inventory control
Besides the scheduling, inventory control plays an important role in OM. Backordering and lost sales are two main approaches for inventory control [ 119 ]. Hiassat et al. [ 76 ] utilized the location-inventory model to find out the number and location of warehouses. Various design constraints have been added in the objective functions of GA and its variants for solving inventory control problem [].
5.1.4 Forecasting and network design
Forecasting is an important component for OM. Researchers are working on forecasting of financial trading, logistics demand, and tourist arrivals. GA has been hybridized with support vector regression, fuzzy set, and neural network (NN) to improve their forecasting capability [ 22 , 78 , 89 , 178 , 214 ]. Supply network design greatly affect the operations planning and scheduling. Most of the research articles are focused on capacity constraints of facilities [ 45 , 184 ]. Multi-product multi-period problems increases the complexity of supply networks. To resolve the above-mentioned problem, GA has been hybridized with other techniques [ 6 , 45 , 55 , 188 , 189 ]. Multi-objective GAs are also used to optimize the cost, profit, carbon emissions, etc. [ 184 , 189 ].
5.2 Multimedia
GAs have been applied in various fields of multimedia. Some of well-known multimedia fields are encryption, image processing, video processing, medical imaging, and gaming.
5.2.1 Information security
Due to development in multimedia applications, images, videos and audios are transferred from one place to another over Internet. It has been found in literature that the images are more error prone during the transmission. Therefore, image protection techniques such as encryption, watermarking and cryptography are required. The classical image encryption techniques require the input parameters for encryption. The wrong selection of input parameters will generate inadequate encryption results. GA and its variants have been used to select the appropriate control parameters. Kaur and Kumar [ 96 ] developed a multi-objective genetic algorithm to optimize the control parameters of chaotic map. The secret key was generated using beta chaotic map. The generated key was use to encrypt the image. Parallel GAs were also used to encrypt the image [ 97 ].
5.2.2 Image processing
The main image processing tasks are preprocessing, segmentation, object detection, denoising, and recognition. Image segmentation is an important step to solve the image processing problems. Decomposing/partitioning an image requires high computational time. To resolve this problem, GA is used due to their better search capability [ 26 , 102 ]. Enhancement is a technique to improve the quality and contrast of an image. The better image quality is required to analyze the given image. GAs have been used to enhance natural contrast and magnify image [ 40 , 64 , 99 ]. Some researchers are working on hybridization of rough set with adaptive genetic algorithm to merge the noise and color attributes. GAs have been used to remove the noise from the given image. GA can be hybridized with fuzzy logic to denoise the noisy image. GA based restoration technique can be used to remove haze, fog and smog from the given image [ 8 , 110 , 146 , 200 ]. Object detection and recognition is a challenging issue in real-world problem. Gaussian mixture model provides better performance during detection and recognition process. The control parameters are optimized through GA [ 93 ].
5.2.3 Video processing
Video segmentation has been widely used in pattern recognition, and computer vision. There are some critical issues that are associated with video segmentation. These are distinguishing object from the background and determine accurate boundaries. GA can be used to resolve these issues [ 9 , 105 ]. GAs have been implemented for gesture recognition successfully by Chao el al. [ 81 ] used GA for gesture recognition. They applied GAs and found an accuracy of 95% in robot vision. Kaluri and Reddy [ 91 ] proposed an adaptive genetic algorithm based method along with fuzzy classifiers for sign gesture recognition. They reported an improved recognition rate of 85% as compared to the existing method that provides 79% accuracy. Beside the gesture recognition, face recognition play an important role in criminal identification, unmanned vehicles, surveillance, and robots. GA is able to tackle the occlusion, orientations, expressions, pose, and lighting condition [ 69 , 95 , 109 ].
5.2.4 Medical imaging
Genetic algorithms have been applied in medical imaging such as edge detection in MRI and pulmonary nodules detection in CT scan images [ 100 , 179 ]. In [ 120 ], authors used a template matching technique with GA for detecting nodules in CT images. Kavitha and Chellamuthu [ 179 ] used GA based region growing method for detecting the brain tumor. GAs have been applied on medical prediction problems captured from pathological subjects. Sari and Tuna [ 176 ] used GA used to solve issues arises in biomechanics. It is used to predict pathologies during examination. Ghosh and Bhattachrya [ 62 ] implemented sequential GA with cellular automata for modelling the coronavirus disease 19 (COVID-19) data. GAs can be applied in parallel mode to find rules in biological datasets [ 31 ]. The authors proposed a parallel GA that runs by dividing the process into small sub-generations and evaluating the fitness of each individual solution in parallel. Genetic algorithms are used in medicine and other related fields. Koh et al. [ 61 ] proposed a genetic algorithm based method for evaluation of adverse effects of a given drug.
5.2.5 Precision agriculture
GAs have been applied on various problems that are related to precision agriculture. The main issues are crop yield, weed detection, and improvement in farming equipment. Pachepsky and Acock [ 145 ] implemented GA to analyze the water capacity in soil using remote sensing images. The crop yield can be predicted through the capacity of water present in soil. The weed identification was done through GA in [ 142 ]. They used aerial image for classification of plants. In [ 124 ], color image segmentation was used to discriminate the weed and plant. Peerlink et al. [ 154 ] determined the appropriate rate of fertilizer for various portions of agriculture field. They GA for determining the nitrogen in wheat field. The energy requirements in water irrigation systems can be optimized by viewing it as a multi-objective optimization problem. The amount of irrigation required and thus power requirements change continuously in a SMART farm. Therefore, GA can be applied in irrigation systems to reduce the power requirements [ 33 ].
5.2.6 Gaming
GAs have been successfully used in games such as gomoku. In [ 202 ], the authors shown that the GA based approach finds the solution having the highest fitness than the normal tree based methods. However, in real-time strategy based games, GA based solutions become less practical to implement [ 82 ]. GAs have been implemented for path planning problems considering the environment constraints as well as avoiding the obstacles to reach the given destination. Burchardt and Salomon [ 18 ] described an implementation for path planning for soccer games. GA can encode the path planning problems via the coordinate points of a two-dimensional playing field, hence resulting in a variable length solution. The fitness function in path planning considers length of path as well as the collision avoiding terms for soccer players.
5.3 Wireless networking
Due to adaptive, scalable, and easy implementation of GA, it has been used to solve the various issues of wireless networking. The main issues of wireless networking are routing, quality of service, load balancing, localization, bandwidth allocation and channel assignment [ 128 , 134 ]. GA has been hybridized with other metaheuristics for solving the routing problems. Hybrid GA not only producing the efficient routes among pair of nodes, but also used for load balancing [ 24 , 212 ].
5.3.1 Load balancing
Nowadays, multimedia applications require Quality-of-Service (QoS) demand for delay and bandwidth. Various researchers are working on GAs for QoS based solutions.GA produces optimal solutions for complex networks [ 49 ]. Roy et al. [ 172 ] proposed a multi-objective GA for multicast QoS routing problem. GA was used with ACO and other search algorithms for finding optimal routes with desired QoS metrics. Load balancing is another issue in wireless networks. Scully and Brown [ 177 ] used MicroGAs and MacroGAs to distribute the load among various components of networks. He et al. [ 73 ] implemented GA to determine the balance load in wireless sensor networks. Cheng et al. [ 25 ] utilized distributed GA with multi-population scheme for load balancing. They used load balancing metric as a fitness function in GA.
5.3.2 Localization
The process of determining the location of wireless nodes is called as localization. It plays an important role in disaster management and military services. Yun et al. [ 216 ] used GA with fuzzy logic to find out the weights, which are assigned according to the signal strength. Zhang et al. [ 218 ] hybridized GA with simulated annealing (SA) to determine the position of wireless nodes. SA is used as local search to eliminate the premature convergence.
5.3.3 Bandwidth and channel allocation
The appropriate bandwidth allocation is a complex task. GAs and its variants have been developed to solve the bandwidth allocation problem [ 92 , 94 , 107 ]. GAs were used to investigate the allocation of bandwidth with QoS constraints. The fitness function of GAs may consists of resource utilization, bandwidth distribution, and computation time [ 168 ]. The channel allocation is an important issue in wireless networks. The main objective of channel allocation is to simultaneously optimize the number of channels and reuse of allocated frequency. Friend et al. [ 59 ] used distributed island GA to resolve the channel allocation problem in cognitive radio networks. Zhenhua et al. [ 221 ] implemented a modified immune GA for channel assignment. They used different encoding scheme and immune operators. Pinagapany and Kulkarni [ 157 ] developed a parallel GA to solve both static and dynamic channel allocation problem. They used decimal encoding scheme. Table 10 summarizes the applications of GA and its variants.
6 Challenges and future possibilities
In this section, the main challenges faced during the implementation of GAs are discussed followed by the possible research directions.
6.1 Challenges
Despite the several advantages, there are some challenges that need to be resolved for future advancements and further evolution of genetic algorithms. Some major challenges are given below:
6.1.1 Selection of initial population
Initial population is always considered as an important factor for the performance of genetic algorithms. The size of population also affects the quality of solution [ 160 ]. The researchers argue that if a large population is considered, then the algorithm takes more computation time. However, the small population may lead to poor solution [ 155 ]. Therefore, finding the appropriate population size is always a challenging issue. Harik and Lobo [ 71 ] investigated the population using self-adaption method. They used two approaches such as (1) use of self-adaption prior to execution of algorithm, in which the size of population remains the same and (2) in which the self-adaption used during the algorithm execution where the population size is affected by fitness function.
6.1.2 Premature convergence
Premature convergence is a common issue for GA. It can lead to the loss of alleles that makes it difficult to identify a gene [ 15 ]. Premature convergence states that the result will be suboptimal if the optimization problem coincides too early. To avoid this issue, some researchers suggested that the diversity should be used. The selection pressure should be used to increase the diversity. Selection pressure is a degree which favors the better individuals in the initial population of GA’s. If selection pressure (SP1) is greater than some selection pressure (SP2), then population using SP1 should be larger than the population using SP2. The higher selection pressure can decrease the population diversity that may lead to premature convergence [ 71 ].
Convergence property has to be handled properly so that the algorithm finds global optimal solution instead of local optimal solution (see Fig. 8 ). If the optimal solution lies in the vicinity of an infeasible solution, then the global nature of GA can be combined with local nature of other algorithms such as Tabu search and local search. The global nature of genetic algorithms and local nature of Tabu search provide the proper balance between intensification and diversification.
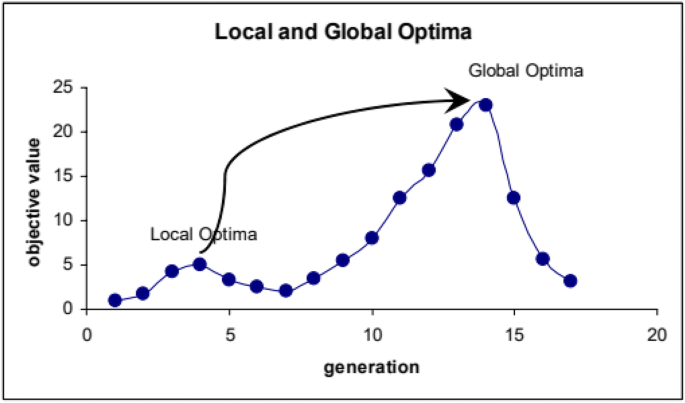
Local and global optima [ 149 ]
6.1.3 Selection of efficient fitness functions
Fitness function is the driving force, which plays an important role in selecting the fittest individual in every iteration of an algorithm. If the number of iterations are small, then a costly fitness function can be adjusted. The number of iterations increases may increase the computational cost. The selection of fitness function depends upon the computational cost as well as their suitability. In [ 46 ], the authors used Davies-Bouldin index for classification of documents.
6.1.4 Degree of mutation and crossover
Crossover and mutation operators are the integral part of GAs. If the mutation is not considered during evolution, then there will be no new information available for evolution. If crossover is not considered during evolution, then the algorithm can result in local optima. The degree of these operators greatly affect the performance of GAs [ 72 ]. The proper balance between these operators are required to ensure the global optima. The probabilistic nature cannot determine the exact degree for an effective and optimal solution.
6.1.5 Selection of encoding schemes
GAs require a particular encoding scheme for a specific problem. There is no general methodology for deciding whether the particular encoding scheme is suitable for any type of real-life problem. If there are two different problems, then two different encoding schemes are required. Ronald [ 171 ] suggested that the encoding schemes should be designed to overwhelm the redundant forms. The genetic operators should be implemented in a manner that they are not biased towards the redundant forms.
6.2 Future research directions
GAs have been applied in different fields by modifying the basic structure of GA. The optimality of a solution obtained from GA can be made better by overcoming the current challenges. Some future possibilities for GA are as follows:
There should be some way to choose the appropriate degree of crossover and mutation operators. For example Self-Organizing GA adapt the crossover and mutation operators according to the given problem. It can save computation time that make it faster.
Future work can also be considered for reducing premature convergence problem. Some researchers are working in this direction. However, it is suggested that new methods of crossover and mutation techniques are required to tackle the premature convergence problem.
Genetic algorithms mimic the natural evolution process. There can be a possible scope for simulating the natural evolution process such as the responses of human immune system and the mutations in viruses.
In real-life problems, the mapping from genotype to phenotype is complex. In this situation, the problem has no obvious building blocks or building blocks are not adjacent groups of genes. Hence, there is a possibility to develop novel encoding schemes to different problems that does not exhibit same degree of difficulty.
7 Conclusions
This paper presents the structured and explained view of genetic algorithms. GA and its variants have been discussed with application. Application specific genetic operators are discussed. Some genetic operators are designed for representation. However, they are not applicable to research domains. The role of genetic operators such as crossover, mutation, and selection in alleviating the premature convergence is studied extensively. The applicability of GA and its variants in various research domain has been discussed. Multimedia and wireless network applications were the main attention of this paper. The challenges and issues mentioned in this paper will help the practitioners to carry out their research. There are many advantages of using GAs in other research domains and metaheuristic algorithms.
The intention of this paper is not only provide the source of recent research in GAs, but also provide the information about each component of GA. It will encourage the researchers to understand the fundamentals of GA and use the knowledge in their research problems.
Abbasi M, Rafiee M, Khosravi MR, Jolfaei A, Menon VG, Koushyar JM (2020) An efficient parallel genetic algorithm solution for vehicle routing problem in cloud implementation of the intelligent transportation systems. Journal of cloud Computing 9(6)
Abdelghany A, Abdelghany K, Azadian F (2017) Airline flight schedule planning under competition. Comput Oper Res 87:20–39
MathSciNet MATH Google Scholar
Abdulal W, Ramachandram S (2011) Reliability-aware genetic scheduling algorithm in grid environment. International Conference on Communication Systems and Network Technologies, Katra, Jammu, pp 673–677
Google Scholar
Abdullah J (2010) Multiobjectives ga-based QoS routing protocol for mobile ad hoc network. Int J Grid Distrib Comput 3(4):57–68
Abo-Elnaga Y, Nasr S (2020) Modified evolutionary algorithm and chaotic search for Bilevel programming problems. Symmetry 12:767
Afrouzy ZA, Nasseri SH, Mahdavi I (2016) A genetic algorithm for supply chain configuration with new product development. Comput Ind Eng 101:440–454
Aiello G, Scalia G (2012) La, Enea M. A multi objective genetic algorithm for the facility layout problem based upon slicing structure encoding Expert Syst Appl 39(12):10352–10358
Alaoui A, Adamou-Mitiche ABH, Mitiche L (2020) Effective hybrid genetic algorithm for removing salt and pepper noise. IET Image Process 14(2):289–296
Alkhafaji BJ, Salih MA, Nabat ZM, Shnain SA (2020) Segmenting video frame images using genetic algorithms. Periodicals of Engineering and Natural Sciences 8(2):1106–1114
Al-Oqaily AT, Shakah G (2018) Solving non-linear optimization problems using parallel genetic algorithm. International Conference on Computer Science and Information Technology (CSIT), Amman, pp. 103–106
Alvesa MJ, Almeidab M (2007) MOTGA: A multiobjective Tchebycheff based genetic algorithm for the multidimensional knapsack problem. Comput Oper Res 34:3458–3470
MathSciNet Google Scholar
Arakaki RK, Usberti FL (2018) Hybrid genetic algorithm for the open capacitated arc routing problem. Comput Oper Res 90:221–231
Arkhipov DI, Wu D, Wu T, Regan AC (2020) A parallel genetic algorithm framework for transportation planning and logistics management. IEEE Access 8:106506–106515
Azadeh A, Elahi S, Farahani MH, Nasirian B (2017) A genetic algorithm-Taguchi based approach to inventory routing problem of a single perishable product with transshipment. Comput Ind Eng 104:124–133
Baker JE, Grefenstette J (2014) Proceedings of the first international conference on genetic algorithms and their applications. Taylor and Francis, Hoboken, pp 101–105
Bolboca SD, JAntschi L, Balan MC, Diudea MV, Sestras RE (2010) State of art in genetic algorithms for agricultural systems. Not Bot Hort Agrobot Cluj 38(3):51–63
Bonabeau E, Dorigo M, Theraulaz G (1999) Swarm intelligence: from natural to artificial systems. Oxford University Press, Inc
MATH Google Scholar
Burchardt H, Salomon R (2006) Implementation of path planning using genetic algorithms on Mobile robots. IEEE International Conference on Evolutionary Computation, Vancouver, BC, pp 1831–1836
Burdsall B, Giraud-Carrier C (1997) Evolving fuzzy prototypes for efficient data clustering," in second international ICSC symposium on fuzzy logic and applications. Zurich, Switzerland, pp. 217-223.
Burkowski FJ (1999) Shuffle crossover and mutual information. Proceedings of the 1999 Congress on Evolutionary Computation-CEC99 (Cat. No. 99TH8406), Washington, DC, USA, 1999, pp. 1574–1580
Chaiyaratana N, Zalzala AM (2000) "Hybridisation of neural networks and a genetic algorithm for friction compensation," in the 2000 congress on evolutionary computation, vol 1. San Diego, USA, pp 22–29
Chen R, Liang C-Y, Hong W-C, Gu D-X (2015) Forecasting holiday daily tourist flow based on seasonal support vector regression with adaptive genetic algorithm. Appl Soft Comput 26:434–443
J.R. Cheng and M. Gen (2020) Parallel genetic algorithms with GPU computing. Impact on Intelligent Logistics and Manufacturing.
Cheng H, Yang S (2010) Multi-population genetic algorithms with immigrants scheme for dynamic shortest path routing problems in mobile ad hoc networks. Applications of evolutionary computation. Springer, In, pp 562–571
Cheng H, Yang S, Cao J (2013) Dynamic genetic algorithms for the dynamic load balanced clustering problem in mobile ad hoc net-works. Expert Syst Appl 40(4):1381–1392
Chouhan SS, Kaul A, Singh UP (2018) Soft computing approaches for image segmentation: a survey. Multimed Tools Appl 77(21):28483–28537
Chuang YC, Chen CT, Hwang C (2016) A simple and efficient real-coded genetic algorithm for constrained optimization. Appl Soft Comput 38:87–105
Coello CAC, Pulido GT (2001) A micro-genetic algorithm for multiobjective optimization. In: EMO, volume 1993 of lecture notes in computer science, pp 126–140. Springer
Das, K. N. (2014). Hybrid genetic algorithm: an optimization tool. In global trends in intelligent computing Research and Development (pp. 268-305). IGI global.
Das AK, Pratihar DK (2018) A direction-based exponential mutation operator for real-coded genetic algorithm. IEEE International Conference on Emerging Applications of Information Technology.
Dash SR, Dehuri S, Rayaguru S (2013) Discovering interesting rules from biological data using parallel genetic algorithm, 3rd IEEE International Advance Computing Conference (IACC), Ghaziabad,, pp. 631–636.
Datta D, Amaral ARS, Figueira JR (2011) Single row facility layout problem using a permutation-based genetic algorithm. European J Oper Res 213(2):388–394
de Ocampo ALP, Dadios EP (2017) "Energy cost optimization in irrigation system of smart farm by using genetic algorithm," 2017IEEE 9th international conference on humanoid. Nanotechnology, Information Technology, Communication and Control, Environment and Management (HNICEM), Manila, pp 1–7
Deb K, Agrawal RB (1995) Simulated binary crossover for continuous search space. Complex Systems 9:115–148
Deb K, Deb D (2014) Analysing mutation schemes for real-parameter genetic algorithms. International Journal of Artificial Intelligence and Soft Computing 4(1):1–28
Deb K, Pratap A, Agarwal S, Meyarivan T (2002) A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans Evol Comput 6(2):182–197
Deep K, Das KN (2008) Quadratic approximation based hybrid genetic algorithm for function optimization. Appl Math Comput 203(1):86–98
Deep K, Thakur M (2007) A new mutation operator for real coded genetic algorithms. Appl Math Comput 193:211–230
Deep K, Thakur M (2007) A new crossover operator for real coded genetic algorithms. Appl Math Comput 188:895–911
Dhal KP, Ray S, Das A, Das S (2018) A survey on nature-inspired optimization algorithms and their application in image enhancement domain. Archives of Computational Methods in Engineering 5:1607–1638
Dhiman G, Kumar V (2017) Spotted hyena optimizer: A novel bio-inspired based metaheuristic technique for engineering applications. Adv Eng Softw 114:48–70
Dhiman G, Kumar V (2018) Emperor penguin optimizer: A bio-inspired algorithm for engineering problems. Knowl-Based Syst 159:20–50
Dhiman G, Kumar V (2019) Seagull optimization algorithm: theory and its applications for large-scale industrial engineering problems. Knowl-Based Syst 165:169–196
Di Fatta G, Hoffmann F, Lo Re G, Urso A (2003) A genetic algorithm for the design of a fuzzy controller for active queue management. IEEE Trans Syst Man Cybern Part C Appl Rev 33(3):313–324
Diabat A, Deskoores R (2016) A hybrid genetic algorithm based heuristic for an integrated supply chain problem. J Manuf Syst 38:172–180
Diaz-Manríquez A, Ríos-Alvarado AB, Barrón-Zambrano JH, Guerrero-Melendez TY, Elizondo-Leal JC (2018) An automatic document classifier system based on genetic algorithm and taxonomy. IEEE Access 6:21552–21559. https://doi.org/10.1109/ACCESS.2018.2815992
Article Google Scholar
Dorigo M, Birattari M, Stutzle T (2006) Ant colony optimization - artificial ants as a computational intelligence technique. IEEE Comput Intell Mag 1(2006):28–39
Ebrahimzadeh R, Jampour M (2013) Chaotic genetic algorithm based on Lorenz chaotic system for optimization problems. I.J. Intelligent Systems and Applications Intelligent Systems and Applications 05(05):19–24
EkbataniFard GH, Monsefi R, Akbarzadeh-T M-R, Yaghmaee M et al. (2010) A multi-objective genetic algorithm based approach for energy efficient qos-routing in two-tiered wireless sensor net-works. In: wireless pervasive computing (ISWPC), 2010 5th IEEE international symposium on. IEEE, pp 80–85
El-Mihoub T, Hopgood A, Nolle L, Battersby A (2004) Performance of hybrid genetic algorithms incorporating local search. In: Horton G (ed) 18th European simulation multi-conference (ESM2004). Germany, Magdeburg, pp 154–160
El-Mihoub TA, Hopgood AA, Lars N, Battersby A (2006) Hybrid genetic algorithms: A review. Eng Lett 13:2
Emmerich MTM, Deutz AH (2018) A tutorial on multiobjective optimization: fundamentals and evolutionary methods. Nat Comput 17(3):585–609
Eshelman LJ, Caruana RA, Schaffer JD (1997) Biases in the crossover landscape.
Espinoza FB, Minsker B, Goldberg D (2003) Performance evaluation and population size reduction for self adaptive hybrid genetic algorithm (SAHGA), in the Genetic and Evolutionary Computation Conference, vol. 2723, Lecture Notes in Computer Science San Francisco, USA: Springer, pp. 922–933.
Farahani RZ, Elahipanah M (2008) A genetic algorithm to optimize the total cost and service level for just-in-time distribution in a supply chain. Int J Prod Econ 111(2):229–243
Fonseca CM, Fleming PJ (1993) Genetic algorithms for multiobjective optimization: formulation, discussion and generalization. In: ICGA, pp 416–423. Morgan Kaufmann
Fox B, McMahon M (1991) Genetic operators for sequencing problems, in Foundations of Genetic Algorithms, G. Rawlins, Ed. Morgan Kaufmann Publishers, San Mateo,CA, Ed. 1991, pp. 284–300.
Freisleben B, Merz P (1996) New genetic local search operators for the traveling salesman problem," in the Fourth Conference on Parallel Problem Solving from Nature vol. 1141, Lectures Notes in Computer Science, H.-M. Voigt, W. Ebeling, I. Rechenberg, and H.-P. Schwefel, Eds. Berlin, Germany: Springer-Verlag, pp. 890–899.
Friend DH, EI Nainay, M, Shi Y, MacKenzie AB (2008) Architecture and performance of an island genetic algorithm-based cognitive network. In: Consumer communications and networking conference,2008. CCNC 2008. 5th IEEE. IEEE, pp 993–997
Fuertes G, Vargas M, Alfaro M, Soto-Garrido R, Sabattin J, Peralta M-A (2019) Chaotic genetic algorithm and the effects of entropy in performance optimization.
Ghaheri A, Shoar S, Naderan M, Hoseini SS (2015) The applications of genetic algorithms in medicine. CJ 30:406–416
Ghosh S, Bhattachrya S (2020) A data-driven understanding of COVID-19 dynamics using sequential genetic algorithm based probabilistic cellular automata. Applied Soft Computing. 96
Ghoshal AK, Das N, Bhattacharjee S, Chakraborty G (2019) A fast parallel genetic algorithm based approach for community detection in large networks. International Conference on Communication Systems & Networks (COMSNETS), Bengaluru, India, pp. 95–101.
Gogna A, Tayal A (2012) Comparative analysis of evolutionary algorithms for image enhancement. Int J Met 2(1)
Goldberg D (1989) Genetic algorithm in search. Optimization and Machine Learning, Addison -Wesley, Reading, MA 1989
Goldberg D, Lingle R (1985) Alleles, loci and the traveling salesman problem. In: Proceedings of the 1st international conference on genetic algorithms and their applications, vol. 1985. Los Angeles, USA, pp 154–159
Guido R, Conforti D (2017) A hybrid genetic approach for solving an integrated multi-objective operating room planning and scheduling problem. Comput Oper Res 87:270–282
Ha QM, Deville Y, Pham QD, Ha MH (2020) A hybrid genetic algorithm for the traveling salesman problem with drone. J Heuristics 26:219–247
HajiRassouliha A, Gamage TPB, Parker MD, Nash MP, Taberner AJ, Nielsen, PM (2013) FPGA implementation of 2D cross-correlation for real-time 3D tracking of deformable surfaces. In Proceedings of the2013 28th International Conference on Image and Vision Computing New Zealand (IVCNZ 2013), Wellington, New Zealand, 27–29 November 2013; IEEE: Piscataway, NJ, USA; pp. 352–357
Harada T, Alba E (2020) Parallel genetic algorithms: a useful survey. ACM Computing Survey 53(4):1–39
Harik GR, Lobo FG (1999) A parameter-less genetic algorithm, in Proceedings of the Genetic and Evolutionary Computation Conference, pp. 258–265.
Hassanat A, Almohammadi K, Alkafaween E, Abunawas E, Hammouri A, Prasath VBS (December 2019) Choosing mutation and crossover ratios for genetic algorithms—A review with a new dynamic approach. Information 10:390. https://doi.org/10.3390/info10120390
He J, Ji S, Yan M, Pan Y, Li Y (2012) Load-balanced CDS construction in wireless sensor networks via genetic algorithm. Int J Sens Netw 11(3):166–178
Hedar A, Fukushima M (2003) Simplex coding genetic algorithm for the global optimization of nonlinear functions, in Multi-Objective Programming and Goal Programming, Advances in Soft Computing, T. Tanino, T. Tanaka, and M. Inuiguchi, Eds.: Springer-Verlag, pp. 135–140.
Helal MHS, Fan C, Liu D, Yuan S (2017) Peer-to-peer based parallel genetic algorithm. International Conference on Information, Communication and Engineering (ICICE), Xiamen, pp 535–538
Hiassat A, Diabat A, Rahwan I (2017) A genetic algorithm approach for location-inventory-routing problem with perishable products. J Manuf Syst 42:93–103
Holland JH (1975) Adaptation in natural and artificial systems. The U. of Michigan Press
Hong W-C, Dong Y, Chen L-Y, Wei S-Y (2011) SVR with hybrid chaotic genetic algorithms for tourism demand forecasting. Appl Soft Comput 11(2):1881–1890
Hong T-P, Lee Y-C, Min-Thai W (2014) An effective parallel approach for genetic-fuzzy data mining. Exp Syst Applic 41(2):655–662
Horn J, Nafpliotis N, Goldberg DE. (1994) A niched Pareto genetic algorithm for multiobjective optimization. Proceedings of the First IEEE Conference on Evolutionary Computation, IEEE World Congress on Computational Intelligence, vol. 1, Piscataway, NJ: IEEE Service Center, p. 67–72.
Hu C, Wang X, Mandal MK, Meng M, Li D (2003) Efficient face and gesture recognition techniques for robot control. Department of Electrical and Computer Engineering University of Alberta, Edmonton, AB, T6G 2V4, Canada. CCECE2003 - CCGEI 2003, Montreal, May/mai 2003 IEEE, pp 1757-1762.
Peng Huo, Simon C. K. Shiu, Haibo Wang, Ben Niu (2009) Application and Comparison of Particle Swarm Optimization and Genetic Algorithm in Strategy Defense Game. Fifth International Conference on Natural Computation, pp 387–392.
Hussain A, Muhammad YS, Nauman Sajid M, Hussain I, Mohamd Shoukry A, Gani S (2017) Genetic algorithm for traveling salesman problem with modified cycle crossover operator. Computational intelligence and neuroscience 2017:1–7
Ishibuchi H, Murata T (1998) A multi-objective genetic local search algorithm and its application to flowshop scheduling. IEEE Trans Syst Man Cybern Part C Appl Rev 28(3):392–403
Jafari A, Khalili T, Babaei E, Bidram A (2020) Hybrid optimization technique using exchange market and GA. IEEE Access 8:2417–2427
Jaszkiewicz A (February 2002) Genetic local search for multi-objective combinatorial optimization. Eur J Oper Res 137(1):50–71
Javidi M, Hosseinpourfard R (2015) Chaos genetic algorithm instead genetic algorithm. Int J Inf Tech 12(2):163–168
Jebari K (2013) Selection methods for genetic algorithms. Abdelmalek Essaâdi University. International Journal of Emerging Sciences 3(4):333–344
Jiang S, Chin K-S, Wang L, Qu G, Tsui KL (2017) Modified genetic algorithm-based feature selection combined with pre-trained deep neural network for demand forecasting in outpatient department. Expert Syst Appl 82:216–230
Jiang M, Fan X, Pei Z, Zhang Z (2018) Research on text feature clustering based on improved parallel genetic algorithm. Tenth International Conference on Advanced Computational Intelligence (ICACI), Xiamen, pp. 235–238
Kaluri R, Reddy P (2016) Sign gesture recognition using modified region growing algorithm and adaptive genetic fuzzy classifier. International Journal of Intelligent Engineering and Systems 9(4):225–233
Kandavanam G, Botvich D, Balasubramaniam S, Jennings B (2010) A hybrid genetic algorithm/variable neighborhood search approach to maximizing residual bandwidth of links for route planning. Artificial evolution. Springer, In, pp 49–60
Kannan S (2020) Intelligent object recognition in underwater images using evolutionary-based Gaussian mixture model and shape matching. SIViP 14:877–885
Karabudak D, Hung C-C, Bing B (2004) A call admission control scheme using genetic algorithms. In: Proceedings of the 2004ACM symposium on applied computing. ACM, pp 1151–1158
Katz P, Aron M, Alfalou A (2001) A face-tracking system to detect falls in the elderly; SPIE newsroom. SPIE, Bellingham, WA, USA, p 201
Kaur M, Kumar V (2018) Beta chaotic map based image encryption using genetic algorithm. Int J Bifurcation Chaos 28(11):1850132
Kaur M, Kumar V (2018) Parallel non-dominated sorting genetic algorithm-II-based image encryption technique. The Imaging Science Journal. 66(8):453–462
Kaur M, Kumar V (2018) Fourier–Mellin moment-based intertwining map for image encryption. Modern Physics Letters B 32(9):1850115
Kaur G, Bhardwaj N, Singh PK (2018) An analytic review on image enhancement techniques based on soft computing approach. Sensors and Image Processing, Advances in Intelligent Systems and Computing 651:255–266
Kavitha AR, Chellamuthu C (2016) Brain tumour segmentation from MRI image using genetic algorithm with fuzzy initialisation and seeded modified region growing (GFSMRG) method. The Imaging Science Journal 64(5):285–297
Kennedy J, Eberhart RC (1995) Particle swarm optimization. In: Proceedings of IEEE international conference on neural networks (1995), pp 1942–1948
Khan, A., ur Rehman, Z., Jaffar, M.A., Ullah, J., Din, A., Ali, A., Ullah, N. (2019) Color image segmentation using genetic algorithm with aggregation-based clustering validity index (CVI). SIViP 13(5), 833–841
Kia R, Khaksar-Haghani F, Javadian N, Tavakkoli-Moghaddam R (2014) Solving a multi-floor layout design model of a dynamic cellular manufacturing system by an efficient genetic algorithm. J Manuf Syst 33(1):218–232
Kim EY, Jung K (2006) Genetic algorithms for video segmentation. Pattern Recogn 38(1):59–73
Kim EY, Park SH (2006) Automatic video segmentation using genetic algorithms. Pattern Recogn Lett 27(11):1252–1265
Kita H, Ono I, Kobayashi S (1999). The multi-parent unimodal normal distribution crossover for real-coded genetic algorithms. Proceedings of the 1999 Congress on Evolutionary Computation, vol. 2, IEEE (1999), pp. 1588–1595
Kobayashi H, Munetomo M, Akama K, Sato Y (2004) Designing a distributed algorithm for bandwidth allocation with a genetic algorithm. Syst Comput Jpn 35(3):37–45
Konak A, Smith AE (1999) A hybrid genetic algorithm approach for backbone design of communication networks, in the 1999 Congress on Evolutionary Computation. Washington D.C, USA: IEEE, pp. 1817-1823.
Kortil Y, Jridi M, Falou AA, Atri M (2020) Face recognition systems: A survey. Sensors. 20:1–34
Krishnan N, Muthukumar S, Ravi S, Shashikala D, Pasupathi P (2013) Image restoration by using evolutionary technique to Denoise Gaussian and impulse noise. In: Prasath R., Kathirvalavakumar T. (eds) mining intelligence and knowledge exploration. Lecture notes in computer science, vol 8284. Springer, Cham.
Kumar A (2013) Encoding schemes in genetic algorithm. Int J Adv Res IT Eng 2(3):1–7
Kumar V, Kumar D (2017) An astrophysics-inspired grey wolf algorithm for numerical optimization and its application to engineering design problems. Adv Eng Softw 112:231–254
Kumar V, Chhabra JK, Kumar D (2014) Parameter adaptive harmony search algorithm for unimodal and multimodal optimization problems. J Comput Sci 5(2):144–155
Kumar C, Singh AK, Kumar P (2017) A recent survey on image watermarking techniques and its application in e-governance. MultiMed Tools Appl.
Kurdi M (2016) An effective new island model genetic algorithm for job shop scheduling problem. Comput Oper Res 67(2016):132–142
Larranaga P, Kuijpers CMH, Murga RH, Yurramendi Y (July 1996) Learning Bayesian network structures by searching for the best ordering with genetic algorithms. in IEEE Transactions on Systems, Man, and Cybernetics - Part A: Systems and Humans 26(4):487–493
Larranaga P, Kuijpers C, Murga R, Inza I, Dizdarevic S (1999) Genetic algorithms for the travelling salesman problem: a review of representations and operators. Artificial Intelligence Review 13:129–170
Chang-Yong Lee (2003) Entropy-Boltzmann selection in the genetic algorithms. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), vol. 33, no. 1, pp. 138–149, Feb. 2003.
Lee CKH (2018) A review of applications of genetic algorithms in operations management. Eng Appl Artif Intell 76:1–12
Lee Y, Hara T, Fujita H, Itoh S, Ishigaki T (July 2001) Automated detection of pulmonary nodules in helical CT images based on an improved template-matching technique. in IEEE Transactions on Medical Imaging 20(7):595–604
Joon-Yong Lee, Min-Soeng Kim, Cheol-Taek Kim and Ju-Jang Lee (2007) Study on encoding schemes in compact genetic algorithm for the continuous numerical problems,SICE Annual Conference 2007, Takamatsu, pp. 2694–2699.
Leng LT (1999) Guided genetic algorithm. University of Essex, Doctoral Dissertation
Li B, Li J, Tang K, Yao X (2015) Many-objective evolutionary algorithms: A survey. ACM Computing surveys
Lie Tang L (2000) Tian and Brian L steward, "color image segmentation with genetic algorithm for in-field weed sensing". Transactions of the ASAE 43(4):1019–1027
Lima S.J.A., de Araújo S.A. (2018) A new binary encoding scheme in genetic algorithm for solving the capacitated vehicle routing problem. In: Korošec P., Melab N., Talbi EG. (eds) Bioinspired Optimization Methods and Their Applications. BIOMA 2018. Lecture notes in computer science, vol 10835. Springer, Cham
Liu D (2019) Mathematical modeling analysis of genetic algorithms under schema theorem. Journal of Computational Methods in Sciences and Engineering 19:S131–S137
Liu Z, Meng Q, Wang S (2013) Speed-based toll design for cordon-based congestion pricing scheme. Transport Res Part C: Emerg Technol 31(2013):83–98
Lorenzo B, Glisic S (2013) Optimal routing and traffic scheduling for multihop cellular networks using genetic algorithm. IEEE Trans Mob Comput 12(11):2274–2288
Lucasius CB, Kateman G (1989) Applications of genetic algorithms in chemometrics. In: Proceedings of the 3rd international conference on genetic algorithms. Morgan Kaufmann, Los Altos, CA, USA, pp 170–176
Luo B, Jinhua Zheng, Jiongliang Xie, Jun Wu. Dynamic crowding distance – a new diversity maintenance strategy for MOEAs. ICNC ‘08, Fourth Int. Conf. on Natural Comp., vol. 1 (2008), pp. 580–585
Maghawry A, Kholief M, Omar Y, Hodhod R (2020) An approach for evolving transformation sequences using hybrid genetic algorithms. Int J Intell Syst 13(1):223–233
Manzoni L, Mariot L, Tuba E (2020) Balanced crossover operators in genetic algorithms. Swarm and Evolutionary Computation 54:100646
Mazinani M, Abedzadeh M, Mohebali N (2013) Dynamic facility layout problem based on flexible bay structure and solving by genetic algorithm. Int J Adv Manuf Technol 65(5–8):929–943
Mehboob U, Qadir J, Ali S, Vasilakos A (2016) Genetic algorithms in wireless networking: techniques, applications, and issues. Soft Comput 20:2467–2501
Michalewicz Z (1992) Genetic algorithms + data structures = evolution programs. Springer-Verlag, New York
Michalewicz Z, Schoenauer M (1996) Evolutionary algorithms for constrained parameter optimization problems. Evol Comput 4(1):1–32
Mishra R, Das KN (2017). A novel hybrid genetic algorithm for unconstrained and constrained function optimization. In bio-inspired computing for information retrieval applications (pp. 230-268). IGI global
Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6(7):e1000097
Mooi S, Lim S, Sultan M, Bakar A, Sulaiman M, Mustapha A, Leong KY (2017) Crossover and mutation operators of genetic algorithms. International Journal of Machine Learning and Computing 7:9–12
Mudaliar DN, Modi NK (2013) Unraveling travelling salesman problem by genetic algorithm using m-crossover operator. International Conference on Signal Processing, Image Processing & Pattern Recognition, Coimbatore, pp 127–130
T. Murata and M. Gen (2000) Cellular genetic algorithm for multi-objective optimization, in Proceedings of the Fourth Asian Fuzzy System Symposium, pp. 538–542
Neto JC, Meyer GE, Jones DD (2006) Individual leaf extractions from young canopy images using gustafsonkessel clustering and a genetic algorithm. Comput Electron Agric 51(1):66–85
NKFC, Viswanatha SDK (2009) Routing algorithm using mobile agents and genetic algorithm. Int J Comput Electr Eng, vol 1, no 3
Ono I, Kobayashi S (1997) A real-coded genetic algorithm for functional optimization using unimodal normal distribution crossover. In: Back T (ed) Proceedings of the 7th international conference on genetic algorithms, ICGA-7. Morgan Kaufmann, East Lansing, MI, USA, pp 246–253
Pachepsky Y, Acock B (1998) Stochastic imaging of soil parameters to assess variability and uncertainty of crop yield estimates. Geoderma 85(2):213–229
Paiva JPD, Toledo CFM, Pedrini H (2016) An approach based on hybrid genetic algorithm applied to image denoising problem. Appl Soft Comput 46:778–791
Palencia AER, Delgadillo GEM (2012) A computer application for a bus body assembly line using genetic algorithms. Int J Prod Econ 140(1):431–438
Palomo-Romero JM, Salas-Morera L, García-Hernández L (2017) An island model genetic algorithm for unequal area facility layout problems. Expert Syst Appl 68:151–162
Pandian S, Modrák V (December 2009) "possibilities, obstacles and challenges of genetic algorithm in manufacturing cell formation," advanced logistic systems, University of Miskolc. Department of Material Handling and Logistics 3(1):63–70
Park Y-B, Yoo J-S, Park H-S (2016) A genetic algorithm for the vendor-managed inventory routing problem with lost sales. Expert Syst Appl 53:149–159
Patel R, Raghuwanshi MM, Malik LG (2012) Decomposition based multi-objective genetic algorithm (DMOGA) with opposition based learning
Pattanaik JK, Basu M, Dash DP (2018) Improved real coded genetic algorithm for dynamic economic dispatch. Journal of electrical systems and information technology. Vol. 5(3):349–362
Payne AW, Glen RC (1993) Molecular recognition using a binary genetic system. J Mol Graph 11(2):74–91
Peerlinck A, Sheppard J, Pastorino J, Maxwell B (2019) Optimal Design of Experiments for precision agriculture using a genetic algorithm. IEEE Congress on Evolutionary Computation.
Pelikan M, Goldberg DE, Cantu-Paz E (2000) Bayesian optimization algorithm, population sizing, and time to convergence, Illinois Genetic Algorithms Laboratory, University of Illinois, Tech. Rep
Pilat ML, White T (2002) Using genetic algorithms to optimize ACS-TSP, in the Third International Workshop on Ant Algorithms, vol. Lecture Notes In Computer Science 2463. Berlin, Germany: Springer-Verlag, pp. 282–287.
Pinagapany S, Kulkarni A (2008) Solving channel allocation problem in cellular radio networks using genetic algorithm. In: Communication Systems software and middleware and workshops, 2008.COMSWARE 2008. 3rd International Conference on. IEEE, pp239–244
Pinel F, Dorronsoro B, Bouvry P (2013) Solving very large instances of the scheduling of independent tasks problem on the GPU. J Parallel Distrib. Comput 73(1):101–110
Pinto G, Ainbinder I, Rabinowitz G (2009) A genetic algorithm-based approach for solving the resource-sharing and scheduling problem. Comput Ind Eng 57(3):1131–1143
Piszcz A, Soule T (2006) Genetic programming: optimal population sizes for varying complexity problems, in Proceedings of the Genetic and Evolutionary Computation Conference, pp. 953–954.
Porta J, Parapar R, Doallo F, Rivera F, Santé I, Crecente R (2013) High performance genetic algorithm for land use planning. Comput Environ Urb Syst 37(2013):45–58
Rafsanjani MK, Riyahi M (2020) A new hybrid genetic algorithm for job shop scheduling problem. International Journal of Advanced Intelligence Paradigms 16(2):157–171
Rathi R, Acharjya DP (2018) A framework for prediction using rough set and real coded genetic algorithm. Arab J Sci Eng 43(8):4215–4227
Rathi R, Acharjya DP (2018) A rule based classification for vegetable production using rough set and genetic algorithm. International Journal of Fuzzy System Applications (IJFSA) 7(1):74–100
Rathi R, Acharjya DP (2020) A comparative study of genetic algorithm and neural network computing techniques over feature selection, In advances in distributed computing and machine learning (pp. 491–500). Springer, Singapore
Ray SS, Bandyopadhyay S, Pal SK (2004) New operators of genetic algorithms for traveling salesman problem," Proceedings of the 17th International Conference on Pattern Recognition, 2004. ICPR 2004., Cambridge pp 497-500
Richter JN, Peak D (2002) Fuzzy evolutionary cellular automata, in international conference on artificial neural networks in engineering, vol 12. USA, Saint Louis pp. 185-191
Riedl A (2002) A hybrid genetic algorithm for routing optimization in ip networks utilizing bandwidth and delay metrics. In: IP operations and management, 2002 IEEE Workshop on. IEEE, pp 166–170
Ripon KSN, Siddique N, Torresen J (2011) Improved precedence preservation crossover for multi-objective job shop scheduling problem. Evolving Systems 2:119–129
Roberge V, Tarbouchi M, Okou F (2014) Strategies to accelerate harmonic minimization in multilevel inverters using a parallel genetic algorithm on graphical processing unit. IEEE Trans Power Electron 29(10):5087–5090
Ronald S (1997) Robust encoding in genetic algorithms: a survey of encoding issues. IEEE international conference on evolutionary computation, pp. 43-48
Roy A, Banerjee N, Das SK (2002) An efficient multi-objective qos-routing algorithm for wireless multicasting. In:Vehicular technology conference, 2002. VTC Spring 2002. IEEE 55th, vol 3., pp 1160–1164
Sadrzadeh A (2012) A genetic algorithm with the heuristic procedure to solve the multi-line layout problem. Comput Ind Eng 62(4):1055–1064
Sahingoz OK (2014) Generation of Bezier curve-based flyable trajectories for multi-UAV systems with parallel genetic algorithm. J Intell Robot Syst 74(1):499–511
Saini N (2017) Review of selection methods in genetic algorithms. International Journal of Engineering and Computer Science 6(12):22261–22263
Sari M, Can T (2018) Prediction of pathological subjects using genetic algorithms. Computational and Mathematical Methods in Medicine 2018:1–9
Scully T, Brown KN (2009) Wireless LAN load balancing with genetic algorithms. Knowl Based Syst 22(7):529–534
Sermpinis G, Stasinakis C, Theofilatos K, Karathanasopoulos A (2015) Modeling, forecasting and trading the EUR exchange rates with hybrid rolling genetic algorithms–support vector regression forecast combinations. European J. Oper. Res. 247(3):831–846
Shabankareh SG, Shabankareh SG (2019) Improvement of edge-tracking methods using genetic algorithm and neural network, 2019 5th Iranian conference on signal processing and intelligent systems (ICSPIS). Shahrood, Iran, pp 1–7. https://doi.org/10.1109/ICSPIS48872.2019.9066026
Book Google Scholar
Sharma S, Gupta K (2011) Solving the traveling salesman problem through genetic algorithm with new variation order crossover. International Conference on Emerging Trends in Networks and Computer Communications (ETNCC), Udaipur, pp. 274–276
Sharma N, Kaushik I, Rathi, R, Kumar S (2020) Evaluation of accidental death records using hybrid genetic algorithm. Available at SSRN: https://ssrn.com/abstract=3563084 or https://doi.org/10.2139/ssrn.3563084
Shayeghi A, Gotz D, Davis JBA, Schafer R, Johnston RL (2015) Pool-BCGA: A parallelised generation-free genetic algorithm for the ab initio global optimisation of nano alloy clusters. Phys Chem Chem Phys 17(3):2104–2112
Guoyong Shi, H. Iima and N. Sannomiya (1996) A new encoding scheme for solving job shop problems by genetic algorithm, Proceedings of 35th IEEE Conference on Decision and Control, Kobe, Japan, 1996, pp. 4395–4400 vol.4.
Shi J, Liu Z, Tang L, Xiong J (2017) Multi-objective optimization for a closed-loop network design problem using an improved genetic algorithm. Appl Math Model 45:14–30
Shukla AK, Singh P, Vardhan M (2019) A new hybrid feature subset selection framework based on binary genetic algorithm and information theory. International Journal of Computational Intelligence and Applications 18(3):1950020(1–10)
Singh A, Deep K (2015) Real coded genetic algorithm operators embedded in gravitational search algorithm for continuous optimization. Int J Intell Syst Appl 7(12):1
Sivanandam SN, Deepa SN (2008) Introduction to genetic algorithm, 1st edn. Springer-Verlag, Berlin Heidelberg
Soleimani H, Kannan G (2015) A hybrid particle swarm optimization and genetic algorithm for closed-loop supply chain network design in large-scale networks. Appl Math Model 39(14):3990–4012
Soleimani H, Govindan K, Saghafi H, Jafari H (2017) Fuzzy multi-objective sustainable and green closed-loop supply chain network design. Comput Ind Eng 109:191–203
Soon GK, Guan TT, On CK, Alfred R, Anthony P (2013) "A comparison on the performance of crossover techniques in video game," 2013 IEEE international conference on control system. Computing and Engineering, Mindeb, pp 493–498
Srinivas N, Deb K (1995) Multi-objective function optimization using non-dominated sorting genetic algorithms. Evol Comput 2(3):221–248
Subbaraj P, Rengaraj R, Salivahanan S (2011) Enhancement of self-adaptive real-coded genetic algorithm using Taguchi method for economic dispatch problem. Appl Soft Comput 11(1):83–92
Tahir M, Tubaishat A, Al-Obeidat F, et al. (2020) A novel binary chaotic genetic algorithm for feature selection and its utility in affective computing and healthcare. Neural Comput & Appl
Tam V, Cheng K-Y, Lui K-S (2006) Using micro-genetic algorithms to improve localization in wireless sensor networks. J Commun 1(4):1–10
Tan KC, Li Y, Murray-Smith DJ, Sharman KC (1995) System identification and linearisation using genetic algorithms with simulated annealing, in First IEE/IEEE Int. Conf. on GA in Eng. Syst.: Innovations and Appl. Sheffield, UK, pp. 164–169.
Tang PH, Tseng MH (2013) Adaptive directed mutation for real-coded genetic algorithms. Appl Soft Comput 13(1):600–614
Tiong SK, Yap DFW, Koh SP (2012) A comparative analysis of various chaotic genetic algorithms for multimodal function optimization. Trends in Applied Sciences Research 7:785–791
Toutouh J, Alba E (2017) Parallel multi-objective metaheuristics for smart communications in vehicular networks. Soft Comput 21(8):1949–1961
Umbarkar A, Sheth P (2015) Crossover operators in genetic algorithms: a review. Journal on Soft Computing 6(1)
Verma D, Vishwakarma VP, Dalal S (2020) A hybrid self-constrained genetic algorithm (HSGA) for digital image Denoising based on PSNR improvement. Advances in Bioinformatics, Multimedia, and Electronics Circuits and Signals, In, pp 135–153
Vitayasak S, Pongcharoen P, Hicks C (2016) A tool for solving stochastic dynamic facility layout problems with stochastic demand using either a genetic algorithm or modified backtracking search algorithm. Int J Prod Econ
Junru Wang and Lan Huang (2014) Evolving gomoku Solver by Genetic Algorithm. IEEE Workshop on Advanced Research and Technology in Industry Applications (WARTIA) pp 1064–1067.
Wang L, Kan MS, Shahriar Md R, Tan ACC (2014) Different approaches of applying single-objective binary genetic algorithm on the wind farm design. In World Congress on Engineering Asset Management.
Wang N, Li Q, Abd El-Latif AA, Zhang T, Niu X (2014) Toward accurate localization and high recognition performance for noisy iris images. Multimed Tools Appl 71(3):1411–1430
Wang JQ, Ersoy OK, He MY et al (2016) Multi-offspring genetic algorithm and its application to the traveling salesman problem. Appl Soft Comput 43:415–423
Wang FL, Fu XM, Zhu HX et al (2016) Multi-child genetic algorithm based on two-point crossover. J Northeast Agric Univ 47(3):72–79
Wang JQ, Cheng ZW, Ersoy OK et al (2018) Improvement analysis and application of real-coded genetic algorithm for solving constrained optimization problems. Math Probl Eng 2018:1–16
Wang J, Zhang M, Ersoy OK, Sun K, Bi Y (2019) An improved real-coded genetic algorithm using the Heuristical Normal distribution and direction-based crossover. Computational Intelligence and Neuroscience 2019:1–17
Wen Z, Yang R, Garraghan P, Lin T, Xu J, Rovatsos M (2017) Fog orchestration for internet of things services. IEEE Internet Comput 21(2) (Mar. 2017):16–24
Wright AH (1991) Genetic algorithms for real parameter optimization. In Foundations of genetic algorithms I,G. J. E. Rawlins, Ed., Morgan Kaufmann, San Mateo, CA,USA
Wu X, Chu C-H, Wang Y, Yan W (2007) A genetic algorithm for cellular manufacturing design and layout. European J Oper Res 181(1):156–167
Yang S, Cheng H, Wang F (2010) Genetic algorithms with immigrants and memory schemes for dynamic shortest path routing problems in mobile ad hoc networks. IEEE Trans Syst Man Cybern Part C Appl Rev 40(1):52–63
Yang C, Li H, Rezgui Y, Petri I, Yuce B, Chen B, Jayan B (2014) High throughput computing based distributed genetic algorithm for building energy consumption optimization. Energy Build 76(2014):92–101
Yu F, Xu X (2014) A short-term load forecasting model of natural gas based on optimized genetic algorithm and improve BR neural network. Appl Energy 134:102–113
Yuce B, Fruggiero F, Packianather MS, Pham DT, Mastrocinque E, Lambiase A, Fera M (2017) Hybrid genetic bees algorithm applied to single machine scheduling with earliness and tardiness penalties. Comput Ind Eng 113:842–858
Yun S, Lee J, Chung W, Kim E, Kim S (2009) A soft computing approach to localization in wireless sensor networks. Expert Syst Appl 36(4):7552–7561
Zhai R (2020) Solving the optimization of physical distribution routing problem with hybrid genetic algorithm. J Phys Conf Ser 1550:1–6
Zhang Q, Wang J, Jin C, Zeng Q (2008) Localization algorithm for wireless sensor network based on genetic simulated annealing algorithm. In: 4th IEEE International Conference on Wireless communications, networking and mobile computing. Pp 1–5
Zhang R, Ong SK, Nee AYC (2015) A simulation-based genetic algorithm approach for remanufacturing process planning and scheduling. Appl Soft Comput 37:521–532
Zhang X-Y, Zhang J, Gong Y-J, Zhan Z-H, Chen W-N, Li Y (2016) Kuhn-Munkres parallel genetic algorithm for the set cover problem and its application to large-scale wireless sensor networks. IEEETrans Evol Comput 20(5):695–710
Zhenhua Y, Guangwen Y, Shanwei L, Qishan Z (2010) A modified immune genetic algorithm for channel assignment problems in cellular radio networks. In: Intelligent system design and engineering application (ISDEA), 2010 International Conference on, vol 2. , pp 823–826
Download references
Author information
Authors and affiliations.
Computer Science and Engineering Department, National Institute of Technology, Hamirpur, India
Sourabh Katoch, Sumit Singh Chauhan & Vijay Kumar
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Vijay Kumar .
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Katoch, S., Chauhan, S.S. & Kumar, V. A review on genetic algorithm: past, present, and future. Multimed Tools Appl 80 , 8091–8126 (2021). https://doi.org/10.1007/s11042-020-10139-6
Download citation
Received : 27 July 2020
Revised : 12 October 2020
Accepted : 23 October 2020
Published : 31 October 2020
Issue Date : February 2021
DOI : https://doi.org/10.1007/s11042-020-10139-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Optimization
- Metaheuristic
- Genetic algorithm
- Find a journal
- Publish with us
- Track your research
Using AI to improve diagnosis of rare genetic disorders
Diagnosing rare Mendelian disorders is a labor-intensive task, even for experienced geneticists. Investigators at Baylor College of Medicine are trying to make the process more efficient using artificial intelligence. The team developed a machine learning system called AI-MARRVEL (AIM) to help prioritize potentially causative variants for Mendelian disorders. The study is published today in NEJM AI .
Researchers from the Baylor Genetics clinical diagnostic laboratory noted that AIM's module can contribute to predictions independent of clinical knowledge of the gene of interest, helping to advance the discovery of novel disease mechanisms. "The diagnostic rate for rare genetic disorders is only about 30%, and on average, it is six years from the time of symptom onset to diagnosis. There is an urgent need for new approaches to enhance the speed and accuracy of diagnosis," said co-corresponding author Dr. Pengfei Liu, associate professor of molecular and human genetics and associate clinical director at Baylor Genetics.
AIM is trained using a public database of known variants and genetic analysis called Model organism Aggregated Resources for Rare Variant ExpLoration (MARRVEL) previously developed by the Baylor team. The MARRVEL database includes more than 3.5 million variants from thousands of diagnosed cases. Researchers provide AIM with patients' exome sequence data and symptoms, and AIM provides a ranking of the most likely gene candidates causing the rare disease.
Researchers compared AIM's results to other algorithms used in recent benchmark papers. They tested the models using three data cohorts with established diagnoses from Baylor Genetics, the National Institutes of Health-funded Undiagnosed Diseases Network (UDN) and the Deciphering Developmental Disorders (DDD) project. AIM consistently ranked diagnosed genes as the No. 1 candidate in twice as many cases than all other benchmark methods using these real-world data sets.
"We trained AIM to mimic the way humans make decisions, and the machine can do it much faster, more efficiently and at a lower cost. This method has effectively doubled the rate of accurate diagnosis," said co-corresponding author Dr. Zhandong Liu, associate professor of pediatrics -- neurology at Baylor and investigator at the Jan and Dan Duncan Neurological Research Institute (NRI) at Texas Children's Hospital.
AIM also offers new hope for rare disease cases that have remained unsolved for years. Hundreds of novel disease-causing variants that may be key to solving these cold cases are reported every year; however, determining which cases warrant reanalysis is challenging because of the high volume of cases. The researchers tested AIM's clinical exome reanalysis on a dataset of UDN and DDD cases and found that it was able to correctly identify 57% of diagnosable cases.
"We can make the reanalysis process much more efficient by using AIM to identify a high-confidence set of potentially solvable cases and pushing those cases for manual review," Zhandong Liu said. "We anticipate that this tool can recover an unprecedented number of cases that were not previously thought to be diagnosable."
Researchers also tested AIM's potential for discovery of novel gene candidates that have not been linked to a disease. AIM correctly predicted two newly reported disease genes as top candidates in two UDN cases.
"AIM is a major step forward in using AI to diagnose rare diseases. It narrows the differential genetic diagnoses down to a few genes and has the potential to guide the discovery of previously unknown disorders," said co-corresponding author Dr. Hugo Bellen, Distinguished Service Professor in molecular and human genetics at Baylor and chair in neurogenetics at the Duncan NRI.
"When combined with the deep expertise of our certified clinical lab directors, highly curated datasets and scalable automated technology, we are seeing the impact of augmented intelligence to provide comprehensive genetic insights at scale, even for the most vulnerable patient populations and complex conditions," said senior author Dr. Fan Xia, associate professor of molecular and human genetics at Baylor and vice president of clinical genomics at Baylor Genetics. "By applying real-world training data from a Baylor Genetics cohort without any inclusion criteria, AIM has shown superior accuracy. Baylor Genetics is aiming to develop the next generation of diagnostic intelligence and bring this to clinical practice."
Other authors of this work include Dongxue Mao, Chaozhong Liu, Linhua Wang, Rami AI-Ouran, Cole Deisseroth, Sasidhar Pasupuleti, Seon Young Kim, Lucian Li, Jill A.Rosenfeld, Linyan Meng, Lindsay C. Burrage, Michael Wangler, Shinya Yamamoto, Michael Santana, Victor Perez, Priyank Shukla, Christine Eng, Brendan Lee and Bo Yuan. They are affiliated with one or more of the following institutions: Baylor College of Medicine, Jan and Dan Duncan Neurological Research Institute at Texas Children's Hospital, Al Hussein Technical University, Baylor Genetics and the Human Genome Sequencing Center at Baylor.
This work was supported by the Chang Zuckerberg Initiative and the National Institute of Neurological Disorders and Stroke (3U2CNS132415).
- Diseases and Conditions
- Parkinson's Research
- Personalized Medicine
- Computers and Internet
- Computer Modeling
- Neural Interfaces
- Personality disorder
- Computer vision
- Psychopathology
- Toxic shock syndrome
- Artificial intelligence
- Computational neuroscience
- Nutrition and pregnancy
Story Source:
Materials provided by Baylor College of Medicine . Note: Content may be edited for style and length.
Journal Reference :
- Dongxue Mao, Chaozhong Liu, Linhua Wang, Rami AI-Ouran, Cole Deisseroth, Sasidhar Pasupuleti, Seon Young Kim, Lucian Li, Jill A. Rosenfeld, Linyan Meng, Lindsay C. Burrage, Michael F. Wangler, Shinya Yamamoto, Michael Santana, Victor Perez, Priyank Shukla, Christine M. Eng, Brendan Lee, Bo Yuan, Fan Xia, Hugo J. Bellen, Pengfei Liu, Zhandong Liu. AI-MARRVEL — A Knowledge-Driven AI System for Diagnosing Mendelian Disorders . NEJM AI , 2024; 1 (5) DOI: 10.1056/AIoa2300009
Cite This Page :
Explore More
- Far-Reaching Effects of Exercise
- Hidden Connections Between Brain and Body
- Novel Genetic Plant Regeneration Approach
- Early Human Occupation of China
- Journey of Inhaled Plastic Particle Pollution
- Earth-Like Environment On Ancient Mars
- A 'Cosmic Glitch' in Gravity
- Time Zones Strongly Influence NBA Results
- Climate Change and Mercury Through the Eons
- Iconic Horsehead Nebula
Trending Topics
Strange & offbeat.
Help | Advanced Search
Computer Science > Artificial Intelligence
Title: crispr-gpt: an llm agent for automated design of gene-editing experiments.
Abstract: The introduction of genome engineering technology has transformed biomedical research, making it possible to make precise changes to genetic information. However, creating an efficient gene-editing system requires a deep understanding of CRISPR technology, and the complex experimental systems under investigation. While Large Language Models (LLMs) have shown promise in various tasks, they often lack specific knowledge and struggle to accurately solve biological design problems. In this work, we introduce CRISPR-GPT, an LLM agent augmented with domain knowledge and external tools to automate and enhance the design process of CRISPR-based gene-editing experiments. CRISPR-GPT leverages the reasoning ability of LLMs to facilitate the process of selecting CRISPR systems, designing guide RNAs, recommending cellular delivery methods, drafting protocols, and designing validation experiments to confirm editing outcomes. We showcase the potential of CRISPR-GPT for assisting non-expert researchers with gene-editing experiments from scratch and validate the agent's effectiveness in a real-world use case. Furthermore, we explore the ethical and regulatory considerations associated with automated gene-editing design, highlighting the need for responsible and transparent use of these tools. Our work aims to bridge the gap between beginner biological researchers and CRISPR genome engineering techniques, and demonstrate the potential of LLM agents in facilitating complex biological discovery tasks.
Submission history
Access paper:.
- HTML (experimental)
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Research Briefing
- Published: 24 April 2024
Three patterns link brain organization to genes in health and disease
Nature Neuroscience ( 2024 ) Cite this article
280 Accesses
1 Altmetric
Metrics details
- Development of the nervous system
- Genetics of the nervous system
Gene expression in the human cortex is shown to exhibit a generalizable three-component architecture that reflects neuronal, metabolic, and immune programmes of healthy brain development. The three components have distinct associations with autism spectrum disorder and schizophrenia, revealing connections between previously unrelated results from studies of case–control neuroimaging, differential gene expression, and genetic risk.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489 , 391–399 (2012). The original paper presenting the AHBA, in which principal components of cortical gene expression were suggested to reflect brain organization.
Article CAS PubMed PubMed Central Google Scholar
Burt, J. B. et al. Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat. Neurosci. 21 , 1251–1259 (2018). This paper characterizes the first component of cortical gene expression, C1, as reflecting a neuronal hierarchy defined by tract-tracing and indexed by structural neuroimaging.
Sydnor, V. J. et al. Neurodevelopment of the association cortices: patterns, mechanisms, and implications for psychopathology. Neuron 109 , 2820–2846 (2021). This review proposes that neurodevelopment involves a ‘sensorimotor–association axis’ defined by ten brain maps, of which one is the cortical gene expression component C1.
Merikangas, A. K. et al. What genes are differentially expressed in individuals with schizophrenia? A systematic review. Mol. Psychiatry 27 , 1373–1383 (2022). This review demonstrates the lack of consistency in genes linked to schizophrenia across differential expression studies, which are also inconsistent with GWAS.
Article PubMed PubMed Central Google Scholar
Johnson, M. B. & Hyman, S. E. A critical perspective on the synaptic pruning hypothesis of schizophrenia pathogenesis. Biol. Psychiatry 92 , 440–442 (2022). This commentary calls for an understanding of synaptic pruning in schizophrenia compared with healthy adolescent neurodevelopment.
Article PubMed Google Scholar
Download references
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Dear, R. et al. Cortical gene expression architecture links healthy neurodevelopment to the imaging, transcriptomics and genetics of autism and schizophrenia. Nat. Neurosci . https://doi.org/10.1038/s41593-024-01624-4 (2024).
Rights and permissions
Reprints and permissions
About this article
Cite this article.
Three patterns link brain organization to genes in health and disease. Nat Neurosci (2024). https://doi.org/10.1038/s41593-024-01625-3
Download citation
Published : 24 April 2024
DOI : https://doi.org/10.1038/s41593-024-01625-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
Genetics is the branch of science concerned with genes, heredity, and variation in living organisms. ... Research Highlights 29 Apr 2024 ... Kirstyn Brunker highlights two papers published in 2017 ...
Maintenance of proteostasis by Drosophila Rer1 is essential for competitive cell survival and Myc-driven overgrowth. Loss of Rer1 induces proteotoxic stress, leading to cell competition and elimination, while increased Rer1 levels provide cytoprotection and support Myc-driven overgrowth. Image credit: pgen.1011171. 02/28/2024.
Genomic research has evolved from seeking to understand the fundamentals of the human genetic code to examining the ways in which this code varies among people, and then applying this knowledge to ...
See all (1,568) Learn more about Research Topics. The most cited genetics and heredity journal, which advances our understanding of genes from humans to plants and other model organisms. It highlights developments in the function and variability o...
Neurogenetics lies at the intersection of Neuroscience and Genetics, where genetic approaches are applied to the study of nervous system development, function, and plasticity. Overseen by Series Editors Oliver Hobert, Cecilia Moens, and Kate O'Connor Giles, this new series aims to make the GSA Journals a home for cutting-edge, robust research ...
In the early 1900s, focusing on the evolution of genetic variants in the population, R. A. Fisher, S. Wright, and J. B. S. Haldane made fundamental theoretical contributions to population genetics (Provine 1971), Fisher in his 1922 paper (Fisher 1922), which was the first to introduce diffusion equations into population genetics, and Haldane in ...
We present selected topics of population genetics and molecular phylogeny. As several excellent review articles have been published and generally focus on European and American scientists, here, we emphasize contributions by Japanese researchers. Our review may also be seen as a belated 50-year celebration of Motoo Kimura's early seminal paper on the molecular clock, published in 1968.
Homologous recombination in animal cells was rapidly exploited by the mouse genetics research community for the production of gene-modified mouse ES cells, and thus gene-modified whole animals [9,10]. ... [47,48,49], was a watershed event to modern science. Moreover, the introduction of CRISPR/Cas9 methodology has revolutionized transgenic ...
For the first time, new large-scale datasets afford sufficient statistical power to identify genetic variants associated with same-sex sexual behavior (ever versus never had a same-sex partner), estimate the proportion of variation in the trait accounted for by all variants in aggregate, estimate the genetic correlation of same-sex sexual behavior with other traits, and probe the biology and ...
The genetic variant selection for the GRM was as follows: missingness < 0.01, HWE < 1 × 10 −08, MAF > 0.01, and imputation info-score > 0.8. The remaining genetic variants were hard called using a threshold of 0.25 and pruned at r 2 < 0.2. Some conflictive genomic regions such as the lactase locus on chromosome 2, the major ...
05 Feb 2024. 23 Jan 2024. Genetics Research is a fully open access journal providing a key forum for original research on all aspects of human and animal genetics, reporting key findings on genomes, genes, mutations, developmental, evolutionary, and population genetics as well as ethical, legal and social aspects.
Journal of Genetics and Genomics (JGG), founded in 1974 and previously known as Acta Genetica Sinica, is an international journal publishing peer-reviewed articles of novel and significant discoveries in all areas of biology and biomedicine. JGG publishes papers reporting findings of general interest that provide significant insights into important biological processes in any living organisms ...
Cancer Genetics is proud to be affiliated with the. The aim of Cancer Genetics is to publish high quality scientific papers on the cellular, genetic and molecular aspects of cancer, including cancer predisposition and clinical diagnostic applications. Specific areas of interest include descriptions of new chromosomal, molecular or epigenetic ….
Center for Cancer Research, Genetics Branch, National Cancer Institute. Bioethics Department, National Institutes of Health. Search for articles by this author ... inaccurate practices for collecting data and conducting genomic research have adversely influenced genomic science and can contribute to the stigmatization of people whose sex and/or ...
Data science allows the extraction of practical insights from large-scale data. Here, we contextualize it as an umbrella term, encompassing several disparate subdomains. We focus on how genomics fits as a specific application subdomain, in terms of well-known 3 V data and 4 M process frameworks (volume-velocity-variety and measurement-mining-modeling-manipulation, respectively). We further ...
A recent report by the National Academies of Sciences, Engineering, and Medicine (NASEM) has urged scientists to scrutinize and justify their use of "race, ethnicity, and genetic ancestry" as population descriptors in genetics and genomics research.That report calls for a shift in scientific practice to engage with the sociopolitical nature of genetics research in terms of how populations ...
There is a lot of debate regarding suicidal behavior and its relationship with psychiatric disorders, but the extent to which they share the same genetic architecture is unknown. This Research Topic was investigated by Kootbodien et al. through the use of genomic structural equation modeling and Mendelian randomization with a large genomic ...
This paper presents results of an issue-spotting exercise conducted by experts in the ethics, law, and science of genetics and infectious diseases (ID). The exercise focused on the collection, storage and sharing of genetic data relating to ID, highlighting ELSIs that differ in important ways from issues in genetics and non-transmissible disease.
Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben, 06466 Seeland, Germany ... This paper reports the characterisation a barley mutation in the HvPIN1a auxin efflux carrier encoding gene which ... 294729), Biotechnology and Biological Sciences Research Council of the United Kingdom and The Wolfson Foundation. HL was ...
In this paper, the analysis of recent advances in genetic algorithms is discussed. The genetic algorithms of great interest in research community are selected for analysis. This review will help the new and demanding researchers to provide the wider vision of genetic algorithms. The well-known algorithms and their implementation are presented with their pros and cons. The genetic operators and ...
The role of rice genomics in breeding progress is becoming increasingly important. Deeper research into the rice genome will contribute to the identification and utilization of outstanding functional genes, enriching the diversity and genetic basis of breeding materials and meeting the diverse demands for various improvements. Here, we review the significant contributions of rice genomics ...
The research is featured on the cover of the April 2024 issue of GENETICS, a peer-reviewed, open access, scientific journal. The work was co-led by two University of Minnesota mechanical ...
Diagnosing rare Mendelian disorders is a labor-intensive task, even for experienced geneticists. Investigators at Baylor College of Medicine are trying to make the process more efficient using ...
The introduction of genome engineering technology has transformed biomedical research, making it possible to make precise changes to genetic information. However, creating an efficient gene-editing system requires a deep understanding of CRISPR technology, and the complex experimental systems under investigation. While Large Language Models (LLMs) have shown promise in various tasks, they ...
This paper characterizes the first component of cortical gene expression, C1, as reflecting a neuronal hierarchy defined by tract-tracing and indexed by structural neuroimaging.
Abstract In this paper, ... two-dimensional, and three-dimensional parameter space. We found that the genetic algorithm (GA) operation has a typical converging trend as the epoch increasing of groups in ultra-fast pulse generation. ... and 2002.7 nm, repetition rate of 9.4, 10.4, and 5.4 MHz, and typical spike duration of ~224 fs. Our research ...
Engaging a century-long debate about the role of race in science. In the wake of the sequencing of the human genome in the early 2000s, genome pioneers and social scientists alike called for an end to the use of race as a variable in genetic research ( 1, 2 ). Unfortunately, by some measures, the use of race as a biological category has ...